Abstract
The formation of carbon–carbon bonds via an acyl-enzyme intermediate plays a central role in fatty acid, polyketide, and isoprenoid biosynthesis. Uniquely among condensing enzymes, 3-hydroxy-3-methylglutaryl (HMG)–CoA synthase (HMGS) catalyzes the formation of a carbon–carbon bond by activating the methyl group of an acetylated cysteine. This reaction is essential in Gram-positive bacteria, and represents the first committed step in human cholesterol biosynthesis. Reaction kinetics, isotope exchange, and mass spectroscopy suggest surprisingly that HMGS is able to catalyze the “backwards” reaction in solution, where HMG–CoA is cleaved to form acetoacetyl-CoA (AcAc–CoA) and acetate. Here, we trap a complex of acetylated HMGS from Staphylococcus aureus and bound acetoacetyl-CoA by cryo-cooling enzyme crystals at three different times during the course of its back-reaction with its physiological product (HMG–CoA). This nonphysiological “backwards” reaction is used to understand the details of the physiological reaction with regards to individual residues involved in catalysis and substrate/product binding. The structures suggest that an active-site glutamic acid (Glu-79) acts as a general base both in the condensation between acetoacetyl-CoA and the acetylated enzyme, and the hydrolytic release of HMG–CoA from the enzyme. The ability to trap this enzyme–intermediate complex may suggest a role for protein dynamics and the interplay between protomers during the normal course of catalysis.
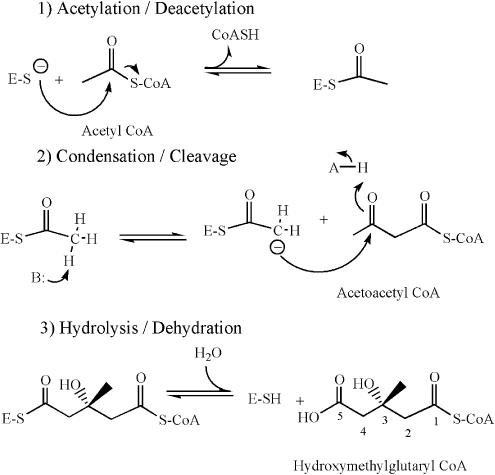
HMGS (3-hydroxy-3-methylglutaryl–CoA synthase) catalyzes the condensation of acetoacetyl–CoA (AcAc–CoA) and acetyl–CoA (Ac-CoA) to form 3-hydroxy-3-methylglutaryl (HMG)–CoA in the first committed, transcriptionally regulated step in cholesterol and isoprenoid biosynthesis. The cholesterol biosynthetic pathway is a proven target for the regulation of serum cholesterol (1, 2), and the mevalonate pathway is essential for many Gram-positive bacteria (3). As described in detail elsewhere (4), the overall three-dimensional structure of HMGS provides structural evidence that the enzyme is a member of the thiolase-like fold (condensing enzyme) superfamily. Mechanistically, the condensation reaction catalyzed by HMGS is reminiscent of the condensation reaction catalyzed by the fatty acid and polyketide condensing enzymes thiolase, β-keto-acyl carrier protein synthases, and chalcone synthase. However, in the second (condensation) step of the reaction, HMGS and the HMGS-like polyketide synthesizing enzymes (e.g., JamH; ref. 5) are distinct in that the methyl group of the acetylated enzyme is activated and attacks the incoming β-keto thioester, whereas the remaining members of the family activate a carbon on the second substrate to attack the enzyme-bound thioester. Overall, the reaction proceeds by means of a ping–pong mechanism as delineated in Scheme 1 (6). It has been thought that the reaction is irreversible because equilibrium greatly favors the products. On the enzyme, the first step in the reverse reaction is that the carboxylic acid of HMG–CoA must be attacked by a cysteine to form a thioester and a water molecule. In the second step of the reverse reaction, the hydroxyl group of HMG–CoA is deprotonated and collapses to form a carbonyl group, and cleaves the carbon–carbon bond. In solution, each of these reactions would be energetically unfavorable. The third step in the reverse reaction, a transthiolation reaction with CoASH to form Ac–CoA and liberate the cysteine thiol, is energetically reasonable and has been observed (7). Mutagenesis and protein chemistry studies have shown that Cys-129 is the active site cysteine and Glu-95 is likely the catalytic base in the avian enzyme (corresponding to Cys-111 and Glu-79 in the Staphylococcus aureus enzyme, respectively; refs. 7–10).
Scheme 1.
Reactions catalyzed by HMGS.
Mapping the trajectories of substrate atoms as they move along a reaction coordinate to elucidate the precise source of an enzyme's catalytic power remains a primary goal of structural enzymology. To that end, crystallographic approaches including cryogenic flow cells, caged substrates, photolysis, and Laue diffraction in various combinations have been used to add to our understanding of how enzymes work (11–13). Here, we take advantage of a very slow catalytic rate to describe, at high resolution, the structure of the S. aureus HMG–CoA synthase cryogenically trapped at three different times as it proceeds with its physiological product “backwards” along its reaction coordinate. Additionally, we demonstrate that this nonphysiological reaction occurs in solution. Together, these data provide insights into the details of HMGS catalysis.
Materials and Methods
Protein Purification. Non-His-tagged protein was expressed and purified as reported (4). Thirty cycles of PCR were performed by using an upstream primer (5′-TAAAACCCCATGGCAATAGGTATCGACAAAATAAACTTTTAC-3′), a downstream primer (5′-AATACACCTCGAGCTCTGGTCTGTGATATTCGCGAACG-3′), and genomic S. aureus DNA to amplify the gene. The DNA, cut with NcoI and XhoI and cloned into pET23d vector, was used to transform BL21(DE3) Escherichia coli (Novagen). The protein containing a C-terminal 6-His-tag was expressed in cells grown at a temperature of 30°C to an optical density of 2.0 at 600 nm and then induced for 4 h with 1 mM isopropyl β-d-thiogalactoside. The protein was purified after lysing cells suspended in 50 mM NaH2PO4 (pH 6.8) and 0.1 mM PMSF by using a French Pressure Cell. After centrifugation for 1 h at 39,000 × g, the supernatant was dialyzed against a buffer containing 50 mM NaH2PO4 (pH 7.0), 300 mM sodium chloride, 5 mM 2-mercaptoethanol, and 20 mM imidazole and loaded onto a Ni-NTA column, where it was eluted by using a 20–300 mM imidazole (pH 7.5) gradient. The protein was dialyzed into storage buffer containing 20 mM NaH2PO4 (pH 6.8), 0.5 mM EDTA, and 1 mM DTT mixed with 25% glycerol and frozen at –80°C before further study. Glycerol was removed by dialysis against either reaction or crystallization buffer.
Reaction Conditions. The HMG–CoA cleavage reaction was performed at 22°C in 100 mM NaH2PO4 (pH 6.5) containing 1 mM DTT and 7.5% glycerol with an initial HMG–CoA concentration of 520 μM (≈20-fold greater than Kd) over the course of 6 days. All proteins (1×) were used at 6 mg/ml (≈100 μM), except when 2× concentration was used. These conditions were intended to mimic crystallization conditions, where the enzyme is saturated with HMG–CoA.
Mass Spectroscopy. Matrix-assisted laser desorption ionization/time-of-flight (MALDI-TOF) mass spectroscopy of modified E95A synthase. Wild-type (molecular mass, 57,916 Da) and E95A (molecular mass, 57,858 Da) avian synthase (6.5 nmol) was incubated with HMG–CoA (31 nmol) at room temperature in a 20-μl final volume, in the presence of 50 mM NaH2PO4 (pH 6.5). After 5 days of incubation a 2-μl aliquot was diluted (1:50) in 3,5-dimethoxy-4-hydroxycinnamic acid matrix (10 mg/ml in 50% acetonitrile/0.1% trifluoroacetic acid). The sample was subjected to MALDI-TOF mass spectrometry on a Voyager DE-PRO (Per-Septive Biosystems) at the Medical College of Wisconsin Protein and Nucleic Acid Core facility.
Exchange of 18O oxygen from water into HMG–CoA. Avian wild-type and E95A synthase enzyme preparations were dialyzed against 20 mM NaH2PO4 buffer (pH 6.6) containing 0.1 mM DTT. After lyophilization, duplicate samples of enzymes (69 nmol) were dissolved in 50 mM NaH2PO4 (pH 6.6) prepared in 18O or 16O water. HMG–CoA was added to the enzyme mixture to a final concentration of 550 μM and incubated at room temperature. Aliquots (100 μl) were centrifuged in a Biomax concentrator (Millipore, 10-kDa molecular mass exclusion limit) to remove protein. A Biogel P2 centrifugal column was equilibrated with 20 mM ammonium acetate and used to further purify the HMG–CoA. The filtrate was lyophilized and redissolved in 20 μl of methanol and subjected to analysis by MALDI-TOF mass spectrometry.
Crystallization and Data Collection. Either His-tagged or native HMGS was purified and concentrated to 10–12 mg/ml (≈250 μM) and dialyzed vs. 20 mM Tris·HCl, pH 7.5/1 mM DTT. HMG–CoA was added to a concentration of 600 μM, and the protein was crystallized by vapor diffusion at 19°C against 1.75–2.5 M ammonium sulfate/50 mM Tris, pH 7.5. Crystals (up to 800 × 400 × 50 μm) formed after 2–5 days. A crystal of nontagged HMGS was cryocooled in paratone oil and exposed to copper Kα radiation (focused and monochromated with multilayer Osmic optics) from a Rigaku rotating anode generator 5 days after the initiation of the vapor diffusion process. X-ray diffraction intensities were measured with an R-AXIS IIc detector and processed with the hkl program suite (14). His-tagged monoclinic crystals were cryocooled in oil (3:1 vol/vol, NVH oil/paraffin) 14 and 31 days after the initiation of the vapor diffusion process. X-ray diffraction intensities were collected at beamlines 22-ID and 19-BM of the Advanced Photon Source (Chicago). Scaled, averaged data were converted to structure factors with the ccp4 suite (15); 5% of the reflections were reserved for calculation of Rfree (16).
Phasing and Refinement. His-tagged HMGS was found to crystallize in a monoclinic (P21) space group, whereas crystals of nontagged protein were triclinic (P1). Phasing was accomplished by molecular replacement using beast (17) and a search model derived from the structure of the abortive AcAc–CoA enzyme complex (Protein Data Bank ID code 1TXT) (4). The asymmetric units of both crystal forms contain two dimers. The structures were manually built in o (18), alternating with automated refinement in cns (19). At later stages of refinement, sulfate, water, and CoA derivative molecules were modeled, as well as alternate protein conformations, and individual atomic B-factors were refined. For the active site of each protomer, up to three overlapping ligation states were modeled: apo (no ligand), HMG–CoA, and AcAc–CoA with both acetylated and unacetylated Cys-111. The occupancy of each possible ligation state was manually adjusted to provide the best fit to the electron density while maintaining comparable temperature factors. The unacetylated Cys-111 shows clear evidence of electron density for a sulfhydryl without corresponding acetyl density. This sulfhydryl position is incompatible with bound HMG–CoA, overlaps with the position of the sulfhydryl found in the apo enzyme, and is only modeled when no apo enzyme is detected. If the occupancy was <10% for any given ligation state, that state was not considered relevant to that protomer and was removed from refinement. At no stage of the refinement were noncrystallographic symmetry (NCS) restraints applied, because of the differences in ligand occupancy. Trial refinements with NCS restraints showed no significant differences in Rfree.
Figures depicting protein structures were generated with molscript (20), povscript+ (21), and pov-ray (www.povray.org).
Results
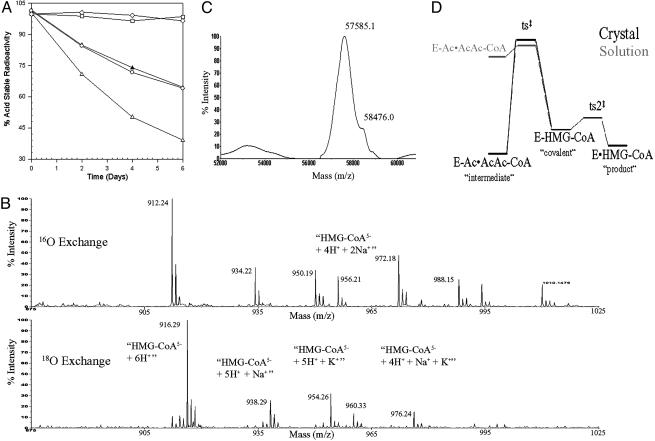
In vitro, S. aureus HMGS catalyzes the overall cleavage of HMG–CoA to AcAc–CoA and acetate (the partial-reverse or backwards reaction) with a first-order rate constant of 0.3 day–1 (Fig. 1A). The avian enzyme shows an enzyme concentration-dependent rate similar to the S. aureus enzyme. The wild-type avian enzyme also catalyzes the rapid exchange of the HMG–CoA carboxylate oxygens with 18O water (Fig. 1B), as evidenced by the 4-atomic mass unit (amu) increase of each peak in the HMG–CoA mass spectrum after a 30-min incubation. In the isotope exchange reaction, HMG–CoA forms a covalent bond with the active site cysteine, the liberated water molecule exchanges with an 18O water molecule, and, subsequently, 18O carboxylate-labeled HMG–CoA is hydrolyzed off the enzyme. Thus, the rate-limiting step for this nonphysiological reverse reaction is the carbon–carbon cleavage step.
Fig. 1.
Monitoring different aspects of the “backwards” reaction catalyzed by HMGS. (A) HMGS catalyzes the cleavage of HMG–CoA; solid triangles and open circles represent wild-type avian and S. aureus enzymes, respectively; open squares and open diamonds represent the avian C129S mutated enzyme and BSA, respectively; and open triangles represent a 2× concentration of wild-type avian enzyme. (B) Mass spectra of HMG–CoA incubated for 30 min with 16O(Upper) or 18O(Lower) water in the presence of wild-type enzyme. This example shows that nearly complete exchange of the carboxylate oxygens has occurred. (C) Mass spectra of the E95A mutated enzyme after a 5-day incubation with HMG–CoA shows that this enzyme is capable of forming a covalent intermediate. (D) Energy profile for the latter half of the reaction pathway in the crystal and in solution.
The catalytic activity of the E95A mutated avian enzyme (corresponding to S. aureus Glu-79) is reduced by 105-fold relative to the wild-type enzyme, which led to the conclusion that the glutamic acid is involved in carbon–carbon bond formation (9). Additional evidence for this role can be found by studying the backwards reaction using MALDI-TOF mass spectroscopy (Fig. 1C). After incubating with HMG–CoA for 5 days, the spectrum of the E95A mutated avian enzyme shows a high molecular mass (+891 atomic mass units) shoulder corresponding to a covalent HMG–CoA intermediate. The mass spectra for the wild-type enzyme only showed the 57.6-kDa protein peak, indicating the normally transient nature of the covalent intermediate. It is important to note that the E95A mutated avian enzyme does not show any 18O exchange, even after 24 h (data not shown), indicating a decreased rate of covalent bond formation and hydrolysis. Thus, this catalytic amino acid is involved in both the formation and hydrolysis of the HMG–CoA covalent intermediate.
Each crystal structure was determined as described in Materials and Methods and gave statistics described in Table 1. The “day 5” crystal structure, determined to a resolution of 2.0 Å, reveals mixtures of both the HMG–CoA-enzyme (product) complex and the AcAc–CoA-acetylated enzyme (intermediate) complex. In one of the active sites, only AcAc–CoA is modeled; in another, only HMG–CoA is modeled, and in the remaining two active sites, there is a 2:1 mixture of AcAc–CoA to HMG–CoA. The origin of the variation of occupancies is unknown; however, it is likely to be due to crystal packing forces. The “day 14” crystal structure, determined to a resolution of 2.0 Å, reveals protomers that are nearly uniformly bound to AcAc–CoA and a mixture of acetylated and unacetylated Cys-111. The observed unacetylated cysteine (≈30%) is likely caused by the nonphysiological hydrolysis of acetylated cysteine. The “day 31” crystal structure, determined to a resolution of 1.6 Å, reveals nearly equal mixtures of HMG–CoA–enzyme complex, AcAc–CoA–acetylated enzyme complexes, and apo-enzyme. The apoenzyme structure observed in the day 31 crystal is consistent with that observed when crystals were grown in the absence of ligands (Protein Data Bank ID code 1TVZ; ref. 4). The CoA-bound forms and the apo-form of the enzyme show widespread differences in the conformation of individual residues both near and away from the CoA-binding site. However, there are no domain movements seen. Residues affected include: 197–205, 209, 232–240, and 245–251. The electron density for one of these day 31 protomers is shown in Fig. 2. The resolution is sufficient to confidently model all three of these species with approximate occupancies.
Table 1. Data refinement and crystallography information.
| Day
|
|||
|---|---|---|---|
| 5 | 14 | 31 | |
| Crystal | |||
| Space group | P1 | P21 | P21 |
| a, Å | 64.27 | 65.33 | 65.12 |
| b, Å | 84.61 | 118.41 | 118.77 |
| c, Å | 94.63 | 121.63 | 121.30 |
| α, ° | 84.25 | 90.00 | 90.00 |
| β, ° | 72.52 | 90.03 | 90.08 |
| γ, ° | 70.44 | 90.00 | 90.00 |
| Data collection | |||
| Resolution | 35-2.0 | 50-2.0 | 25-1.6 |
| Rsym, % | 2.8 (13.2) | 6.2 (32.5) | 8.2 (60.2) |
| Completeness, % | 78.8 (32.2) | 87.0 (78.7) | 89.8 (77.7) |
| I/σ | 27.0 (4.66) | 13.2 (2.46) | 28.3 (2.73) |
| Observed reflections | 163,029 (6,462) | 213,547 (18,750) | 1,877,193 (152,731) |
| Unique reflections | 94,526 (3,865) | 105,350 (9,475) | 217,551 (18,778) |
| Redundancy | 1.7 (1.7) | 2.0 (2.0) | 8.6 (8.1) |
| Refinement | |||
| Resolution | 35-2.0 | 50-2.0 | 15-1.6 |
| σ cutoff | 0 | 0 | 0 |
| Protein atoms | 11,692 | 12,176 | 15,222 |
| Nonprotein atoms | 842 | 782 | 1,553 |
| Total atoms | 12,534 | 12,958 | 13,669 |
| Working reflections | 87,765 | 100,054 | 205,819 |
| Data/parameter | 1.75 | 1.93 | 3.38 |
| Test reflections | 4,609 | 5,269 | 10,669 |
| Rwork, % | 20.4 | 20.6 | 21.1 |
| Rfree, % | 23.9 | 23.9 | 22.7 |
| rmsd bonds, Å | 0.006 | 0.006 | 0.005 |
| rmsd angles, ° | 1.3 | 1.3 | 1.3 |
| Protein Data Bank ID | 1XPK | 1XPL | 1XPM |
Values in parentheses are for the highest-resolution shell. rmsd, rms deviation.
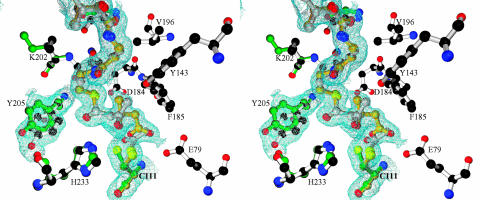
Fig. 2.
Stereo diagram of the electron density map (2Fo – Fc) surrounding the CoA moiety, Cys-111, and Tyr-205 from one of the active sites in the day 31 crystal superimposed on the final atomic model. Residues modeled with green bonds correspond specifically to the apo enzyme, residues modeled with gold bonds correspond to the HMG–CoA complex, residues modeled with silver bonds correspond to the AcAc–CoA complex, and residues modeled with white bonds correspond to common conformations. The position of the sulfur of Cys-111 that is nearest to the observer corresponds to the position when HMG–CoA is bound, whereas the position to the left corresponds to the apo enzyme conformation. The apo enzyme conformation is sterically incompatible with the binding of HMG–CoA. Note how the modeled alternative conformation of Tyr-205 helps to explain the observed electron density.
Our decision to not use noncrystallographic symmetry restraints during refinement (which would make these structures dependent on each other) is justified because the statistical errors for distances within the active site are smaller than what would have been anticipated from the corresponding Luzatti plot (22). Seven instances of the product complex, 11 instances of the acetylated intermediate complex, and four instances of the apo-enzyme were independently observed. Excluding the apoenzyme conformations, the overall rms deviation (rmsd) for all of the protomers was 0.39 Å for all protein atoms, whereas the α carbons, the HMG moiety, AcAc moiety, and apo-enzyme residue conformations have rmsds of 0.22 Å, 0.12 Å, 0.11 Å, and 0.34 Å, respectively. The errors appear to be random, and show neither systematic crystal nor protomer dependencies. Thus, the hydrogen bond and close contact distances shown in Fig. 3 are presented with statistical errors. Together, the three crystal structures of HMGS cocrystallized with HMG–CoA yield structural details for the HMG–CoA complex, the acetylated cysteine-AcAc–CoA complex, and structure of the apo-enzyme (Fig. 4).
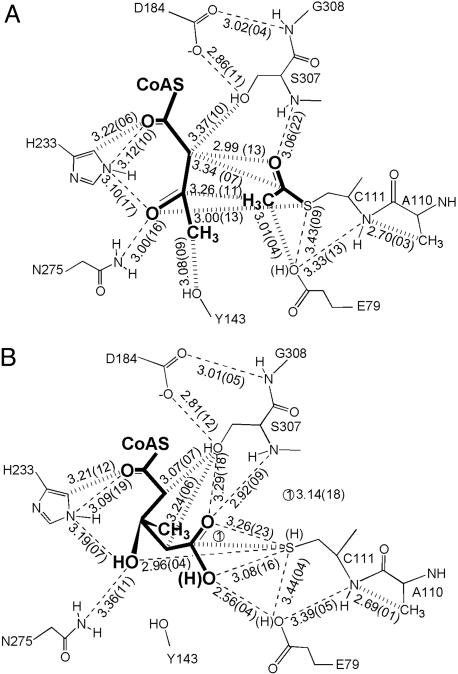
Fig. 3.
Schematic representation of the contacts in the active site with AcAc–CoA (A) and the acetylcysteine and HMG–CoA (B). The wide dashed lines indicate close contacts that violate van der Waals distance constraints, and the narrow dashed lines indicate potential hydrogen bonds. The average distances are given in angstroms (10–10 m) based on either 11 or 7 structures, and the numbers in parentheses are the standard deviations of the distance multiplied by 100.
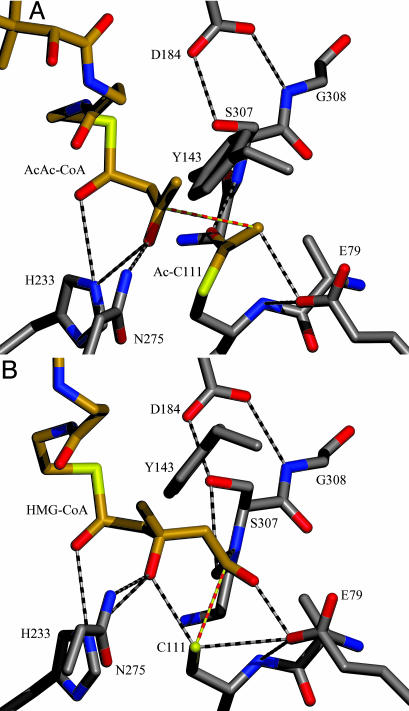
Fig. 4.
Models of the AcAc–CoA-acetylated enzyme complex (A) and the HMG–CoA enzyme complex (B). The atoms are colored according to their CPK atom type, and the bonds are shown in either white (for protein) or gold (for the acetyl moiety and the CoA molecules). Yellow-and-red dashed bonds tie reacting atoms together.
Discussion
The complex of AcAc–CoA with HMGS acetylated at Cys-111 (Figs. 3A and 4A) represents the stage of the forward reaction just before condensation. Dissociation of the ligand from enzyme is hindered by crystal-packing contacts against the adenine ring. The acetoacetyl moiety is constrained by interactions with Tyr-143, Phe-185, His-233, Asn-275, Ser-307, and the acetylcysteine group. Although catalysis of the H264A mutated avian enzyme (corresponding to His-233 in S. aureus) is down only 20-fold, the Michaelis constant for the AcAc–CoA is 100-fold greater than that of the wild-type enzyme (11). Thus, we believe that both this histidine and Asn-275 are responsible for the observed stereochemistry of the reaction and that His-233 acts as the general acid during the condensation step of the reaction.
Activation of the acetyl group is accomplished by a Glu-79 carboxylate oxygen that is situated to abstract a proton from the acetylcysteine C-2 (methyl) carbon (the O–C distance is 3.0 Å). This methyl carbon, in turn, is positioned to attack the C-3 (carbonyl) carbon of AcAc–CoA (the C–C distance is 3.3 Å). The conformational flexibility inherent in the acetoacetyl moiety could bring the carbonyl carbon within bonding distance to a newly formed carbanion of the acetylcysteine. This structure, combined with earlier mutagenesis, strongly suggests that Glu-79 is, in fact, the catalytic base during the condensation step of the reaction. This is the first instance of a condensing enzyme positioning a carboxylate as a general base to initiate the formation of a carbon–carbon bond. In thiolase, which abstracts a proton from the incoming (second) acetyl-CoA, a cysteine general base is positioned near the incoming acetyl-CoA rather than near the acetylated cysteine (23). Furthermore, although the β-keto-acyl carrier protein synthase enzymes have a histidine (corresponding to His-233 of HMGS) positioned near the incoming second substrate, it is proposed that this histidine acts as a general base to assist the decarboxylation reaction rather than as a general acid as is proposed here (24, 25). Thus, the active site of HMGS is uniquely arranged to use an activated acetylcysteine within the thiolase superfamily.
The orientation of the acetylated cysteine suggests a hydrogen bond between the carbonyl oxygen of the thioester and the backbone amide of Ser-307 (3.1 Å). This hydrogen bond could stabilize both the oxyanion formed during the initial acetylation reaction (step 1 in Scheme 1), as noted for other condensing enzymes (e.g., β-keto-acyl carrier protein synthase) (24), and the partial negative charge resulting from resonance after a proton is abstracted from the methyl group during the condensation step (step 2 in Scheme 1). Ser-307 is at the amino terminus of a β-turn that includes a conserved cis–peptide bond between Gly-308 and Ser-309. The conformation of this β-turn is stabilized by a hydrogen bond to both the amide nitrogen of Gly-308 and the hydroxyl oxygen of Ser-307 from the carboxylate of Asp-184. The importance of this hydrogen bond has been demonstrated by mutation of this Asp to Ala in the avian enzyme, where the overall activity is reduced by 104-fold and the rate of acetylation is reduced by 102-fold (10). Thus, this backbone amide nitrogen not only plays the same critical role in HMGS as it does in the other condensing enzymes, but also plays a role in the condensation reaction.
The HMG–CoA-enzyme complex is observed in some of the protomers of the day 5 and day 31 crystals (Figs. 3B and 4B). As with the acetoacetyl portion of AcAc–CoA, the HMG moiety of HMG–CoA is conformationally locked within the active site by protein residues (Asp-184, Phe-185, His-233, Ser-307, and Cys-111). A short distance (2.6 Å) between a Glu-79 carboxylate oxygen and a HMG–CoA carboxylate oxygen suggests that one or both oxygens are protonated and form a hydrogen bond. The proximity of Glu-79 to the carboxylic acid of HMG–CoA is consistent with rapid formation and cleavage of the covalent HMG–CoA-enzyme adduct, as evidenced by the rate of 18O exchange (Fig. 1B) and by the previously determined apoenzyme and abortive binary complex structures (Protein Data Bank ID codes 1TVZ and 1TXT; ref. 4), where a water molecule is found in nearly the same position as the carboxylate oxygen of the bound HMG–CoA.
Comparing this structure to the AcAc–CoA complex shows that nearly all of the atoms in the keto moiety along with the atoms in the cysteine acetyl group undergo significant changes in position upon their conversion to the HMG moiety (Fig. 2). In the acetyl moiety, the attacking methyl group must move 2.1 Å during the course of the reaction, whereas the acetyl oxygen only moves 0.8 Å and maintains its hydrogen bond to the amide nitrogen of Ser-307. On the acetoacetyl moiety, the attacked carbonyl carbon atom moves 0.7 Å away from the Cys-111 sulfur, whereas the carbonyl oxygen atom moves 0.7 Å, which reduces the strength of its hydrogen bonds to His-233 and Asn-275 and permits tetrahedral geometry at the attacked central carbon. The methyl carbon moves 0.7 Å away from Tyr-143, and the bridging methylene carbon atoms also move 0.5 Å to form the tetrahedral geometry of the central carbon atom.
It is possible to construct an energy profile of the enzyme as initially observed in the crystal and in solution (Fig. 1D). The observation of only acetylated enzyme at day 14 implies that the equilibrium between the product complex and the intermediate complex is shifted toward the intermediate complex. By contrast, in solution, the equilibrium between the product and intermediate complexes must lie toward the product complex given the relative rates of the forwards and backwards reactions (it can be estimated that kcat,f/Km,AcAc–CoA is ≈104-fold greater than kcat,b/Km,HMG–CoA; ref. 26). However, the energy of the transition state corresponding to conversion of the covalent intermediate to the product must be lower than that for the cleavage of the carbon–carbon bond because the rate of 18O isotope exchange is at least 100-fold greater than the overall rate of the backwards reaction (Fig. 1B).
We posit that, when the crystal first forms with HMG–CoA bound to each active site, the enzyme is “trapped” in a particular conformation where the intermediate complex is favored (thus the observed transformation of product to intermediate), although there are virtually no changes between the protein structures with HMG–CoA bound at day 5 and AcAc–CoA bound at day 14. By contrast, the apo-enzyme has a profound and widespread effect on the structure over a long distance when compared to that of the CoA-bound enzymes. The apo-enzyme is only seen in the day-31 crystal, and its presence coincides with the reemergence of HMG–CoA, where none was seen at day 14. It is possible that the relative energies of the product and intermediate complexes in the crystal shift back toward those in solution when the active site of the neighboring protomer is no longer occupied by a CoA molecule. This dependence on the state of the neighboring protomer has previously been suggested by the maximum stoichiometry of enzyme acetylation, which never exceeds a value of 0.7 (7). NMR spectroscopy has demonstrated the importance of the CoA moiety on active site reactivity, where acetylated cysteine may be observed for hours when a near stoichiometric amount of acetyl-CoA is added (9). On the other hand, under turnover conditions, where the amount of acetyl-CoA vastly exceeds the amount of enzyme, the nonphysiological hydrolysis of acetyl-CoA occurs in a matter of minutes (10). Because there is no obvious structural change to explain the apparent difference in the relative energies of the two bound species, although small structural changes cannot be ruled out, we posit that protein dynamics, as has seen in dihydrofolate reductase (27), play a significant role in catalysis.
Acknowledgments
We thank Drs. Younchang Kim and Frank Rotella for assistance in synchrotron x-ray data collection. This work has been supported by National Institutes of Health Grant DK21491 (to H.M.M.) and “startup” funds from Rosalind Franklin University of Medicine and Science (to D.H.T.H.). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. Data were collected at both the SER-CAT 22-ID and the SBC-CAT 19-BM beamlines.
Author contributions: H.M.M. and D.H.T.H. designed research; M.J.T., I.M., D.S., and D.H.T.H. performed research; N.C. contributed new reagents/analytic tools; M.J.T., I.M., H.M.M., and D.H.T.H. analyzed data; and M.J.T. and D.H.T.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HMG, 3-hydroxy-3-methylglutaryl; HMGS, HMG–CoA synthase; AcAc–CoA, acetoacetyl–CoA; MALDI-TOF, matrix-assisted laser desorption ionization/time-of-flight.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1XPk, 1XPL, and 1XPM).
See Commentary on page 16399.
References
- 1.Greenspan, M. D., Yudkovitz, J. B., Lo, C. Y., Chen, J. S., Alberts, A. W., Hunt, V. M., Chang, M. N., Yang, S. S., Thompson, K. L., Chiang, Y. C., et al. (1987) Proc. Natl. Acad. Sci. USA 84, 7488–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teo, K. K. & Burton, J. R. (2002) Drugs 62, 1707–1715. [DOI] [PubMed] [Google Scholar]
- 3.Wilding, E. I., Brown, J. R., Bryant, A. P., Chalker, A. F., Holmes, D. J., Ingraham, K. A., Iordanescu, S., So, C. Y., Rosenberg, M. & Gwynn, M. N. (2000) J. Bacteriol. 182, 4319–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campobasso, N., Patel, M., Wilding, E. I., Kallender, H., Rosenberg, M. & Gwynn, M. (2004) J. Biol. Chem. 279, 44883–44888. [DOI] [PubMed] [Google Scholar]
- 5.Edwards, D. J., Marquez, B. L., Nogle, L. M., McPhail, K., Goeger, D. E., Roberts, M. A. & Gerwick, W. H. (2004) Chem. Biol. 11, 817–833. [DOI] [PubMed] [Google Scholar]
- 6.Miziorko, H. M. & Lane, M. D. (1977) J. Biol. Chem. 252, 1414–1420. [PubMed] [Google Scholar]
- 7.Miziorko, H. M., Clinkenbeard, K. D., Reed, W. D. & Lane, M. D. (1975) J. Biol. Chem. 250, 5768–5773. [PubMed] [Google Scholar]
- 8.Misra, I., Narasimhan, C. & Miziorko, H. M. (1993) J. Biol. Chem. 268, 12129–12135. [PubMed] [Google Scholar]
- 9.Chun, K. Y., Vinarov, D. A., Zajicek, J. & Miziorko, H. M. (2000) J. Biol. Chem. 275, 17946–17953. [DOI] [PubMed] [Google Scholar]
- 10.Chun, K. Y., Vinarov, D. A. & Miziorko, H. M. (2000) Biochemistry 39, 14670–14681. [DOI] [PubMed] [Google Scholar]
- 11.Schlichting, I., Berendzen, J., Phillips, G. N., Jr., & Sweet, R. M. (1994) Nature 371, 808–812. [DOI] [PubMed] [Google Scholar]
- 12.Schlichting, I., Almo, S. C., Rapp, G., Wilson, K., Petratos, K., Lentfer, A., Wittinghofer, A., Kabsch, W., Pai, E. F., Petsko, G. A., et al. (1990) Nature 345, 309–315. [DOI] [PubMed] [Google Scholar]
- 13.Stoddard, B. L., Bruhnke, J., Koenigs, P., Porter, N., Ringe, D. & Petsko, G. A. (1990) Biochemistry 29, 8042–8051. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 15.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 16.Brünger, A. T. (1992) Nature 355, 472–475. [DOI] [PubMed] [Google Scholar]
- 17.Read, R. J. (2001) Acta Crystallogr. D 57, 1373–1382. [DOI] [PubMed] [Google Scholar]
- 18.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 19.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 20.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 21.Fenn, T. (2003) J. Appl. Crystallogr. 36, 944–947. [Google Scholar]
- 22.Luzatti, P. V. (1952) Acta Crystallogr. A 5, 802–810. [Google Scholar]
- 23.Modis, Y. & Wierenga, R. K. (2000) J. Mol. Biol. 297, 1171–1182. [DOI] [PubMed] [Google Scholar]
- 24.Qiu, X., Janson, C. A., Konstantinidis, A. K., Nwagwu, S., Silverman, C., Smith, W. W., Khandekar, S., Lonsdale, J. & Abdel-Meguid, S. S. (1999) J. Biol. Chem. 274, 36465–36471. [DOI] [PubMed] [Google Scholar]
- 25.Davies, C., Heath, R. J., White, S. W. & Rock, C. O. (2000) Struct. Fold. Des. 8, 185–195. [DOI] [PubMed] [Google Scholar]
- 26.Menahan, L. A., Hron, W. T., Hinkelman, D. G. & Miziorko, H. M. (1981) Eur. J. Biochem. 119, 287–294. [DOI] [PubMed] [Google Scholar]
- 27.Agarwal, P. K., Billeter, S. R., Rajagopalan, P. T., Benkovic, S. J. & Hammes-Schiffer, S. (2002) Proc. Natl. Acad. Sci. USA 99, 2794–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]