Abstract
Background
Duckweed (family Lemnaceae) has recently been recognized as an ideal biomass feedstock for biofuel production due to its rapid growth and high starch content, which inspired interest in improving their productivity. Since microbes that co-exist with plants are known to have significant effects on their growth according to the previous studies for terrestrial plants, this study has attempted to understand the plant–microbial interactions of a duckweed, Lemna minor, focusing on the growth promotion/inhibition effects so as to assess the possibility of accelerated duckweed production by modifying co-existing bacterial community.
Results
Co-cultivation of aseptic L. minor and bacterial communities collected from various aquatic environments resulted in changes in duckweed growth ranging from −24 to +14% compared to aseptic control. A number of bacterial strains were isolated from both growth-promoting and growth-inhibitory communities, and examined for their co-existing effects on duckweed growth. Irrespective of the source, each strain showed promotive, inhibitory, or neutral effects when individually co-cultured with L. minor. To further analyze the interactions among these bacterial strains in a community, binary combinations of promotive and inhibitory strains were co-cultured with aseptic L. minor, resulting in that combinations of promotive–promotive or inhibitory–inhibitory strains generally showed effects similar to those of individual strains. However, combinations of promotive–inhibitory strains tended to show inhibitory effects while only Aquitalea magnusonii H3 exerted its plant growth-promoting effect in all combinations tested.
Conclusion
Significant change in biomass production was observed when duckweed was co-cultivated with environmental bacterial communities. Promotive, neutral, and inhibitory bacteria in the community would synergistically determine the effects. The results indicate the possibility of improving duckweed biomass production via regulation of co-existing bacterial communities.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-017-0746-8) contains supplementary material, which is available to authorized users.
Keywords: Duckweed, Plant growth-promoting bacteria, Plant growth-inhibiting bacteria, Biomass production
Background
Duckweed is a tiny floating aquatic plant that is characterized by a rapid growth, high tolerance to polluted water, global distribution, and high starch content [1]. For decades, duckweed was considered as an industrially versatile plant that could be used for animal feed [2, 3], organic fertilizer [4], and chemical toxicity tests [5, 6]. In recent years, duckweed has been recognized as an ideal feedstock for biofuel production, because their soft and starch-rich biomass enables larger yield of fuel ethanol, butanol, and biogas [7, 8]. Xu et al. [9] calculated that bioethanol production from duckweed is 1.5 times greater than that from maize, when considering all parts of the cultivation and fermentation processes. Further, since duckweed can efficiently remove nitrogen, phosphorus, and heavy metals from water during growth, it has also been used in low-cost and low-energy wastewater treatment systems [10–12]. Thus, co-beneficial systems that combine biofuel production and water purification using duckweed have been proposed [13].
The attractive features of duckweed as a biomass resource have inspired interest in improving their productivity via selection of species/strains with higher growth rates [14] and optimizing the design and operational parameters, such as harvest period and water depth, of cultivation systems [15–17]. The effects of nutrient strength [18, 19], light intensity, photoperiod [20], and temperature [19] on duckweed growth and starch accumulation have also been examined to improve the production.
In addition, microbes that co-exist with duckweed are believed to have significant effects on growth in natural cultivation systems. In the terrestrial sphere, plants are widely recognized to develop intimate interactions with microbes that are critical for their growth or survival [21]. Some symbiotic bacteria called plant growth-promoting bacteria (PGPB) are known to enhance host plant growth by increasing nutrient acquisition or alleviating biotic and abiotic stresses [22]. In the last few decades, considerable efforts have been dedicated to isolate and characterize PGPB for important terrestrial agricrops, and it is clear that an extremely wide range of the plants harbor beneficial bacteria such as PGPB.
Although there have been several studies to understand plant-associated microbial communities and to engineer them for optimal production of crops [23, 24], such studies on aquatic plants, including duckweed, have just started lately [25]. Crump et al. [26], Xie et al. [27], and Matsuzawa et al. [28] have recently found that aquatic plants, including duckweed, also harbor diverse and specific bacterial communities. Yamaga et al. [29] isolated the first PGPB recognized to promote duckweed growth in a sterile synthetic medium. Another bacterium was recently found to promote the growth of the duckweed Lemna minor in a medium containing chromium [30]. These confirmed that bacteria living with duckweed can exert significant effects on host plant growth, similar to those seen in terrestrial crops. Thus, extended studies on duckweed–microbe interactions, especially those affecting duckweed growth, are needed to realize efficient and sustainable cultivation of duckweed species utilizing beneficial bacteria.
The aim of the current research was to evaluate the effects of diverse environmental bacteria on the growth of the duckweed L. minor. Fifteen native bacterial communities collected from various aquatic environments were investigated for their effects on duckweed growth. Bacterial strains in communities that strongly enhanced or repressed the growth of duckweed were isolated for a more profound understanding of duckweed–microbe interactions.
Results
Effects of bacterial communities on duckweed growth
The growth of duckweed cultivated with fifteen environmental bacterial communities is shown in Fig. 1. Because all of the experiments were performed in the same axenic culture conditions, both promotive and inhibitory effects on duckweed growth should be a function of the bacterial communities. Many bacterial communities were found to have promotive or neutral effects on duckweed growth, with bacterial community H, which showed the greatest growth promotion, increasing the number of fronds by +14% over aseptically cultured L. minor. On the other hand, bacterial communities M and N decreased the number of duckweed fronds by −24 and −14%, respectively, compared to that of the controls. No remarkable difference in frond size, shape, color, or disease symptoms was observed in plants grown with the different bacterial communities in this series of experiments (Fig. 2).
Fig. 1.
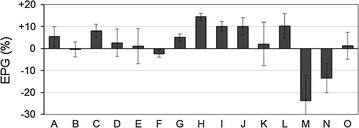
Effects on plant growth (EPGs) of bacterial communities collected from ponds or rivers. A–O indicate bacterial communities recovered from water samples. EPGs were evaluated based on the number of fronds after 7 days of cultivation compared to that of an aseptic control. There were 91.33 (±4.50), 85.67 (±2.05), and 81.33 (±6.60) fronds at the end of control experiments for bacterial communities A–E, F–J, and K–O. Error bars show the standard errors and include errors among treatments performed in triplicate and the control
Fig. 2.

Images of Lemna minor after 7 days of cultivation with bacterial communities H (a) and M (b) and the aseptic control (c)
Isolation of bacterial strains from duckweed
From the plant bodies cultivated with bacterial communities H and M, which conferred the highest and lowest L. minor growth in the previous experiment, 10 and 12 morphologically distinct bacterial strains were isolated, respectively, and used for the further investigations. Table 1 shows the results of a BLAST search for the 16S rDNA sequences of all 22 isolates. All isolates showed at least 97% sequence identity with known strains. All isolates were found to be members of the alpha, beta, and gamma subclasses of Proteobacteria, except for H8, which belonged to the phylum Actinobacteria. In addition, both communities contained members of the order Rhizobiales (H1, H2, H5, M2), Pseudomonadales (H4, H6, M10, M12), Burkholderiales (H7, H9, M7, M8, M9), and Sphingomonadales (H10, M5, M11), which often comprise the large fraction of the rhizobacterial communities of terrestrial plants [31, 32].
Table 1.
Nucleotide BLAST search of bacterial strains isolated from Lemna minor cultivated with communities H and M
| Strain | Most similar species | Accession number of the closest match | Nucleotide similarity (%) |
|---|---|---|---|
| H1 (++) | Starkeya koreensis | NR_113962.1 | 98 |
| H2 (−) | Rhizobium daejeonense | NR_114121.1 | 97 |
| H3 (++) | Aquitalea magnusonii | NR_043475.1 | 99 |
| H4 (+) | Pseudomonas geniculata | NR_024708.1 | 99 |
| H5 (+/−) | Methylobacterium fujisawaense | NR_112232.1 | 99 |
| H6 (−) | Pseudomonas oryzihabitans | NR_114041.1 | 99 |
| H7 (+) | Aquabacterium commune | NR_024875.1 | 98 |
| H8 (++) | Leucobacter alluvii | NR_042426.1 | 99 |
| H9 (+/−) | Acidovorax radicis | NR_117776.1 | 99 |
| H10 (+/−) | Sphingomonas ursincola | NR_119243.1 | 99 |
| M1 (++) | Azospirillum irakense | NR_044949.1 | 99 |
| M2 (++) | Ensifer adhaerens | NR_113893.1 | 98 |
| M3 (− −) | Acinetobacter ursingii | NR_025392.1 | 100 |
| M4 (+/−) | Enterobacter ludwigii | NR_042349.1 | 99 |
| M5 (−−) | Blastomonas natatoria | NR_113794.1 | 99 |
| M6 (−−) | Asticcacaulis excentricus | NR_074137.1 | 99 |
| M7 (+/−) | Ideonella dechloratans | NR_026108.1 | 99 |
| M8 (+) | Aquincola tertiaricarbonis | NR_043913.1 | 98 |
| M9 (+/−) | Rubrivivax gelatinosus | NR_025841.1 | 98 |
| M10 (+/−) | Pseudomonas psychrotolerans | NR_042191.1 | 99 |
| M11 (+/−) | Sphingomonas parapaucimobilis | NR_113729.1 | 99 |
| M12 (++) | Pseudomonas otitidis | NR_043289.1 | 99 |
Symbols in parentheses indicate the effects on plant growth of strains (Fig. 3): ++, greater than 10%; +, between +5 and +10%; +/−, between −5 and +5%; −, between −5 and −10%; −−, less than −10%
Cultivation of L. minor with single bacterial isolates
A total of 22 isolates were evaluated for their effects on duckweed growth by co-culture with sterilized L. minor. In this experiment, frond numbers and dry weights were highly correlated (r = 0.93), so only the EPGs calculated from the dry weight were used in further analyses. As shown in Fig. 3, duckweed growth was affected both positively and negatively by the inoculation of isolates. The EPGs of members of bacterial community H varied from −6.3 to +21%, whereas in community M, they ranged from −14.4 to +17.5%. Bacterial strains were classified into five groups: those showed EPG values greater than 10% (++), between +5 and +10% (+), between −5 and +5% (+/−), between −5 and −10% (−), and less than −10% (−−). According to this classification scheme, there were 3, 2, 3, 2, and 0 isolates from community H, corresponding to (++), (+), (+/−), (−), and (−−), respectively, whereas there were 3, 1, 5, 0, and 3 isolates from community M. Bacterial communities H and M, which were associated the greatest and least growth in the first experiment, contained both promotive bacteria (++ or +) and inhibitory bacteria (− or −−). However, it is worth noting that inhibitory bacteria with EPG values less than −10% (−−) were isolated only from community M.
Fig. 3.
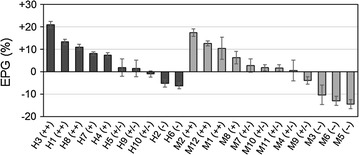
Effects on plant growth (EPGs) of single bacteria isolated from communities H (black bars) and M (gray bars). EPGs were evaluated by the change in dry weight of Lemna minor relative to that of the aseptic control, which had 119.67 (±5.19) fronds at the end. Error bars show the standard errors (n = 3)
Cultivation of L. minor with mixtures of two bacterial isolates
Based on the results of the previous experiment and ease of cultivation, promotive strains H1 (++), H3 (++), and M12 (++) and inhibitory strains H6 (−), M3 (−−), M5 (−−), and M6 (−−) were selected for further experiments, and all binary combinations of these isolates (7C2 = 21 patterns) were examined for their effects on duckweed growth. Simultaneously, single cultures of seven strains were tested again as positive controls. Because the frond number and dry weight showed a strong correlation (r = 0.95), only the dry weights were used to calculate EPGs (%). As shown in Table 4, all positive controls showed similar effects as those seen in the previous experiment, although the effects of H1 and H6 in this experiment were regarded as (+) and (−−), respectively. Generally, binary combinations of promotive bacteria showed promotive or neutral effects on duckweed growth, and combinations of inhibitory bacteria showed inhibitory effects. Combinations of promotive and inhibitory strains, however, resulted in both growth promotion and growth inhibition. Specifically, combinations of strain H3 (++) and an inhibitory bacterium tended to show promotive effects, whereas all combinations of strain H1 (+) and M12 (++) with inhibitory strains resulted in a negative effect.
Table 4.
The effects on plant growth (EPGs, %) based on dry weight of a mixed inoculation of two species of bacteria
| H3 | M12 | H1 | H6 | M6 | M3 | M5 | |
|---|---|---|---|---|---|---|---|
| H3 | +24.8 (±1.7) | ||||||
| M12 | +15.5 (±3.2) | +11.6 (±4.2) | |||||
| H1 | +23.7 (±2.6) | +1.8 (±3.6) | +5.1 (±1.9) | ||||
| H6 | +6.7 (±4.5) | −7.4 (±2.2) | −11.3 (±5.1) | −10.6 (±2.5) | |||
| M6 | +15.7 (±1.6) | −16.7 (±4.0) | −7.0 (±5.3) | −7.7 (±1.9) | −13.2 (±4.1) | ||
| M3 | +3.1 (±2.9) | −15.9 (±3.1) | −3.8 (±5.4) | −20.2 (±3.6) | −15.0 (±6.5) | −14.0 (±3.0) | |
| M5 | +10.9 (±2.7) | −19.5 (±3.5) | −12.0 (±4.7) | −10.5 (±4.4) | −6.8 (±3.9) | −13.9 (±3.4) | −19.2 (±3.1) |
Rows and columns indicate the isolates used. The cells with rows and columns that indicate the same strain show the results of single inoculations as positive controls. There were 110.33 (±4.50) fronds at the end of control experiments. Values in parenthesis represent the mean ± standard error of the mean (n = 3)
Evaluation of bacterial isolates for plant growth-affecting traits
The isolates were examined for traits that affect plant growth in a series of assays. The results for the 22 isolates are shown in Table 2. Of the 22 isolates, 10 could synthesize IAA, 11 could solubilize insoluble phosphate, and 12 could produce siderophores. In addition, 12 isolates were positive for hydrogen cyanide production, which is believed to be involved in plant growth inhibition. All of the isolates excepting for H2 (–) and H5 (+/−) exhibited at least one of these traits. Strains H3 (++), M1 (++), and M12 (++) were positive for all traits. The occurrence of these traits was similar between the isolates from communities H and M, except that the siderophore-producing isolates were more frequently found in community M than H. Table 3 shows the results of the multiple-way ANOVA test to detect the contribution of these traits to the effects on duckweed growth. Of the four traits, only phosphate solubilization was found to correlate (p < 0.05) with duckweed growth promotion, whereas the other traits correlated with neither growth promotion nor growth inhibition.
Table 2.
Indole acetic acid (IAA) production, phosphate (P) P solubilization, siderophore production, and hydrogen cyanide (HCN) production by bacterial strains
| Strain | IAA production | P solubilization | Siderophore production | HCN production |
|---|---|---|---|---|
| H1 (++) | − | + | − | − |
| H2 (−) | − | − | − | − |
| H3 (++) | + | + | + | + |
| H4 (+) | − | − | + | + |
| H5 (+/−) | − | − | − | − |
| H6 (−) | − | + | + | + |
| H7 (+) | + | + | − | − |
| H8 (++) | − | − | − | + |
| H9 (+/−) | + | − | − | + |
| H10 (+/−) | − | − | − | + |
| M1 (++) | + | + | + | + |
| M2 (++) | − | + | + | − |
| M3 (−−) | − | + | + | − |
| M4 (+/−) | − | + | + | + |
| M5 (−−) | − | − | − | − |
| M6 (−−) | + | − | + | + |
| M7 (+/−) | + | − | − | − |
| M8 (+) | + | − | − | − |
| M9 (+/−) | + | − | − | − |
| M10 (+/−) | − | + | + | + |
| M11 (+/−) | − | + | + | + |
| M12 (++) | + | + | + | + |
Symbols in parentheses indicate the effects on plant growth of strains (Fig. 3): ++, greater than 10%; +, between +5 and +10%; +/−, between −5 and +5%; −, between −5 and −10%; −−, less than −10%
Table 3.
The result of multiple-way analysis of variance (ANOVA)
| Factor | Mean square | F value | Coefficients | Significance |
|---|---|---|---|---|
| IAA | 141.79 | 1.986 | 4.88 | 0.177 |
| P solub. | 354.82 | 4.969 | 11.34 | 0.040 |
| Sidero. | 118.20 | 1.665 | −7.41 | 0.216 |
| HCN | 39.29 | 0.550 | 3.12 | 0.468 |
IAA indole acetic acid production, P solub. phosphate solubilization, sidero. siderophore production, HCN hydrogen cyanide production were analyzed as factors related to the effects on plant growth (%) of 22 isolates
Discussion
This study revealed that bacterial communities in freshwater environments can both enhance and repress the growth of duckweed L. minor (Fig. 1). The effects of bacterial communities on the 7-day growth of L. minor ranged from −24 to +14%, which indicates that a 1.5-fold difference in duckweed yield can be controlled by selecting a bacterial community. Approximately, the same change in relative growth rate of L. minor was observed with light intensities of 400 and 110 µmol/m2/s [20], and ammonium (as a sole nitrogen source) concentrations of 28 and 2 mg/L [33]. Considering these facts, environmental bacterial communities were shown to be a critical factor that affects duckweed growth, with effects that are comparable with other important environmental factors such as light and nutrients. Enhancement of crop yields by optimizing co-existing bacteria has long been a goal for sustainable agriculture. Here, our results show its feasibility, even for the hydroculture of duckweed. This strategy should be fascinating choice if attained, since co-existing bacteria can potentially be modulated with lower energy than light, nutrient, and temperature.
Although culture-dependent methods have clear limitations for analyzing bacterial communities, we believe that it is useful to isolate bacterial strains in order to characterize their functions, since it is expected that readily culturable bacteria comprise larger fraction in duckweed rhizoplane than that in other natural environments according to Matsuzawa et al. [28]. Moreover, to rationally design co-existent bacteria for enhanced duckweed biomass production, understanding which bacterial strains promote or inhibit duckweed growth is indispensable. In this study, we isolated and characterized representative bacterial strains from both promotive and inhibitory bacterial communities H and M, respectively (Fig. 3). Taxonomically, large part of the bacterial isolates belonged to taxa that are known inhabitants of the terrestrial plant rhizosphere. It might suggest that duckweed rhizobacteria share the same characteristics with those of terrestrial plants to a certain extent. Interestingly, both promotive and inhibitory communities contained bacterial strains that expressed promotive, inhibitory, and neutral effects on duckweed growth, and their isolation frequencies were not significantly different between the two communities. The only notable difference was that the activities of inhibitory bacterial strains isolated from the inhibitory community were stronger than those of the strains isolated from the promotive community. We conclude that promotive, inhibitory, and neutral bacteria are ubiquitous in duckweed-associated bacterial communities, and that the activities of these bacteria likely determine, synergistically, the net effect of a bacterial community on duckweed growth.
As far as we know, Acinetobacter calcoaceticus P23 [29] and Exiguobacterium sp. MH3 [30] are the only PGPB that have been reported for duckweed species. In this study, we discovered six new bacterial strains that promoted the growth of duckweed by more than 10% with 7 days of cultivation. Sequencing of 16S rRNA genes revealed that these strains belong to diverse genera that were different from previously isolated PGPB, suggesting that PGPB for duckweed are distributed across a wider range of taxa. Interestingly, strains M1 (++) and M12 (++) were identified as Azospirillum and fluorescent Pseudomonas, respectively, both of which are common PGPB for terrestrial plants, except for Pseudomonas syringae, which is a plant pathogen. On the other hand, the most efficient PGPB strain H3 (++) was identified as belonging to the genus Aquitalea, which has been discovered only in freshwater environments. Quisehuatl-Tepexicuapan et al. [34] isolated one strain of Aquitalea from the rhizoplane of duckweed L. gibba. Therefore, strain H3 may be a PGPB specific for aquatic plants, including duckweed, which has evolved in freshwater environments.
Bacterial strains that suppress the plant growth without any apparent pathogenic symptoms are known as plant growth-inhibiting bacteria (PGIB) or deleterious rhizobacteria (DRB) in the field of agriculture. Although these bacteria are difficult to detect, a number of studies indicate that PGIB and DRB can be regulated to improve crop production [35, 36] and to control weeds [37]. We isolated these bacteria for the first time from duckweed or aquatic plants in this study. Because these bacteria can significantly lower the efficiency of duckweed production, attention should be paid to PGIB, as well as PGPB. Interestingly, we found PGIB in the genus Acinetobacter, which is the same taxonomic group as that of the first PGPB identified in duckweed [29]. Therefore, culture-independent metagenomic analysis of the 16S rRNA gene is not sufficient to detect and distinguish between duckweed PGPB and PGIB in bacterial communities, and further isolation-based research, such as this study, will contribute to not only a deeper understanding of duckweed–microbe interactions but also the construction of a relevant bacterial database.
Many studies have been dedicated to elucidating the mechanisms by which PGPB affect plant growth [22]. In this study, we examined the correlation between EPGs assessed by co-cultivation and the presence of four physiological traits that are known to be associated with plant growth promotion or inhibition. Although many bacteria were found to have more than one of these traits (Table 2), no clear-cut correlation was found between the possession of these traits and duckweed growth promotion/inhibition effects of the bacterial strains in a multiple ANOVA analysis (Table 3). Therefore, multiple mechanisms, probably including unknown ones, are related to bacterial promotion/inhibition of duckweed growth. Among the four tested traits, only the ability to solubilize phosphate was shown to be slightly correlated with duckweed growth promotion. Although bacterial phosphate solubilization is widely recognized to contribute to phosphorus availability in soil environments [38], this result was unexpected, because all of the phosphorus was added in soluble form at the start of an experiment. However, it is possible that phosphate supply via degradation of dead bacterial cells, plant exudates, and phosphate salts formed in the medium was influenced by phosphate-solubilizing activity of duckweed-associated bacteria. Since aquatic environments also contain a variety of unavailable phosphorus [39], the effects of bacterial phosphate supply to plants should be evaluated in real hydroculture.
In contrast to our relatively substantial knowledge on the mechanisms of PGPB, reports on the mechanisms by which PGIB inhibit the growth of plants are quite limited. Cyanide production is virtually the only proposed mechanism with enough supporting data [40, 41], whereas other studies have suggested the benefit of hydrogen cyanide based on antifungal activity [42]. The current study did not show a significant correlation between cyanide production and plant growth. Further studies are required to understand duckweed growth inhibition associated with bacteria or bacterial community.
To better understand the complex effects of bacterial communities, effects of binary combinations of selected isolates on duckweed growth were tested as simple artificial bacterial community models. In contrast with results of a previous study conducted for terrestrial plant [43], synergistic effects were generally not observed with promotive–promotive or inhibitory–inhibitory bacterial combinations. Interestingly, the results of promotive–inhibitory bacterial combinations showed that promotive strains H1 (+) and M12 (++) were not effective in the presence of any of the inhibitory strains (Table 4). This suggests that not all PGPB are able to function in their native environments, and that inhibitory bacteria may have a stronger influence on the effects of the bacterial community as a whole. This observation shows the difficulty of using PGPB in non-sterilized conditions as reported in Liu et al. [44]. It also indicates that regulation of PGIB may be effective for maximizing PGPB activity in a bacterial community. In contrast to strains H1 (+) and M12 (++), promotive strain H3 (++) was less susceptible to the deleterious effects of inhibitory strains, and was found to exert a promotive effect or at least negate the inhibitory effects of other bacteria. From this point of view, strain Aquitalea magnusonii H3 can be regarded as a PGPB for potential use in open environments.
There are many possible explanations for what determines the result of the conflicting effects of duckweed PGPB and PGIB described above. For example, competition between bacteria on root exudates and spaces, inactivation of promotive or inhibitory mechanisms, and masking effects are likely. Elucidating such bacterial interactions is an important next step for optimizing duckweed hydroculture systems via the design of beneficial bacterial communities. For this reason, bacterial strains obtained in this study may be useful as model PGPB and PGIB for duckweed.
Conclusion
This study reported that (1) bacterial community strongly influences the production speed of duckweed biomass; (2) duckweed harbors bacteria which have promotive, neutral, or inhibitory effects for their growth; (3) promotive effects of PGPB strains can sustain or cannot sustain in the presence of other bacteria, depending on the kind of PGPB strain and some unknown mechanisms; and (4) many of isolates from duckweed-associated bacterial communities have some common characteristics in their taxa and ability to influence plant growth with terrestrial rhizobacteria. From these, it can be concluded that modulating bacterial community is the possible choice for improving biomass production from duckweed hydroculture. Further, it may be applicable to the other aquatic feedstocks such as water lettus, water hyacinth, and Azolla plants which have similar morphology to duckweed.
Methods
Plant material
Common duckweed (Lemna minor, RDSC #5512), obtained from a small pond in a botanical garden of Hokkaido University (Sapporo, Japan), was used in the experiments. The plants were sterilized by washing with 0.5% sodium hypochlorite for 7 min, followed by washing with sterilized water twice. The sterilized plants were successively cultured in flasks containing Hoagland medium (36.1 mg/L KNO3, 293 mg/L K2SO4, 3.87 mg/L NaH2PO4, 103 mg/L MgSO4·7H2O, 147 mg/L CaCl2·H2O, 3.33 mg/L FeSO4·7H2O, 0.95 mg/L H3BO3, 0.39 mg/L MnCl2·4H2O, 0.03 mg/L CuSO4·5H2O, 0.08 mg/L ZnSO4·7H2O, and 0.254 mg/L H2MoO4·4H2O; pH 7.0) in an incubation chamber at 28 °C, an irradiance of 80 µmol/m2/s, and a photoperiod of 16 h/8 h day/night.
Cultivation of duckweed with environmental bacterial communities
Water samples were taken from the surfaces of 15 freshwater ponds and rivers located in the northern part of Osaka, Japan in August 24, 27, and 30 of 2015. Descriptions of sampled sites are shown in Additional file 1: Figure S1. The native bacterial communities in the samples were recovered and used for duckweed cultivation experiments as follows. First, coarse particles, including fungi and microalgae, were removed from the water samples using filters with a pore size of 3.0 µm (SSWP, MF-Millipore, Merck Millipore, Darmstadt, Germany), followed by centrifugation (10,000×g, 4 °C, 10 min) to collect bacterial cells from the native bacterial communities. The collected bacterial cells were washed twice with sterilized Hoagland medium and re-suspended in the original volume of Hoagland medium. Ten fronds of L. minor were transferred to flasks filled with 60 mL of the medium containing the bacteria and cultivated for 7 days in the above-mentioned conditions. During cultivation, duckweed growth was monitored by counting the frond number. The effects of the bacterial communities on duckweed growth were evaluated in comparison with growth of a control without introduced bacteria (sterile Hoagland medium).
Isolation of bacterial strains attached to duckweed
At the end of 7 days of cultivation of duckweed with bacterial communities, whole plant bodies in each flask were collected and washed with 20 mL of sterilized 5 mg/L sodium tripolyphosphate (TPP). Then, the duckweed samples were homogenized in TPP using a BioMasher II (Nippi, Tokyo, Japan). The homogenates were spread onto solid 1:10 LB medium in TPP containing 1.5% agar and incubated at 28 °C for 7 days. All morphologically distinct colonies were picked and purified using the same medium.
Identification of bacterial strains
Isolated bacterial strains were identified based on their 16S rRNA gene sequences. A single colony of each bacterial strain was picked and added to PCR reagent containing primers 27F (5′-AGAGTTTGATCTGGCTCAG-3′) [45] and 1392R (5′-ACGGGCGGTGTGTACA-3′) [46] and Ex Taq DNA polymerase (TaKaRa Bio Inc., Shiga, Japan). PCR amplification of the 16S rRNA gene fragments was performed as described previously [47] using a T100 Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA). The amplicons were sequenced by Hokkaido System Science Co., Ltd (Hokkaido, Japan). The NCBI Nucleotide BLAST tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was employed for taxonomic identification of strains H1–H10 and M1–12 using the obtained sequences as queries. The nucleotide sequences of the partial 16S rRNA gene from strains H1–H10 and M1–12 were submitted to the DNA Data Bank of Japan (DDBJ) under accession number LC191965–LC191986.
Cultivation of duckweed with isolated bacterial strains
To cultivate bacterial isolates used in the experiments, a loop of bacterial colony was inoculated into 20 or 100 mL of liquid LB medium in a vial or flask that was held overnight at 28 °C with shaking at 120 rpm. Cells were harvested by centrifugation (10,000×g, 4 °C, 10 min), washed twice with sterilized Hoagland medium, and then re-suspended in the same medium with cells at an optical density at 600 nm (OD600) = 0.1. To allow bacterial strains to attach to the plants, aseptic L. minor were placed on each bacterial suspension for 24 h. Then, 10 duckweed fronds were transferred to a flask filled with fresh bacteria-free medium at the start of cultivation. This method minimized the effect of nutrient leakage from dead bacterial cells and enabled an evaluation of the direct physiological effects of bacteria on the duckweed [48]. After 7 days of cultivation, the number of duckweed fronds and dry weight (12 h drying at 80 °C) were measured. Cultivation experiments using a combination of two bacterial strains were performed using the same procedure, except that equal amounts (30 mL each) of two separately prepared bacterial suspensions (OD600 = 0.1) were mixed and allowed to attach to plants. Control experiments were performed using sterile Hoagland medium without the introduction of bacterial strains.
Evaluation of bacterial isolates for traits that affect plant growth
Indole acetic acid (IAA) production
Indole acetic acid production in the presence of L-Trp was tested according to a method described by Orlando [49], with some modifications. First, a single bacterial colony was inoculated into a vial containing 20 mL of LB medium with 0.05% (w/v) of L-Trp. After 5 days of incubation with shaking (28 °C, 120 rpm), the culture was centrifuged (2000×g, 30 min, 24 °C), and 2 mL of the supernatant was added to 2 mL of Salkowski reagent (98 mL of 35% HClO4, 2 mL of 0.5 M FeCl3). Then, the mixture was placed at room temperature for 30 min for observation. Development of a pink color indicated the production of IAA.
Phosphate-solubilizing ability
The ability to solubilize insoluble phosphate was evaluated using Pikovskaya’s agar, which contains calcium phosphate as an insoluble phosphate [50]. Then, each bacterial colony was streaked onto an agar plate and incubated at 28 °C for 7 days. Results were considered positive when clear zones developed around a colony.
Siderophore production
Bacterial siderophore production was detected using the method of Schwyn and Neilands [51]. In this assay, each bacterial colony was streaked on a chrome azurol S (CAS) agar plate containing blue dye. Plates were incubated at 28 °C for 7 days and then examined for a yellow or orange halo around the colonies, which would indicate the production of a siderophore.
Hydrogen cyanide (HCN) production
The assay of bacterial cyanide production was performed according to a method described by Saber et al. [52]. In short, the bacterial strains were grown in 5 mL of LB medium in a test tube with Whatman No. 1 filter paper (GE Healthcare Life Science, Buckinghamshire, UK) soaked in cyanide reagent. Cyanide-producing bacteria were detected when the Whatman paper changed color from yellow to orange or red.
Statistical analyses
Duckweed cultivation experiments were performed in triplicate for all treatments and controls. In all duckweed cultures, the effects on plant growth (EPG) of each bacterial community or each bacterial strain were calculated as follows:
where G(T) is the mean growth of duckweeds in the presence of microbes, which was evaluated by the frond number or dry weight of the duckweed after 7 days of cultivation, and G(C) is that in the aseptic controls. Here, the standard errors (SE) for EPG were calculated using the following formula:
Multiple-way analysis of variance (ANOVA) was performed to test whether bacterial IAA production, phosphate-solubilizing ability, siderophore production, or hydrogen cyanide production correlated with growth promotion or inhibition of L. minor. In this analysis, the results of four assays were treated as qualitative factors, and the EPG (%) based on dry weights in the duckweed cultivation experiments with single bacterial strains were used as the response variable. All statistical analyses were performed in R v3.2.3. (http://www.r-project.org).
Authors’ contributions
HI designed and performed the experiments, interpreted the results, and drafted the manuscript. MK designed the experiments, interpreted the results, drafted and revised the manuscript. MM interpreted the results, critically revised the manuscript, and supervised the study. MI designed the experiments, interpreted the results, drafted and revised the manuscript, and supervised the study. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Tokitaka Oyama and Dr. Shogo Ito for giving valuable information about the plant material.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Funding
This study was supported by the Advanced Low Carbon Technology Research and Development Program (ALCA) of the Japan Science and Technology Agency (JST), and partially supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number 26281041.
Abbreviations
- PGPB
plant growth-promoting bacteria
- BLAST
The Basic Local Alignment Search Tool
- DNA
deoxyribonucleic acid
- EPG
effects on plant growth
- IAA
indole acetic acid
- RNA
ribonucleic acid
- PGIB
plant growth-inhibiting bacteria
- DRB
deleterious rhizobacteria
- NCBI
The National Center for Biotechnology Information
- PCR
polymerase chain reaction
- CAS
chrome azurol S
- SE
standard error
Additional file
Additional file 1: Figure S1. Locations and descriptions of water sampled sites.
Contributor Information
Hidehiro Ishizawa, Email: ishizawa@wb.see.eng.osaka-u.ac.jp.
Masashi Kuroda, Email: kuroda@see.eng.osaka-u.ac.jp.
Masaaki Morikawa, Email: morikawa@ees.hokudai.ac.jp.
Michihiko Ike, Phone: +81-(0)6-6879-7672, Email: ike@see.eng.osaka-u.ac.jp.
References
- 1.Landolt E. Biosystematic investigations in the family of duckweed (Lemnaceae) Stiftung Rubel, Zúrich: Geobotanischen Inst ETH; 1986. [Google Scholar]
- 2.Islam MS, Kabir MS, Khan SI, Ekramullah M, Nair GB, Sack RB, Sack DA. Wastewater-grown duckweed may be safely used as fish feed. Can J Microbiol. 2004;50:51–56. doi: 10.1139/w03-102. [DOI] [PubMed] [Google Scholar]
- 3.Cheng JJ, Stomp AM. Growing duckweed to recover nutrients from wastewaters and for production of fuel ethanol and animal feed. Clean. 2009;37:17–26. [Google Scholar]
- 4.Ahmad Z, Hossain NS, Hussain SG, Khan AH. Effect of duckweed (Lemna minor) as complement to fertilizer nitrogen on the growth and yield of rice. Int J Trop Agric. 1990;8:72–79. [Google Scholar]
- 5.Lahive E, O’Halloran J, Jansen MAK. Differential sensitivity of four Lemnaceae species to zinc sulphate. Environ Exp Bot. 2011;71:25–33. doi: 10.1016/j.envexpbot.2010.10.014. [DOI] [Google Scholar]
- 6.Ziegler P, Sree KS, Appenroth KJ. Duckweeds for water remediation and toxicity testing. Toxicol Environ Chem. 2016;98:1127–1154. doi: 10.1080/02772248.2015.1094701. [DOI] [Google Scholar]
- 7.Cui W, Cheng JJ. Growing duckweed for biofuel production: a review. Plant Biol. 2015;17:16–23. doi: 10.1111/plb.12216. [DOI] [PubMed] [Google Scholar]
- 8.Yu C, Sun C, Yu L, Zhu M, Xu H, Zhao J, Ma Y, Zhou G. Comparative analysis of duckweed cultivation with sewage water and SH media for production of fuel ethanol. PLoS ONE. 2014 doi: 10.1371/journal.pone.0115023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Cui W, Cheng JJ, Stomp AM. Production of high-starch duckweed and its conversion to bioethanol. Biosyst Eng. 2011;110:67–72. doi: 10.1016/j.biosystemseng.2011.06.007. [DOI] [Google Scholar]
- 10.El-Shafai SA, El-Gohary FA, Nasr FA, van der Steen NP, Gijzen HJ. Nutrient recovery from domestic wastewater using a UASB-duckweed ponds system. Bioresour Technol. 2007;98:798–807. doi: 10.1016/j.biortech.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Stout L, Nüsslein K. Biotechnological potential of aquatic plant–microbe interactions. Curr Opin Biotechnol. 2010;21:339–345. doi: 10.1016/j.copbio.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Alaerts GJ, Rahman Mahbubar MD, Kelderman P. Performance analysis of a full-scale duckweed-covered sewage lagoon. Water Res. 1996;30:843–852. doi: 10.1016/0043-1354(95)00234-0. [DOI] [Google Scholar]
- 13.Fujita M, Mori K, Kodera T. Nutrient removal and starch production through cultivation of Wolffia arrhiza. J Biosci Bioeng. 1999;87:194–198. doi: 10.1016/S1389-1723(99)89012-4. [DOI] [PubMed] [Google Scholar]
- 14.Bergmann BA, Cheng J, Classen J, Stomp AM. In vitro selection of duckweed geographical isolates for potential use in swine lagoon effluent renovation. Bioresour Technol. 2000;73:13–20. doi: 10.1016/S0960-8524(99)00137-6. [DOI] [Google Scholar]
- 15.Zhao Y, Fang Y, Jin Y, Huang J, Bao S, He Z, Wang F, Zhao H. Effects of operation parameters on nutrient removal from wastewater and high-protein biomass production in a duckweed-based (Lemma aequinoctialis) pilot-scale system. Water Sci Technol. 2014;70:1195–1204. doi: 10.2166/wst.2014.334. [DOI] [PubMed] [Google Scholar]
- 16.Verma R, Suthar S. Impact of density loads on performance of duckweed bioreactor: a potential system for synchronized wastewater treatment and energy biomass production. Environ Prog Sustain Energy. 2015;34:1596–1604. doi: 10.1002/ep.12157. [DOI] [Google Scholar]
- 17.Oron G, de-Vegt A, Porath D. Nitrogen removal and conversion by duckweed grown on waste-water. Water Res. 1988;22:179–184. doi: 10.1016/0043-1354(88)90076-0. [DOI] [Google Scholar]
- 18.Caicedo JR, Steen NP, Arce O, Gijzen HJ. Effect of total ammonia nitrogen concentration and pH on growth rates of duckweed (Spirodela polyrrhiza) Water Res. 2000;34:3829–3835. doi: 10.1016/S0043-1354(00)00128-7. [DOI] [Google Scholar]
- 19.Lasfar S, Monette F, Millette L, Azzouz A. Intrinsic growth rate: a new approach to evaluate the effects of temperature, photoperiod and phosphorus-nitrogen concentrations on duckweed growth under controlled eutrophication. Water Res. 2007;41:2333–2340. doi: 10.1016/j.watres.2007.01.059. [DOI] [PubMed] [Google Scholar]
- 20.Yin Y, Yu C, Yu L, Zhao J, Sun C, Ma Y, Zhou G. The influence of light intensity and photoperiod on duckweed biomass and starch accumulation for bioethanol production. Bioresour Technol. 2015;187:84–90. doi: 10.1016/j.biortech.2015.03.097. [DOI] [PubMed] [Google Scholar]
- 21.Berg G, Grube M, Schloter M, Smalla K. Unraveling the plant microbiome: looking back and future perspectives. Front Microbiol. 2014;5:1–7. doi: 10.3389/fmicb.2014.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glick BR. Plant growth-promoting bacteria: mechanisms and applications. Scientifica. 2012 doi: 10.6064/2012/963401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson M, Habiger J. Characterization and identification of productivity-associated rhizobacteria in wheat. Appl Environ Microbiol. 2012;78:4434–4446. doi: 10.1128/AEM.07466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendes R, Kruijt M, de Bruijn I, Dekker E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Anderson GL, Bakker PAHM, Raaijmakers JM. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 25.Appenroth KJ, Ziegler P, Sree KS. Duckweed as a model organism for investigating plant–microbe interactions in an aquatic environment and its applications. Endocytobiosis Cell Res. 2016;27:94–106. [Google Scholar]
- 26.Crump BC, Koch EW. Attached bacterial populations shared by four species of aquatic angiosperms. Appl Environ Microbiol. 2008;74:5948–5957. doi: 10.1128/AEM.00952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie WY, Su JQ, Zhu YG. Phyllosphere bacterial community of floating macrophytes in paddy soil environments as revealed by Illumina high-throughput sequencing. Appl Environ Microbiol. 2015;81:522–532. doi: 10.1128/AEM.03191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuzawa H, Tanaka Y, Tamaki H, Kamagata Y, Mori K. Culture-dependent and independent analyses of the microbial communities inhabiting the giant duckweed (Spirodela polyrrhiza) rhizoplane and isolation of a variety of rarely cultivated organisms within the phylum Verrucomicrobia. Microbes Environ. 2010;25:302–308. doi: 10.1264/jsme2.ME10144. [DOI] [PubMed] [Google Scholar]
- 29.Yamaga F, Washio K, Morikawa M. Sustainable biodegradation of phenol by Acinetobacter calcoaceticus P23 isolated from the rhizosphere of duckweed Lemna aoukikusa. Environ Sci Technol. 2010;44:6470–6474. doi: 10.1021/es1007017. [DOI] [PubMed] [Google Scholar]
- 30.Tang J, Zhang Y, Cui Y, Ma J. Effects of a rhizobacterium on the growth of and chromium remediation by Lemna minor. Environ Sci Pollut Res. 2015;22:9686–9693. doi: 10.1007/s11356-015-4138-y. [DOI] [PubMed] [Google Scholar]
- 31.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Glavina del Rio T, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Green Tringe S, Dangl JL. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buée M, de Boer W, Martin F, van Overbeek L, Jurkevitch E. The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil. 2009;321:189–212. doi: 10.1007/s11104-009-9991-3. [DOI] [Google Scholar]
- 33.Wang W, Yang C, Tang X, Gu X, Zhu Q, Pan K, Hu Q, Ma D. Effects of high ammonium level on biomass accumulation of common duckweed Lemna minor L. Environ Sci Pollut Res. 2014;21:14202–14210. doi: 10.1007/s11356-014-3353-2. [DOI] [PubMed] [Google Scholar]
- 34.Quisehuatl-Tepexicuapan E, Ferrera-Cerrato R, Silva-Rojas HV, Rodriguez-Zaragoza S, Alarcón A, Almaraz-Suárez JJ. Free-living culturable bacteria and protozoa from the rhizoplanes of three floating aquatic plant species. Plant Biosyst. 2014;2014:1–11. [Google Scholar]
- 35.Suslow TV, Schroth MN. Role of deleterious rhizobacteria as minor pathogens in reducing crop growth. Dis Control Pest Manag. 1981;72:111–115. [Google Scholar]
- 36.Jagadeesh KS, Krishnaraj PU, Kulkarni JH. Suppression of deleterious bacteria by rhizobacteria and subsequent improvement of germination and growth of tomato seedlings. Curr Sci. 2006;91:1458–1459. [Google Scholar]
- 37.Li J, Kremer RJ. Growth response of weed and crop seedlings to deleterious rhizobacteria. Biol Cont. 2006;39:58–65. doi: 10.1016/j.biocontrol.2006.04.016. [DOI] [Google Scholar]
- 38.Gyaneshwar P, Naresh Kumar G, Parekh LJ, Poole PS. Role of soil microorganisms in improving P nutrition of plants. Plant Soil. 2002;245:83–93. doi: 10.1023/A:1020663916259. [DOI] [Google Scholar]
- 39.Baldwib DS. Organic phosphorus in the aquatic environment. Environ Chem. 2013;10:439–454. doi: 10.1071/EN13151. [DOI] [Google Scholar]
- 40.Blom D, Fabbri C, Eberl L, Weisskopf L. Volatile-mediated killing of Arabidopsis thaliana by bacteria is mainly due to hydrogen cyanide. Appl Environ Microbiol. 2011;77:1000–1008. doi: 10.1128/AEM.01968-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Astrom B. Role of bacterial cyanide production in differential reaction of plant cultivars to deleterious rhizosphere pseudomonads. Plant Soil. 1991;133:93–100. doi: 10.1007/BF00011903. [DOI] [Google Scholar]
- 42.Voisard C, Keel C, Haas D, Défago G. Cyanide production by Pseurdomonas fluorescence helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 1989;8:351–358. doi: 10.1002/j.1460-2075.1989.tb03384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh M, Awasthi A, Soni SK, Singh R, Verma RK, Kalra A. Complementarity among plant growth promoting traits in rhizospheric bacterial communities promotes plant growth. Sci Rep. 2015 doi: 10.1038/srep15500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W, Yang C, Shu W. Effects of plant growth-promoting bacteria isolated from copper tailings on plants in sterilized and non-sterilized tailings. Chemosphere. 2014;97:47–53. doi: 10.1016/j.chemosphere.2013.10.089. [DOI] [PubMed] [Google Scholar]
- 45.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lane DJ, Pace B, Olsen G, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 48.Suzuki W, Sugawara M, Miwa K, Morikawa M. Plant growth-promoting bacterium Acinetobacter calcoaceticus P23 increases the chlorophyll content of the monocot Lemna minor (duckweed) and the dicot Lactuca sativa (lettuce) J Biosci Bioeng. 2014;118:41–44. doi: 10.1016/j.jbiosc.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Orland OA, Aida VRT, Eugenia LL, Angelica RD. Characterization of indole acetic acid endophyte producers in autochthonous Lemna gibba plants from Xochimilco Lake. Afr J Biotechnol. 2015;14:604–611. doi: 10.5897/AJB2014.14309. [DOI] [Google Scholar]
- 50.Goteti PK, Desai S, Emmanuel LDA, Taduri M, Sultana U. Phosphate solubilization potential of fluorescent Pseudomonas spp. isolated from diverse agro-ecosystems of India. Int J Soil Sci. 2014;9:101–110. doi: 10.3923/ijss.2014.101.110. [DOI] [Google Scholar]
- 51.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 52.Saber FMA, Abdelhafez AA, Hassan EA, Ramadan EM. Characterization of fluorescent pseudomonads isolates and their efficiency on the growth promotion of tomato plant. Ann Agric Sci. 2015;60:131–140. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.


