Abstract
Based on the formation of the XY body at pachytene and expression studies of a few X-linked genes, the X and Y chromosomes seem to undergo transcriptional inactivation during mammalian spermatogenesis. However, the extent and the mechanism of X and Y inactivation are not known. Here, we show that both the X and Y chromosomes undergo sequential changes in their histone modifications beginning at the pachytene stage of meiosis. These changes usually are associated with transcriptional inactivation in somatic cells, and they coincide with the exclusion of the phosphorylated (active) form of RNA polymerase II from the XY body. Both sex chromosomes undergo extensive deacetylation at histones H3 and H4 and (di)methylation of lysine (K)9 on histone H3; however, there are no changes in H3–K4 methylation. These changes persist even when the XY body disappears in late pachytene, and the X and Y chromosomes segregate from one another after the first meiotic division. By the spermatid stage, histone modifications of the X and Y chromosomes revert to those of active chromatin and RNA polymerase II reengages with both chromosomes. Our observations indicate that X and Y inactivation is extensive and persists even when the X and Y chromosomes are separated in secondary spermatocytes. These findings provide insights into epigenetic programming and chromatin dynamics in the male germ line.
Keywords: epigenetic, meiosis, X inactivation, chromatin, methylation
The process of spermatogenesis in mammals consists of the sequential stages shown in Fig. 1, starting with diploid spermatogonia and ending in haploid sperm. At the pachytene stage (Fig. 1), when the pairing of the homologous autosomal chromosomes occurs, the X and Y chromosomes pair only at their pseudoautosomal regions and form the sex, or XY, body (1–6). In cytological preparations, the sex body is heteropycnotic, staining more intensely than the autosomes (7). Previous studies demonstrated that several genes on the X chromosome are subject to transcriptional repression during meiosis, beginning at pachytene with reactivation by the spermatid stage (8, 9). Furthermore, RNA polymerase II has been shown to be excluded from the XY body at pachytene (10, 11). However, the mechanism and even the extent of X and Y inactivation (XYi) during spermatogenesis are not known.
Fig. 1.
Stages of spermatogenesis showing the X and Y chromosomes. Chromosomal DNA (blue) is stained with 4′,6-diamidino-2-phenylindole (DAPI). Sex chromosomes are shown with DNA whole-chromosome paints for X (pink) and Y (green). The X and Y chromosomes pair at their pseudoautosomal regions to form the sex body at pachytene. After the first meiotic division (metaphase I), the X and Y chromosomes are in separate secondary spermatocytes (metaphase II). Four spermatids are formed from each spermatogonium. The duration of spermatogenesis, from spermatogonia to mature sperm, is ≈14–26 days in mice and 64–74 days in humans (1).
Unlike the inactive X in female somatic cells, the promoters of housekeeping genes on the X chromosome remain unmethylated throughout spermatogenesis in both humans and mice (8, 12). Xist, the X-inactive-specific transcript gene associated with X inactivation (Xi) in female cells, is transiently transcribed in the mouse testes at pachytene and repressed again in spermatids (13, 14). However, Xist transcription is not essential, given that male mice with an ablated Xist gene have normal spermatogenesis (15) and XY bodies (9, 16). Thus, the mechanisms of Xi in female somatic cells and XYi during spermatogenesis appear to have fundamental differences.
Recent evidence that the XY body at pachytene is enriched for histone H3–lysine (K)9 dimethylation in the mouse (17) and in Caenorhabditis elegans (18) suggested that histone modifications are involved in XYi during mammalian spermatogenesis, despite previous evidence to the contrary (19). Therefore, we have examined this possibility by using immunocytochemistry and DNA hybridization in situ during the various stages of mouse spermatogenesis. We find that the histones of both the X and Y chromosomes undergo sequential modifications (both acetylation and methylation) beginning at pachytene. However, in spermatids, histone modifications of X and Y were reversed to their status in early spermatogenesis. Furthermore, RNA polymerase II, which becomes excluded from the XY body at pachytene, reengages the X and Y chromosomes in round spermatids. These results indicate that both chromosomes undergo extensive inactivation from pachytene to the early round spermatid stage, but reactivation of the X and Y chromosomes occurs before the end of spermatogenesis.
Materials and Methods
Immunocytochemistry. Seminiferous tubules from adult mouse (age, ≥21 days) testes were minced into small fragments with small scissors to release germ cells. Immunocytochemistry was carried out as described in ref. 20. Briefly, germ cells were cytospun onto microscope slides, incubated with a primary antibody for 1 h, washed in KCM buffer (120 mM KCl/20 mM NaCl/10 mM Tris·HCl, pH 7.5/0.5 mM EDTA) and then incubated with a secondary antibody for 30 min. Cells were fixed by incubating the slides in 10% formalin for 10 min. Chromosomal DNA was stained by propidium iodide. Images were captured by using an Olympus microscope connected to an Applied Imaging (San Jose, CA) CytoVision system. The minimum numbers of cells examined with each antibody were 10 spermatogonial metaphases, 20 leptotenes, 20 zygotenes, 50 pachytenes, 10 diplotenes, 15 diakinesis, 15 metaphase I, 10 metaphase II, and 50 spermatids. All primary antibodies for histone modifications were purchased from Upstate Biotechnology (Lake Placid, NY) with the exception of H3-trimethyl-K4 (Abcam, Cambridge, U.K.). Antibody for the phosphorylated form of RNA polymerase II at serine 2 (H5) was purchased from Covance (Princeton).
Fluorescence in Situ Hybridization. DNA in situ hybridization was performed on immunostained slides by using DNA whole-chromosome paints for the mouse X and Y chromosomes as recommended by the manufacturer (Cambio, Cambridge, U.K.). Briefly, slides were dehydrated in an ethanol series, denatured in 70% formamide/30% 2× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) for 5 min at 65°C before incubation with denatured chromosome paints overnight at 42°C. The slides were washed in 0.4× SSC at 45°C for 2 min and in 2× SSC/0.1% Nonidet P-40 for 1 min. Chromosomal DNA was stained blue by 4′,6-diamidino-2-phenylindole. Images were captured as described above.
Results
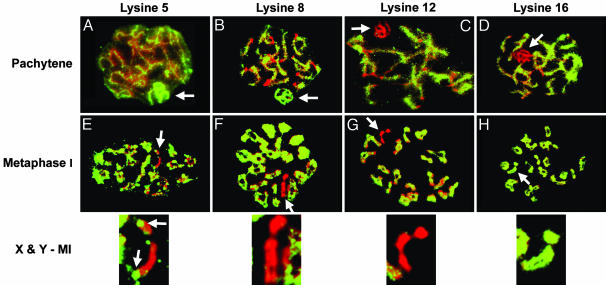
Differences in Histone H4 Acetylation Begin at Pachytene. The histone modifications of the X and Y chromosomes present in early spermatogenesis (spermatogonia, leptotene, and zygotene) are similar to those present in transcriptionally active chromatin. Specifically, there is acetylation at histones H4-K5, H4-K8, H4-K12, and H4-K16. However, at pachytene, the X and Y chromosomes become hyperacetylated at H4-K5 and H4-K8 (Fig. 2 A and B) and underacetylated at H4-K12 and H4-K16 (Fig. 2 C and D).
Fig. 2.
Changes in histone H4 acetylation of X and Y at pachytene and metaphase I. (A–D) At pachytene, the XY body is hyperacetylated at K5 and K8 (A and B) and underacetylated at K12 and K16 (C and D). (E–H) By metaphase I (MI), the X and Y chromosomes have become underacetylated at K5 and K8 (E and F) and are still underacetylated at K12 (G) and reacetylated at K16 (H). (E Lower) Note that the pericentromeric regions (arrows) of X and Y remain acetylated at K5.
By diplotene, the X and Y chromosomes are underacetylated at H4-K5, H4-K8, and H4-K12 (Table 1 and Fig. 2 E–G). Reacetylation of the X and Y chromosomes occurs at H4-K16 by diplotene (Table 1 and Fig. 2H), at H4-K8 by metaphase II, and at H4-K12 and H4-K5 by the round spermatid stage. The changes on the X and Y chromosomes are extensive except for the pericentromeric regions of X and Y, which remain acetylated at H4-K5 at diplotene, diakinesis, and metaphase I (Fig. 2E).
Table 1. Histone modifications of X and Y during mouse spermatogenesis.
| Acetyl
|
Dimethyl
|
Trimethyl
|
||||||
|---|---|---|---|---|---|---|---|---|
| Stage | H4-K5 | H4-K8 | H4-K12 | H4-K16 | H3-K9 | H3-K9 | H3-K4 | H3-K4 |
| Spermatogonia | A | A | A | A | A | UnderM | HyperM | UnderM |
| Leptotene | A | A | A | A | A | UnderM | HyperM | UnderM |
| Zygotene | A | A | A | A | A | UnderM | HyperM | UnderM |
| Early pachytene | HyperA | HyperA | UnderA | UnderA | HyperA | UnderM | HyperM | UnderM |
| Late pachytene | HyperA | HyperA | UnderA | UnderA | UnderA | HyperM | HyperM | UnderM |
| Diplotene | UnderA | UnderA | UnderA | A | UnderA | HyperM | HyperM | UnderM |
| Diakinesis | UnderA | UnderA | UnderA | A | UnderA | HyperM | HyperM | UnderM |
| Metaphase I | UnderA | UnderA | UnderA | A | UnderA | HyperM | HyperM | UnderM |
| Metaphase II | UnderA | A | UnderA | A | UnderA | HyperM | HyperM | UnderM |
| Early spermatids | A | A | A | A | UnderA | HyperM | HyperM | UnderM |
| Mid spermatids | A | A | A | A | A | UnderM | HyperM | UnderM |
A, acetylated; hyperA, hyperacetylated; underA, underacetylated; hyperM, hypermethylated; underM, undermethylated.
Transition from H3-K9 Hyperacetylation to H3-K9 Dimethylation at Pachytene. In the early stages of spermatogenesis (spermatogonia, leptotene, and zygotene), the X and Y chromosomes are acetylated at H3-K9 to the same extent as the autosomes. But by early pachytene, the level of acetylation at H3-K9 increases (Fig. 3A). This increase is not accompanied by a change in methylation (Fig. 3B). Yet by late pachytene, the X and Y chromosomes become underacetylated and hyperdimethylated at H3-K9 (Table 1). Fig. 3 C and D shows that the X and Y chromosomes are extensively underacetylated and hyperdimethylated at H3-K9 at diplotene, except for a small region on the X chromosome. Pericentromeric regions of the autosomes are also enriched with H3-K9 dimethylation (Fig. 5, which is published as supporting information on the PNAS web site). The transition from dimethylation back to acetylation at H3-K9 begins in early round spermatids, where the X and Y chromosomes are still methylated, but to a lesser extent than in previous stages (i.e., late pachytene to metaphase II). By midstage spermatids, the transition is complete and X and Y are undermethylated and reacetylated at H3-K9 (Table 1).
Fig. 3.
Transition from H3-K9 hyperacetylation to H3-K9 hypermethylation. (A and B) In early pachytene, the XY body is hyperacetylated at H3-K9 (A) and undermethylated at H3-dimethyl-K9 (B). (C) At diplotene, the X and Y chromosomes are extensively underacetylated at H3-K9; however, there is a discrete region on the X chromosome that remains acetylated (Lower). (D) X and Y are extensively hypermethylated at H3-dimethyl-K9; however, there is a discrete region on the X chromosome that remains undermethylated (Lower) and appears to correspond to the same location that remains acetylated, as shown in C Lower.
RNA Polymerase II Distribution During Spermatogenesis. To link the changes in histone modifications that we observe during spermatogenesis to the transcription of the X and Y chromosomes, we used an antibody that recognizes the elongating form of RNA polymerase II (phosphorylated at serine 2), which is only present in transcriptionally engaged chromatin (21–25). We found that RNA polymerase II became excluded from the XY body at pachytene (Fig. 4A), the stage at which the X and Y chromosomes become deacetylated at histones H3 and H4, and enriched with H3-K9 dimethylation (Table 1). At diplotene, diakinesis, metaphase I, and metaphase II, RNA polymerase II is largely excluded from all chromosomes. However, by the spermatid stage, RNA polymerase II reengages all chromosomes, including X and Y (Fig. 4B).
Fig. 4.
RNA polymerase II distribution at pachytene and the round spermatid stage. (A) The phosphorylated (active) form of RNA polymerase II (H5) is excluded from XY body (arrow) at pachytene. (B) The phosphorylated form of RNA polymerase II is engaged with the X chromosome in the round spermatid (arrow) but excluded from the chromocenter. PI, propidium iodide.
H3-K4 Methylation Does Not Change Throughout Spermatogenesis. At H3-K4, the X and Y chromosomes are markedly dimethylated but lack H3-K4 trimethylation throughout spermatogenesis (Table 1 and Fig. 6, which is published as supporting information on the PNAS web site). However, at all of the stages, there are discrete regions on the X and Y chromosomes with H3-K4 trimethylation, including the pseudoautosomal regions.
Histone Modifications of X and Y Are Concordant. The histone modifications of the Y chromosome are completely concordant with the X chromosome at every stage of spermatogenesis (Table 1). Even when the chromosomes are in separate metaphase II spermatocytes, the X and Y chromosomes remain concordant (Fig. 7, which is published as supporting information on the PNAS web site).
Discussion
The patterns of histone modifications of the X and Y chromosomes in early spermatogenesis and in round spermatids are consistent with patterns associated with active regions of the genome in somatic cells (i.e., H3 and H4 acetylation and H3-K9 undermethylation). In contrast, the changes beginning at pachytene are those expected for transcriptionally inactive chromatin (i.e., H3 and H4 deacetylation and H3-K9 dimethylation) (26–29). That almost all of the X and Y chromosomes are subject to these changes indicates that the repression is extensive, affecting most of the genes on both chromosomes. The initiation of changes in histone modifications is coincident with the formation of the sex body at pachytene (2–4, 6, 7) and the exclusion of phosphorylated (active) form of RNA polymerase II (H5) from the sex body (10, 11) and is consistent with the limited gene expression data showing repression at pachytene and reactivation by the round spermatid stage (8, 9). Our results also show that, unlike the sex body, which disappears at the diplotene stage, inactivation persists to the end of meiosis. By the round spermatid stage, RNA polymerase II reengages the X and Y chromosomes, indicating an end to their inactivation. Because the changes in histone modifications that we observed are not restricted to the pachytene stage but also occur in stages after pachytene, it does not seem likely that these effects are related to crossing over at pachytene.
Although all lysines of H4 examined underwent sequential changes in acetylation during spermatogenesis, the pattern of change was clearly distinct from one lysine to the other (Table 1). Before deacetylation, the acetylation levels at H4-K5, H4-K8, and H3-K9 on the sex body became greater than on the autosomes. The significance of the hyperacetylation preceding deacetylation at pachytene remains to be determined. Also of interest are the H4-K5 acetylation of the X and Y pericentromeric regions at diplotene, diakinesis, and metaphase I, when the rest of the X and Y chromosomes are underacetylated, and the relatively short period of H4-K16 underacetylation.
Transcriptionally repressed genes in somatic cells and the inactive X chromosome in females are devoid of acetylation at all four lysines of histone H4 (20, 28). The differences in changes at these lysines that we observed on the X and Y chromosomes during spermatogenesis have, to our knowledge, not been previously reported in somatic cells. Also, during the process of Xi in female embryonic stem cells, H3-K9 dimethylation precedes H4 deacetylation (30). By contrast, deacetylation of K12 and K16 of H4 precedes H3-K9 dimethylation during XYi. It is not currently clear how the acetylation and deacetylation at each lysine contribute to the activation and repression of genes, but future studies during spermatogenesis should help elucidate the functional roles of the changes in histone modifications that we observed.
In somatic cells, dimethylation of H3-K4 is seen in both active and repressed chromatin, whereas trimethylation of H3-K4 is only associated with active chromatin (31). We found that throughout spermatogenesis, the X and Y chromosomes were enriched with H3-dimethyl-K4 and largely devoid of H3-K4 trimethylation. Thus the status of H3-K4 di- and tri-methylation was stable throughout spermatogenesis and did not reflect the transient transcriptional repression that occurs between the pachytene and spermatid stages. These modifications may be programmed in primordial germ cells before spermatogenesis. Therefore, it seems that some modifications are programmed for transient repression and derepression, whereas others are programmed for the long run.
Xi in somatic cells is needed to compensate for the sex difference in numbers of X chromosomes (32–36). However, the reason for XYi has not yet been elucidated (3, 4, 6). It could be that XYi is also a means of dosage compensation to assure that X-bearing spermatocytes would not have a selective advantage over those with only a Y chromosome.
It has recently been suggested that Xi in females occurs earlier than has been thought (22, 37). Analysis of histone modifications by Okamoto et al. (22) suggest that the paternal X becomes preferentially inactivated in the female mouse embryo at the four-cell stage of development. However, Huynh and Lee's (37) study of gene expression patterns suggested that inactivation occurs even earlier, perhaps during spermatogenesis before fertilization. Our observations at the spermatid stage, when the histone modifications of the X chromosome revert to those of active chromatin and the phosphorylated form of RNA polymerase II reengages the X chromosome, indicate that the paternal X chromosome is no longer inactive. The data would suggest that the X chromosome is modified epigenetically during spermatogenesis, which may contribute to the preferential inactivation of the paternal X that occurs again only after fertilization.
In summary, our results indicate that the chromatin of both the X and Y chromosomes is transcriptionally inactive throughout the first and second meiotic cell divisions, which suggests that XYi may play a role in facilitating chromosome pairing at prophase I and dosage compensation in secondary spermatocytes (which only have either an X or Y chromosome). That the two chromosomes remain concordant even when separated into individual secondary spermatocytes indicates that the inactivation of one chromosome is not dependent on the inactivation of the other. Our observations also strongly suggest that some but not all of the histone modifications reflect the transient inactivation of the X and Y chromosomes during spermatogenesis. Because mammalian spermatogenesis takes place during an extended period and has distinct stages that can be identified cytologically, it provides a good model to dissect the functional significance of specific histone modifications and their role in chromatin organization.
Supplementary Material
Acknowledgments
We thank Barbara R. Migeon for critically reviewing the manuscript and providing insightful comments, Bryan M. Turner and Hugh Spotswood for providing protocol for immunocytochemistry and helpful suggestions, Bill Dunn and Tom Hollinger for valuable discussions regarding spermatogenesis, and Michelle Whitney for providing assistance with figure art. This work was supported by National Institutes of Health Grant HD36417 and the Hayward Foundation.
Author contributions: A.M.K., F.Z.B., and D.J.D. designed research; A.M.K. and F.Z.B. performed research; A.M.K. and D.J.D. analyzed data; and A.M.K. and D.J.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: XYi, X and Y inactivation; Xi, X inactivation.
References
- 1.Clermont, Y. (1972) Physiol. Rev. 52, 198–236. [DOI] [PubMed] [Google Scholar]
- 2.Solari, A. J. (1974) Int. Rev. Cytol. 38, 273–317. [DOI] [PubMed] [Google Scholar]
- 3.Baarends, W. M. & Grootegoed, J. A. (2003) Cytogenet. Genome Res. 103, 225–234. [DOI] [PubMed] [Google Scholar]
- 4.Handel, M. A. (2004) Exp. Cell Res. 296, 57–63. [DOI] [PubMed] [Google Scholar]
- 5.Handel, M. A. (2004) Nat. Genet. 36, 12–13. [DOI] [PubMed] [Google Scholar]
- 6.Hoyer-Fender, S. (2003) Cytogenet. Genome Res. 103, 245–255. [DOI] [PubMed] [Google Scholar]
- 7.Driscoll, D. J., Palmer, C. G. & Melman, A. (1979) Cytogenet. Cell Genet. 23, 23–32. [DOI] [PubMed] [Google Scholar]
- 8.McCarrey, J. R., Berg, W. M., Paragioudakis, S. J., Zhang, P. L., Dilworth, D. D., Arnold, B. L. & Rossi, J. J. (1992) Dev. Biol. 154, 160–168. [DOI] [PubMed] [Google Scholar]
- 9.McCarrey, J. R., Watson, C., Atencio, J., Ostermeier, G. C., Marahrens, Y., Jaenisch, R. & Krawetz, S. A. (2002) Genesis 34, 257–266. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Capetillo, O., Mahadevaiah, S. K., Celeste, A., Romanienko, P. J., Camerini-Otero, R. D., Bonner, W. M., Manova, K., Burgoyne, P. & Nussenzweig, A. (2003) Dev. Cell 4, 497–508. [DOI] [PubMed] [Google Scholar]
- 11.Ayoub, N., Richler, C. & Wahrman, J. (1997) Chromosoma 106, 1–10. [DOI] [PubMed] [Google Scholar]
- 12.Driscoll, D. J. & Migeon, B. R. (1990) Somatic Cell Mol. Genet. 16, 267–282. [DOI] [PubMed] [Google Scholar]
- 13.McCarrey, J. R. & Dilworth, D. D. (1992) Nat. Genet. 2, 200–203. [DOI] [PubMed] [Google Scholar]
- 14.Salido, E. C., Yen, P. H., Mohandas, T. K. & Shapiro, L. J. (1992) Nat. Genet. 2, 196–199. [DOI] [PubMed] [Google Scholar]
- 15.Marahrens, Y., Panning, B., Dausman, J., Strauss, W. & Jaenisch, R. (1997) Genes Dev. 11, 156–166. [DOI] [PubMed] [Google Scholar]
- 16.Turner, J. M., Mahadevaiah, S. K., Elliott, D. J., Garchon, H. J., Pehrson, J. R., Jaenisch, R. & Burgoyne, P. S. (2002) J. Cell Sci. 115, 4097–4105. [DOI] [PubMed] [Google Scholar]
- 17.Peters, A. H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C., Weipoltshammer, K., Pagani, M., Lachner, M., Kohlmaier, A., et al. (2001) Cell 107, 323–337. [DOI] [PubMed] [Google Scholar]
- 18.Bean, C. J., Schaner, C. E. & Kelly, W. G. (2004) Nat. Genet. 36, 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armstrong, S. J., Hulten, M. A., Keohane, A. M. & Turner, B. M. (1997) Exp. Cell Res. 230, 399–402. [DOI] [PubMed] [Google Scholar]
- 20.Jeppesen, P. & Turner, B. M. (1993) Cell 74, 281–289. [DOI] [PubMed] [Google Scholar]
- 21.Patturajan, M., Wei, X., Berezney, R. & Corden, J. L. (1998) Mol. Cell. Biol. 18, 2406–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto, I., Otte, A. P., Allis, C. D., Reinberg, D. & Heard, E. (2004) Science 303, 644–649. [DOI] [PubMed] [Google Scholar]
- 23.Ahn, S. H., Kim, M. & Buratowski, S. (2004) Mol. Cell 13, 67–76. [DOI] [PubMed] [Google Scholar]
- 24.Sims, R. J., III, Mandal, S. S. & Reinberg, D. (2004) Curr. Opin. Cell Biol. 16, 263–271. [DOI] [PubMed] [Google Scholar]
- 25.Orphanides, G. & Reinberg, D. (2000) Nature 407, 471–475. [DOI] [PubMed] [Google Scholar]
- 26.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 27.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 28.Litt, M. D., Simpson, M., Gaszner, M., Allis, C. D. & Felsenfeld, G. (2001) Science 293, 2453–2455. [DOI] [PubMed] [Google Scholar]
- 29.Turner, B. M. (2002) Cell 111, 285–291. [DOI] [PubMed] [Google Scholar]
- 30.Heard, E., Rougeulle, C., Arnaud, D., Avner, P., Allis, C. D. & Spector, D. L. (2001) Cell 107, 727–738. [DOI] [PubMed] [Google Scholar]
- 31.Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C., Schreiber, S. L., Mellor, J. & Kouzarides, T. (2002) Nature 419, 407–411. [DOI] [PubMed] [Google Scholar]
- 32.Lyon, M. F. (1961) Nature 190, 372–373. [DOI] [PubMed] [Google Scholar]
- 33.Avner, P. & Heard, E. (2001) Nat. Rev. Genet. 2, 59–67. [DOI] [PubMed] [Google Scholar]
- 34.Gartler, S. M. & Goldman, M. A. (2001) Curr. Opin. Pediatr. 13, 340–345. [DOI] [PubMed] [Google Scholar]
- 35.Migeon, B. R. (2002) Cytogenet. Genome Res. 99, 8–16. [DOI] [PubMed] [Google Scholar]
- 36.Plath, K., Mlynarczyk-Evans, S., Nusinow, D. A. & Panning, B. (2002) Annu. Rev. Genet. 36, 233–278. [DOI] [PubMed] [Google Scholar]
- 37.Huynh, K. D. & Lee, J. T. (2003) Nature 426, 857–862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.