Introduction
Numerous hard coral species exhibit substantial intraspecific morphological variation (Foster 1977, Foster 1983, Veron 1995), which is observable at intracolonial, intrapopulation and interpopulation levels (Best et al. 1984). Such variability can arise from genetic differences or phenotypically plastic responses to the surrounding environment (Todd et al. 2004, Pinzón et al. 2013). As coral taxonomy is largely reliant on skeletal morphology (e.g. Veron 2000), such variability can potentially blur species limits and render identification difficult.
The genus Pocillopora Lamarck, 1816, is of special interest as it is characterised by exceptionally high levels of phenotypic variation (Veron 2000, Flot and Tillier 2006, Schmidt-Roach et al. 2014a), and its species are frequently used as models in ecological and experimental studies. Due to a lack of clear morphological traits to distinguish among species, some taxa have historically been synonymised. For example, Pocillopora damicornis (Linnaeus, 1758) has had at least five synonyms (P. acuta Lamarck, 1816; P. brevicornis Lamarck, 1816; P. bulbosa Ehrenberg, 1834; P. favosa Ehrenberg, 1834; and P. caespitosa Dana, 1846), which are all considered to be morphological variants of a single complex associated with different environments (Veron and Pichon 1976, Veron 2000, Schmidt-Roach et al. 2013).
Recently, P. acuta was re-established as an entirely separate species from P. damicornis based on differences in their mitochondrial open reading frame (ORF) sequences (Schmidt-Roach et al. 2014a). Pocillopora acuta also differs from P. damicornis by possessing more elongated, sharper and thinner branchlets, as well as having dark brown pigmentation around the oral openings of the polyps (Schmidt-Roach et al. 2014a).
The morphological characteristics described for P. acuta are exhibited by corals previously identified as Pocillopora damicornis in Singapore. Based on this observation, we hypothesise that most of the Pocillopora colonies on Singapore’s reefs are likely to be P. acuta and not P. damicornis. Here, we examine a range of P. ‘damicornis-like’ (sensu Pinzón et al. 2013) colony morphologies, their mitochondrial ORF sequences, and verify their taxonomic identity as the basis for characterisation of P. acuta or P. damicornis in Singapore.
Results of this study are important for coral diversity records in Singapore and will also help clarify the geographical range limits of morphologically closely-related Pocillopora species. As one of the most widespread corals on Singapore's reefs (Huang et al. 2009), these P. ‘damicornis-like’ colonies are able to settle and grow on natural reefs and even artificial substrates such as seawalls and pontoons (Toh et al. 2017). Consequently, they are widely utilised in reef restoration research (Ng et al. 2016). Their accurate identification, whether they are P. acuta or P. damicornis, is critical for achieving species diversity targets in local and regional restoration efforts (Mace 2004, Wheeler 2004). Similar to other corals, Pocillopora is threatened by habitat loss and climate change, so a clear understanding of species boundaries will facilitate the conservation of species under this genus.
Materials and methods
Sixteen Pocillopora samples were collected from five coral reef sites across the southern offshore islands of Singapore. We focused on a wide range of colony morphologies that, following Veron and Pichon (1976) and Veron (2000), had been identified previously as P. damicornis. We photographed each colony in the field before part or all of it was collected. A 1-cm length of a branch was preserved in 100% ethanol while the rest of the colony sample was soaked in freshwater, bleached in 2% sodium hypochlorite, then rinsed, cleaned and dried. These samples (Fig. 1), along with others loaned from the Zoological Reference Collection (ZRC; Fig. 2) at the Lee Kong Chian Natural History Museum (LKCNHM, Singapore), were photographed and characterised according to the diagnostic characters specified by Schmidt-Roach et al. (2014a).
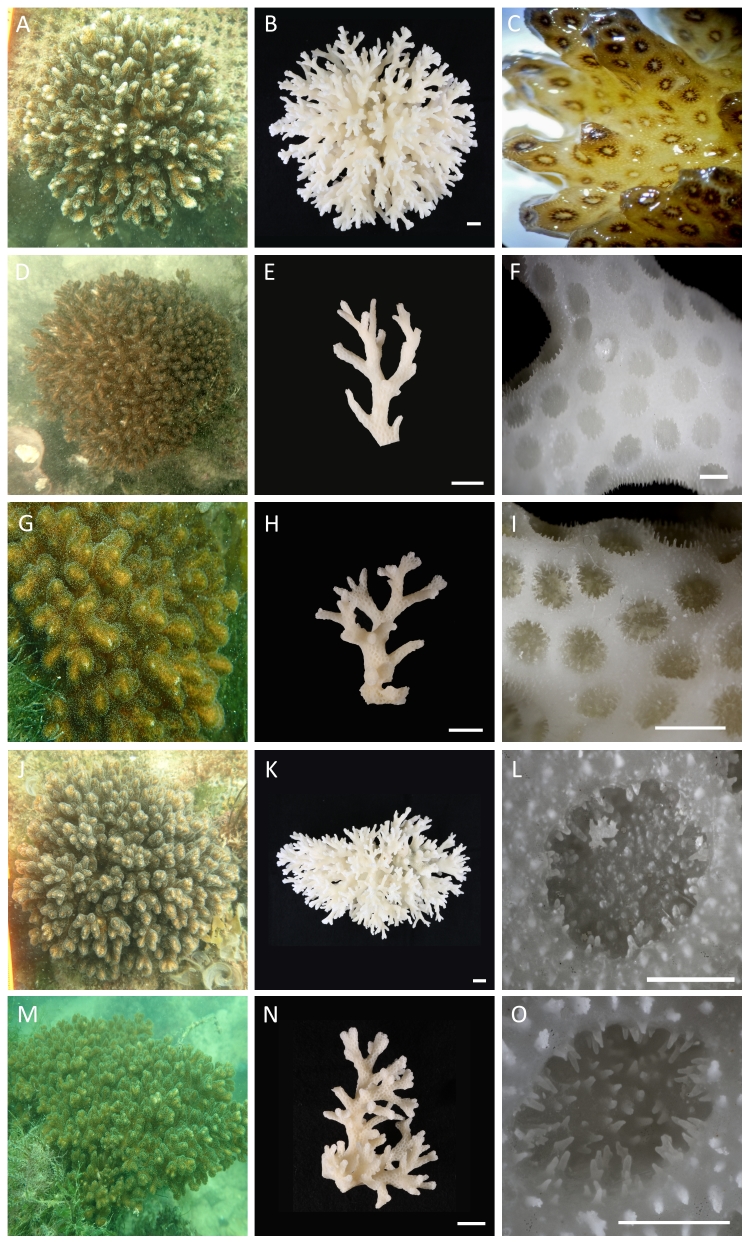
Figure 1.
Pocillopora specimens examined in this study. In situ appearances (A: HD159, D: HD162, G: HD161, J: HD160, M: HD154), with corresponding images of bleached skeletons (B, E, H, K, N). C, live specimen showing brown ring surrounding each oral opening (image by Jenny). F, I, branches from colonies shown in D and G respectively. L, O, calices and septa from colonies shown in J and M respectively. Scale bars represent 1 cm (B, E, H, K, N) and 1 mm (F, I, L, O) respectively.
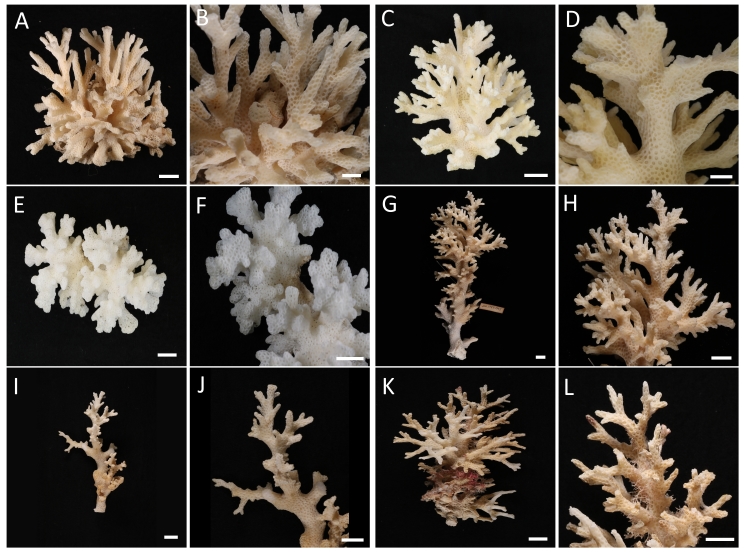
Figure 2.
Pocillopora specimens previously identified as P. damicornis from the Zoological Reference Collection, Lee Kong Chian Natural History Museum, Singapore (A, B: ZRC.1980.20.133; C, D: ZRC.1991.766; E, F: ZRC.1987.1538; G, H: ZRC.1987.1995; I, J: ZRC.1991.763; K, L: ZRC.1987.1537). A–F, colonies with thick branches; G–L, colonies with thinner branches. Scale bars represent 1 cm.
DNA was extracted by overnight digestion in hexadecyltrimethylammonium bromide (CTAB) and proteinase K, followed by phase separation using phenol: chloroform: isoamyl-alcohol (25:24:1). Polymerase chain reactions primed using FATP6.1 and RORF were performed according to Flot et al. (2008) for amplifying the mitochondrial open reading frame (Flot and Tillier 2007). Reaction products were purified using SureClean Plus (Bioline) and sequenced with the 3730xl DNA Analyzer (Thermo Fisher Scientific). Sequences were deposited in GenBank (accession numbers KY587458–KY587472).
We compiled in Mesquite 3.10 (Maddison and Maddison 2016) the sequences obtained here combined with 146 sequences available in GenBank (Schmidt-Roach et al. 2012, Schmidt-Roach et al. 2013, Marti-Puig et al. 2014), which were derived from Pocillopora eydouxi and P. ligulata—designated as outgroups in our analyses—as well as P. damicornis, P. acuta, P. aliciae, P. verrucosa and P. meandrina (Types α, β, δ, γ and m, respectively, according to Schmidt-Roach et al. 2014a). Alignments were carried out in MAFFT 7.205 using default parameters (Katoh and Standley 2013, Katoh and Toh 2008, Katoh et al. 2002), and the 850-bp data matrix was analysed phylogenetically under maximum likelihood and Bayesian optimality criteria.
We used RAxML 8.0.9 (Stamatakis 2006, Stamatakis 2014, Stamatakis et al. 2008) for maximum likelihood analysis, generating 50 alternate runs from distinct parsimony starting trees with the default GTRGAMMA substitution model. Branch supports were assessed via 1000 replicates of bootstrap analysis. For Bayesian analysis, we first selected the most suitable evolutionary model for the data (GTR + I) using jModelTest 2.1.10 (Guindon and Gascuel 2003, Darriba et al. 2012, Posada 2008), following the Akaike information criterion (AIC). Then, Bayesian inference was carried out using MrBayes 3.2.5 (Huelsenbeck and Ronquist 2001, Ronquist and Huelsenbeck 2003, Ronquist et al. 2012), implementing six million generations of Markov chain Monte Carlo in two separate runs and saving a tree every hundredth generation. We used Tracer 1.6 (Rambaut et al. 2014) to assess convergence among the runs, and determined that the first 10001 trees were to be discarded as burn-in.
Results
Our phylogenetic analysis of seven Pocillopora species recovers two moderately-supported monophyletic groups of P. meandrina + P. verrucosa (P. damicornis type γ) and ‘Clade 1’, as defined by Schmidt-Roach et al. (2014a). The latter clade comprises the P. damicornis types α, β and δ, which correspond to the newly-delimited taxa P. damicornis, P. acuta and P. aliciae, respectively (Fig. 3).
Figure 3.
Maximum likelihood tree of seven Pocillopora species based on the mitochondrial open reading frame. Colonies from Singapore are shown in red. Bootstrap values (≥ 50) and Bayesian posterior probabilities (≥ 0.85) are shown for supported clades.
The topology and statistical supports within this clade match those obtained by Schmidt-Roach et al. (2014a), with only the subclade of type α being highly supported. Types β and δ are only minimally supported by maximum likelihood inference and not supported by Bayesian analysis. Nevertheless, each of these sequence types are supported by nucleotide changes that are fixed within the clade. Type α sequences are distinguished from type β and type δ sequences based on six and four variable sites, respectively, while the latter two types are different at two sites across the 850-bp alignment. Therefore, sequencing errors aside, ORF sequences can help separate the three species effectively despite minimal phylogenetic supports.
Pocillopora acuta Lamarck, 1816
Pocillopora acuta Lamarck, 1816, p. 274; Schmidt-Roach et al. 2014a, p. 17; Schmidt-Roach et al. 2014b, p. 11; Kitano et al. 2015, p. 21; Mayfield et al. 2015, p. 1.
Materials examined. MNHN-IK-2010-792 (holotype, Muséum national d’Histoire naturelle de Paris, France; type locality: Indian Ocean); see Table 1 for voucher specimens (ZRC, LKCNHM).
Table 1.
Voucher specimens examined.
| Specimen no. | Catalogue no. | Locality | Latitude, Longitude | Date collected | Collector | Last identification |
| RA1 | - | Raffles Lighthouse | 1.1602°N, 103.7403°E | Oct 2015 | R.C. Poquita-Du | Pocillopora damicornis |
| GA1 | - | Raffles Lighthouse | 1.1602°N, 103.7403°E | Oct 2015 | R.C. Poquita-Du | Pocillopora damicornis |
| SA1 | - | St. John’s Island | 1.2236°N, 103.8452°E | Oct 2015 | R.C. Poquita-Du | Pocillopora damicornis |
| HD154 | ZRC.CNI.1067 | Pulau Subar Darat | 1.2158°N, 103.8314°E | Oct 2016 | D. Huang | Pocillopora acuta |
| HD155 | - | Pulau Subar Darat | 1.2158°N, 103.8314°E | Oct 2016 | D. Huang | Pocillopora acuta |
| HD156 | - | Pulau Subar Darat | 1.2158°N, 103.8314°E | Oct 2016 | Y.C. Tay | Pocillopora acuta |
| HD157 | ZRC.CNI.1068 | Pulau Subar Darat | 1.2158°N, 103.8314°E | Oct 2016 | Y.C. Tay | Pocillopora acuta |
| HD158 | - | Pulau Subar Darat | 1.2158°N, 103.8314°E | Oct 2016 | Y.C. Tay | Pocillopora acuta |
| KA1 | - | Kusu Island | 1.2257°N, 103.8602°E | Oct 2015 | R.C. Poquita-Du | Pocillopora damicornis |
| HD159 | ZRC.CNI.1069 | Kusu Island | 1.2257°N, 103.8602°E | Oct 2016 | C.S.L. Ng | Pocillopora acuta |
| HD160 | ZRC.CNI.1070 | Kusu Island | 1.2257°N, 103.8602°E | Oct 2016 | C.S.L. Ng | Pocillopora acuta |
| HD161 | ZRC.CNI.1071 | Pulau Subar Laut | 1.2126°N, 103.8334°E | Oct 2016 | Y.C. Tay | Pocillopora acuta |
| HD162 | ZRC.CNI.1072 | Pulau Subar Laut | 1.2126°N, 103.8334°E | Oct 2016 | Y.C. Tay | Pocillopora acuta |
| HD163 | - | Pulau Subar Laut | 1.2126°N, 103.8334°E | Oct 2016 | Y.C. Tay | Pocillopora acuta |
| HD164 | - | Pulau Subar Laut | 1.2126°N, 103.8334°E | Oct 2016 | Y.C. Tay | Pocillopora acuta |
| HD165 | - | Pulau Subar Laut | 1.2126°N, 103.8334°E | Oct 2016 | Y.C. Tay | Pocillopora acuta |
| - | ZRC.1980.3.20.133 | Sentosa | - | Sep 1979 | L.T. Chan | Pocillopora damicornis |
| - | ZRC.1987.1537 | Pulau Hantu | - | 1987 | L.M. Chou | Pocillopora damicornis |
| - | ZRC.1987.1538 | Pulau Hantu | - | 1987 | L.M. Chou | Pocillopora damicornis |
| - | ZRC.1991.763 | Pulau Hantu | - | 1991 | Reef Ecology Study Team | Pocillopora damicornis |
| - | ZRC.1991.766 | Singapore | 1991 | Reef Ecology Study Team | Pocillopora damicornis | |
| - | ZRC.1987.1995 | Singapore | - | - | Reef Ecology Study Team | Pocillopora damicornis |
Description. Colonial, densely caespitose (Fig. 1A, D, G, J, M); branches typically round in cross section, but may become flattened at the tips, which are usually sharply pointed. Branches of colonies in exposed sites thicken and have smaller spacing between branches (Fig. 2A–F), while those in sheltered sites are elongate and slender (Fig. 2G–L). Calices typically oval, with the small diameter ranging between 0.6 and 0.8 mm, and the large diameter between 1.0 and 1.2 mm. Columellae flat. Septa poorly developed, in two equal cycles; 12 septa per corallite (Fig. 1L, O). Coenosteum with fine spinules. Living colony pale-greenish in colour with characteristic darker pigmentation surrounding oral opening of polyps (Fig. 1C).
Remarks. Colonies collected from Singapore’s reefs show great variation in branching morphologies, overlapping with those described for Pocillopora acuta and P. damicornis by Schmidt-Roach et al. (2014a). For instance, Pocillopora acuta colonies observed from exposed environments in Singapore have considerably thicker branches (~6 mm) than those shown in Schmidt-Roach et al. (2014a). The pigmentation rings may not be clearly visible for all corallites in every colony; they vary from faint to intense brown. Nevertheless, sequences of the mitochondrial open reading frame from these colonies are all nested within the type-β clade that has been redefined as P. acuta (Schmidt-Roach et al. 2014a). The bootstrap support and posterior probability are low, but our sequences exactly match (100% identity) nearly all the published P. acuta sequences obtained from Australia and Japan (Fig. 3).
Discussion
This study contributes to the limited data that have emerged from the South China Sea region on the identity of Pocillopora species. The recently-revived species P. acuta is described to have a wide distribution reaching from the central Pacific to the Indian Ocean (Schmidt-Roach et al. 2014a). Our results show clearly the presence of Pocillopora acuta in Singapore, one of the southernmost localities in the South China Sea. Here, it is found in nearly every offshore reef (Huang et al. 2009), and colonies are common in the shallows (0–4 m below chart datum) but generally do not exist beyond 6 m depth. They thrive in various types of habitats including on artificial substrates such as seawalls and pontoons (Toh et al. 2017), and serve as habitat for diverse ectosymbiont communities (Lee and Sin 2009). The species is hermaphroditic (Kerr et al. 2011) and has been documented to brood monthly in Singapore, just before or after the new moon (Chou and Quek 1993, Toh et al. 2013).
Although we find no contemporary evidence of P. damicornis on Singapore’s reefs, the variability of colony branch thickness among specimens from past collections held at the LKCNHM (Fig. 2) appears greater than those collected from the field for this study and overlaps with that of P. damicornis as defined by Schmidt-Roach et al. (2014a). Nevertheless, some specimens we sequenced as P. acuta here, which were collected from several environments spread across most of the reef localities in Singapore, also had thick branches and rounded tips (e.g. HD154; Fig. 1M, N) diagnostic of P. damicornis (Schmidt-Roach et al. 2014a). This wide morphological variation appears to be associated with wave and current exposure, with an increase in branch thickness going from sheltered sites to more exposed ones. It is also possible that the actual P. damicornis species had been present before but faced recent losses locally due to pressures from coastal development (Chou 2006, Huang et al. 2009). Hence, a regional investigation with genetic samples from less impacted reefs may help clarify the historical distributions of P. acuta and P. damicornis.
The taxonomy of South China Sea Pocillopora remains poorly understood. A previous study has shown that Pocillopora ‘damicornis-like’ types 4 and 5 are present in Taiwan, while the Gulf of Thailand only hosts the latter type (Pinzón et al. 2013). More recently, these genetic types 4 and 5 have been delimited as Pocillopora damicornis (type α) and P. acuta (type β) respectively by Schmidt-Roach et al. (2013), Schmidt-Roach et al. (2014a). These findings were based on the mitochondrial ORF, the same marker we have sequenced here, thus underlining its utility as a reliable method of identifying the Pocillopora species present in Singapore.
Broadly, Pocillopora acuta is present in both Taiwan and Gulf of Thailand, but P. damicornis appears to be limited to the northern South China Sea as it has thus far only been confirmed from Taiwan using the mitochondrial ORF. Further north at the Yaeyama Islands, Japan, P. damicornis is present but is likely rare relative to P. acuta (Kitano et al. 2015). Despite that, P. damicornis is the only Pocillopora species detected along mainland Japan to date (Pinzón et al. 2013). All these sequencing work, applied on a wide range of colony and corallite morphologies that overlap with both species (Figs 1, 2), suggests that P. damicornis is rare in the South China Sea and southern Japan.
Thus far ranging from the Central Pacific to the Indian Ocean through Singapore, Taiwan and Hawai’i, further sampling may reveal the presence of P. acuta in more localities in the Central Indo-Pacific. Overall, the emerging picture shows that most ‘damicornis-like’ corals in the southwestern South China Sea region are actually P. acuta instead of P. damicornis.
Acknowledgements
This study is funded by the National Research Foundation (Singapore) through the Marine Science R&D Programme (R-154-000-A25-281). We thank Bert Hoeksema and an anonymous reviewer for helping to improve the manuscript, and Chua Keng Soon (LKCNHM) for assistance with loans, imaging and cataloguing of specimens.
References
- Best Maya Borel, Boekschoten G J, Oosterbaan A. Species concept and ecomorph variation in living and fossil Scleractinia. http://archive.org/details/palaeontographic541984pale Palaeontographica Americana. 1984;54:70–79. [Google Scholar]
- Chou Loke Ming. Marine habitats in one of the world’s busiest harbours. In: Wolanski E, editor. The Environment in Asia Pacific Harbours. Springer; The Netherlands: 2006. 377-391. [DOI] [Google Scholar]
- Chou Loke Ming, Quek S T. Planulation in the scleractinian coral Pocillopora damicornis in Singapore waters. Proceedings of the 7th International Coral Reef Symposium. 1993;1:500. [Google Scholar]
- Darriba Diego, Taboada Guillermo L, Doallo Ramón, Posada David. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9(8) doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flot Jean-François, Tillier Simon. Molecular phylogeny and systematics of the scleractinian coral genus Pocillopora in Hawaii. Proceedings of the 10th International Coral Reef Symposium. 2006:24–29.
- Flot Jean-François, Tillier Simon. The mitochondrial genome of Pocillopora (Cnidaria: Scleractinia) contains two variable regions: The putative D-loop and a novel ORF of unknown function. Gene. 2007;401:80–87. doi: 10.1016/j.gene.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Flot Jean-François, Magalon H, Cruaud Corinne, Couloux A, Tillier Simon. Patterns of genetic structure among Hawaiian corals of the genus Pocillopora yield clusters of individuals that are compatible with morphology. Comptes Rendus Biologies. 2008;331:239–247. doi: 10.1016/j.crvi.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Foster Ann Budd. Patterns of small-scale variation of skeletal morphology within the scleractinian corals, Montastrea annularis and Siderastrea siderea. http://www.reefbase.org/resource_center/publication/pub_22034.aspx Proceedings of the Third International Coral Reef Symposium. 1977;2:409–415. [Google Scholar]
- Foster Ann Budd. The relationship between corallite morphology and colony shape in some massive reef-corals. http://dx.doi.org/10.1007/BF00304728. Coral Reefs. 1983;2:19–25. doi: 10.1007/BF00304728. [DOI] [Google Scholar]
- Guindon Stéphane, Gascuel Oliver. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Huang Danwei, Tun Karenne P P, Chou Loke Ming, Todd Peter A. An inventory of zooxanthellate scleractinian corals in Singapore, including 33 new records. http://rmbr.nus.edu.sg/rbz/biblio/s22/s22rbz069-080.pdf Raffles Bulletin of Zoology. 2009;Supplement 22:69–80. [Google Scholar]
- Huelsenbeck John P, Ronquist Fredrik. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Katoh Kazutaka, Standley Daron M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Kazutaka, Toh Hiroyuki. Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics. 2008;9(4):286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Katoh Kazutaka, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr Alexander M, Baird Andrew H, Hughes Terence P. Correlated evolution of sex and reproductive mode in corals (Anthozoa: Scleractinia) http://rspb.royalsocietypublishing.org/cgi/content/abstract/278/1702/75. Proceedings of the Royal Society B-Biological Sciences. 2011;278(1702):75–81. doi: 10.1098/rspb.2010.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano Yuko F, Nagai Satoshi, Ueno Mitsuhiro, Yasuda Nina. Most Pocillopora damicornis around Yaeyama Islands are Pocillopora acuta according to mitochondrial ORF sequences. http://doi.org/10.3755/galaxea.17.21. Galaxea. 2015;17(1):21–22. doi: 10.3755/galaxea.17.21. [DOI] [Google Scholar]
- Lee Ai Chin, Sin Tsai Min. Trapezia septata Dana, 1852 (Brachyura, Trapeziidae): a new record for Singapore with notes on its relationship with the host coral, Pocillopora verrucosa. http://www.jstor.org/stable/20696235. Crustaceana. 2009;82(12):1603–1608. doi: 10.1163/156854009X463838. [DOI] [Google Scholar]
- Mace G. M. The role of taxonomy in species conservation. Philosophical Transactions of the Royal Society B: Biological Sciences. 2004;359(1444):711–719. doi: 10.1098/rstb.2003.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W P, Maddison D R. Mesquite: A Modular System for Evolutionary Analysis (Version 3.10) http://mesquiteproject.org 2016
- Marti-Puig Patricia, Forsman Zac H, Haverkort-Yeh Roxanne D, Knapp Ingrid S S, Maragos James E, Toonen Robert J. Extreme phenotypic polymorphism in the coral genus Pocillopora; micro-morphology corresponds to mitochondrial groups, while colony morphology does not. http://dx.doi.org/10.5343/bms.2012.1080. Bulletin of Marine Science. 2014;90(1):211–231. doi: 10.5343/bms.2012.1080. [DOI] [Google Scholar]
- Mayfield Anderson B, Bruckner Andrew W, Chen Chien-Hsun, Chen Chii-Shiarng. A survey of pocilloporid corals and their endosymbiotic dinoflagellate communities in the Austral and Cook Islands of the South Pacific. Platax. 2015;12:1–17. [Google Scholar]
- Ng Chin Soon Lionel, Toh Tai Chong, Chou Loke Ming. Coral restoration in Singapore’s sediment-challenged sea. Regional Studies in Marine Science. 2016;8:422–429. doi: 10.1016/j.rsma.2016.05.005. [DOI] [Google Scholar]
- Pinzón Jorge H, Sampayo Eugenia, Cox Evelyn, Chauka Leonard J, Chen Chaolun Allen, Voolstra Christian R, LaJeunesse Todd C. Blind to morphology: genetics identifies several widespread ecologically common species and few endemics among Indo-Pacific cauliflower corals (Pocillopora, Scleractinia) http://doi.wiley.com/10.1111/jbi.12110. Journal of Biogeography. 2013;40:1595–1608. doi: 10.1111/jbi.12110. [DOI] [Google Scholar]
- Posada David. jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution. 2008;25(7):1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Suchard M A, Xie D, Drummond A J. Tracer: MCMC Trace Analysis Tool. Version 1.6. http://beast.bio.ed.ac.uk/Tracer 2014
- Ronquist Fredrik, Huelsenbeck John P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ronquist Fredrik, Teslenko M, van der Mark P, Ayres Daniel L, Darling Aaron, Höhna Sebastian, Larget B, Liu Liang, Suchard Marc A, Huelsenbeck John P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. http://sysbio.oxfordjournals.org/cgi/doi/10.1093/sysbio/sys029. Systematic Biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Roach Sebastian, Miller Karen J, Lundgren Petra, Andreakis Nikos. With eyes wide open: a revision of species within and closely related to the Pocillopora damicornis species complex (Scleractinia; Pocilloporidae) using morphology and genetics. http://doi.wiley.com/10.1111/zoj.12092. Zoological Journal of the Linnean Society. 2014;170(1):1–33. doi: 10.1111/zoj.12092. [DOI] [Google Scholar]
- Schmidt-Roach Sebastian, Johnston Erika, Fontana Silvia, Jury Christopher P, Forsman Zac. Daytime spawning of Pocillopora species in Kaneohe Bay, Hawai'i. Galaxea. 2014;16:11–12. doi: 10.3755/galaxea.16.11. [DOI] [Google Scholar]
- Schmidt-Roach Sebastian, Miller Karen J, Woolsey Erika, Gerlach Gabriele, Baird Andrew H. Broadcast spawning by Pocillopora species on the Great Barrier Reef. http://dx.plos.org/10.1371/journal.pone.0050847. PLoS ONE. 2012;7(12) doi: 10.1371/journal.pone.0050847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Roach Sebastian, Lundgren Petra, Miller Karen J, Gerlach Gabriele, Noreen Annika M E, Andreakis Nikos. Assessing hidden species diversity in the coral Pocillopora damicornis from Eastern Australia. http://link.springer.com/10.1007/s00338-012-0959-z. Coral Reefs. 2013;32(1):161–172. doi: 10.1007/s00338-012-0959-z. [DOI] [Google Scholar]
- Stamatakis Alexandros. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis Alexandros. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis Alexandros, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Todd Peter A, Ladle Richard J, Lewin-Koh N J I, Chou Loke Ming. Genotype × environment interactions in transplanted clones of the massive corals Favia speciosa and Diploastrea heliopora. Marine Ecology-Progress Series. 2004;271:167–182. doi: 10.3354/meps271167. [DOI] [Google Scholar]
- Toh Kok Ben, Ng Chin Soon Lionel, Wu Bokai, Toh Tai Chong, Cheo Pei Rong, Tun Karenne, Chou Loke Ming. Spatial variability of epibiotic assemblages on marina pontoons in Singapore. http://dx.doi.org/10.1007/s11252-016-0589-2. Urban Ecosystems. 2017;20:183–197. doi: 10.1007/s11252-016-0589-2. [DOI] [Google Scholar]
- Toh Tai Chong, Peh Jia Wei Kassler, Chou Loke Ming. Heterotrophy in recruits of the scleractinian coral Pocillopora damicornis. http://www.tandfonline.com/doi/abs/10.1080/10236244.2013.832890. Marine and Freshwater Behaviour and Physiology. 2013;46(5):313–320. doi: 10.1080/10236244.2013.832890. [DOI] [Google Scholar]
- Veron John E N. Corals in Space and Time. UNSW Press; Sydney: 1995. 320 [Google Scholar]
- Veron John E N. Corals of the World. Australian Institute of Marine Science; Townsville: 2000. 1381 [Google Scholar]
- Veron John E N, Pichon Michel. Scleractinia of Eastern Australia. Part I. Families Thamnasteriidae, Astrocoeniidae, Pocilloporidae. Vol. 1. Australian Institute of Marine Science; Townsville: 1976. 85. [Google Scholar]
- Wheeler Q. D. Taxonomic triage and the poverty of phylogeny. Philosophical Transactions of the Royal Society B: Biological Sciences. 2004;359(1444):571–583. doi: 10.1098/rstb.2003.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]