Abstract
Background & objectives:
Scrub typhus is a vector-borne zoonotic infection caused by Orientia tsutsugamushi. Local epidemiology of the circulating serotypes of scrub typhus is not available from most parts of India. We conducted this study for the diagnosis of scrub typhus using IgM ELISA and to detect O. tsutsugamushi serotypes circulating in southern Andhra Pradesh, India.
Methods:
Samples were collected from patients clinically suspected to have scrub typhus and were subjected to IgM ELISA to measure IgM antibodies against O. tsutsugamushi. Nested polymerase chain reaction (PCR) was performed targeting strain-specific regions in ELISA-positive samples.
Results:
Of a total of 663 samples, 258 (38.91%) were found to be positive by IgM ELISA. Serotypes could be detected in 230 (34.69%) samples only. Only two serotypes, Karp and Kawasaki, were found in the serum samples, with the former being predominant. The dual infection of Karp and Kawasaki serotypes was found in seven patients. Other serotypes such as Gilliam, Kuroki and Kato were not detected in the samples.
Interpretation & conclusion:
The nested PCR products proved useful in presumptively identifying the endemic O. tsutsugamushi serotypes. The present study could be significant in understanding scrub typhus epidemiology in this region.
Keywords: 56 kDa gene, blood clot, Karp, Kawasaki, nested polymerase chain reaction, Orientia tsutsugamushi, scrub typhus
Scrub typhus is an acute febrile illness caused by the obligate intracellular bacterium Orientia tsutsugamushi. It is transmitted to humans by the bite of larval trombiculid mites, and people who inhabit regions infested with these vectors are at high risk of acquiring scrub typhus1. Scrub typhus is confined geographically to the Asia-Pacific region with more than a billion people at risk and one million new cases each year2.
The clinical features include fever, headache, myalgia, lymphadenopathy, rash and eschar that can be complicated by interstitial pneumonitis, meningitis and myocarditis3. Disease severity and manifestations vary widely from asymptomatic to fatal and show marked geographical differences. The general course of the disease and the prognosis vary considerably depending on the character of the endemic strain4. Originally, the antigenic diversity of the three prototype strains, Gilliam, Karp and Kawasaki was illustrated5. Later on, additional antigenic types were described, with representative strains including the Kawasaki6, Kuroki7, Shimokoshi8 types and other distinct serotypes present in the tsutsugamushi triangle9. Antigenic heterogeneity of the organism may be the reason for frequent outbreaks and reinfection. It has been postulated that the virulence of O. tsutsugamushi differs among the strains depending on their serotype10. Hence, rapid diagnosis of O. tsutsugamushi endemic serotypes is essential to reduce the burden of the disease. There are several countries such as India, Indonesia, Pakistan and Uzbekistan, where scrub typhus is proven or suspected to be endemic and limited data on the circulating serotypes are available1. Outbreaks of scrub typhus have been previously reported from various parts of India3,11,12,13,14. However, there has been no information on circulating serotypes and genotypes of O. tsutsugamushi from southern part of the country. The objective of this study was to diagnose scrub typhus by ELISA on serum samples of patients clinically suspected to have scrub typhus and to detect the O. tsutsugamushi serotypes that are circulating in southern Andhra Pradesh, India.
Material & Methods
This study was a collaborative work between Sri Venkateswara University (SVU) and Sri Venkateswara Institute of Medical Sciences (SVIMS), Tirupati, Andhra Pradesh. The patients with suspected cases of fever of unknown origin (FUO) were included in the study. All government and private health centres in and around Chittoor and other neighbouring districts of southern Andhra Pradesh, India, were requested to send the clinically suspected scrub typhus patients’ blood samples to SVIMS. A total of 663 patients experiencing a febrile illness clinically consistent with scrub typhus were selected as per the standard inclusion and exclusion criteria15.
Patients who presented with an acute febrile illness having at least five out of the following clinical features were included in the study: headache, myalgia, lymphadenopathy, hepatomegaly, splenomegaly, presence of an eschar or presence of a maculopapular rash. A case was excluded, if the cause of fever was known at the time of admission and/or tested positive for typhoid by Widal test or Blood culture test, chikungunya by IgM ELISA and dengue by NSI Ag ELISA.
After recording clinical symptoms, 2 ml of blood sample was collected from the patients and blood was sent to the department of Microbiology, SVIMS, for serological diagnosis of scrub typhus and serotype identification. Blood samples that were collected during 2011-2013 only in the months of September to December were included in the present study. The study was approved by the Institutional Ethics Committee, SVIMS, Tirupati. Serum and clot were separated by centrifugation and preserved at -20°C until processing. Detection of IgM antibodies against O. tsutsugamushi was performed by commercial ELISA kit (InBiOS International Inc. USA) as per the manufactures’ instructions. The patients having IgM antibodies against O. tsutsugamushi were diagnosed as having scrub typhus and their blood samples were used for polymerase chain reaction (PCR) amplification.
Extraction of genomic DNA from blood clot: Genomic DNA was extracted and purified according to the method described by Furuya et al16. About 1.5 ml of homogenized blood clot was mixed with 10 per cent sodium dodecyl sulphate and incubated at 4°C for 16 h. Followed by addition of 0.1 ml of digestion buffer [100 mM Tris-Hcl, 10 mM ethylenediaminetetraacetic acid (EDTA), pH 8.0, 2 mg/ml lysozyme] for 30 min in an ice bath, Proteinase K at a final concentration of 0.2 mg/ml was added and incubated at 55°C for 1 h. The mixture was subjected to phenol, chloroform and isoamyl alcohol (25:24:1 v/v) extraction and ethanol precipitation. The pellet was washed twice with 75 per cent ethanol, air dried and suspended in 50 µl of TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0).
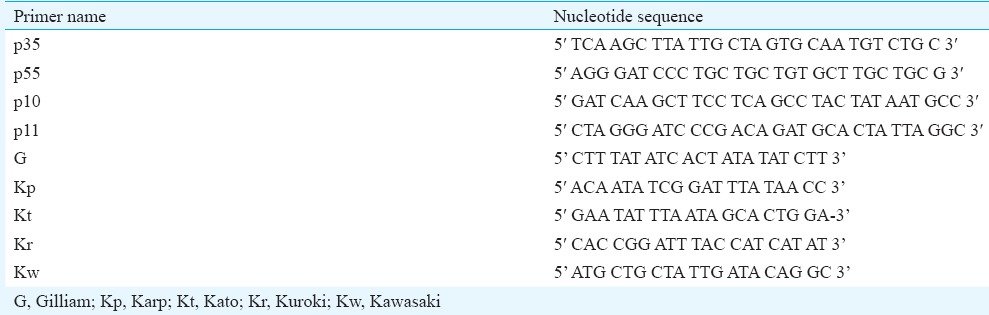
Nested PCR amplification: Oligonucleotide primers for nested PCR were synthesized (Eurofins Genomics, Bengaluru, India) from the previously established nucleotide sequence of the 56-kDa antigen of O. tsutsugamushi by Furuya et al16. Details regarding primer name and primer sequence listed are listed in Table I. The first PCR amplification was carried out using primers p35 and p55 to amplify the 1003 bp region. For identification of Gilliam, Karp, Kato and Kuroki serotypes, second PCR amplification was carried out by combination of primers p10 with G, Kp, Kt and Kr to amplify the 407, 230, 242 and 220 bp regions, respectively. Kawasaki serotyping was conducted by combination of p11 with Kw to amplify 523 bp region.
Table I.
List of primers used for the present study
The first amplification mixture (total volume of 50 µl) contained 5 µl of template DNA, 1.5 mM of MgCl2, ×10 buffer with KCl, 200 µM of dNTPs mix, 10 pmoles of each primer and 1.25 U of Taq DNA polymerase (Fermentas, USA). For the second PCR, 5 µl of the first PCR product was used as template and the remaining master mix was the same as the first PCR mix. The PCR reaction conditions were same for the first and the second PCR: initial denaturation at 94°C for 2 sec; denaturation at 94°C for 30 sec, annealing at 57°C for one minute and extension at 72°C for one minute for 35 cycles and final extension at 72° for 10 min. The amplified DNA fragments were separated on a 1.0 per cent agarose gel containing 0.5 µg/ml ethidium bromide. The PCR products were viewed under UV illumination and documented using a gel documentation system (Bio-Rad, USA).
Purification of amplified products and DNA sequencing: The amplified PCR product of O. tsutsugamushi was purified with the QIAquick kit (QIAgen, USA) as per the manufactures’ instructions. Purified PCR products were sequenced by Sanger's dideoxy method17 on ABI 3730 × 1 automated sequencer (Applied Biosystem, Foster City, CA). Nucleotide sequences obtained from the present study have been deposited in the GenBank data library under accession numbers KJ094996 (Karp serotype) and KJ094997 (Kawasaki serotype).
Statistical analysis:The Statistical analysis was performed using SPSS software, version 16.0 (SPSS Inc., Chicago, IL, USA). The continuous data were expressed as mean ± standard deviation. Descriptive statistics for the categorical variables were performed by computing the frequencies (percentages) in each category. The association between two categorical variables was analyzed by Chi-square test or Fisher's exact test as appropriate.
Results
A total of 663 patients were enrolled in this study hailing from the four districts of Andhra Pradesh, India. This comprised 439 patients from Chittoor district, 126 from Nellore, 62 from Kadapa and 36 from Anantapur. The patients investigated included 433 (65.30%) males and 230 (34.69%) females. The mean age of the patients was 42.88 ± 1.12 yr (range, 2 to 89 yr; median, 45 yr). The mean time interval between onset of symptoms and hospitalization of patients and serum sampling was 8.7 ± 5.9 days (range, 1-30 days; median, 6 days). Of the 663 patients, 258 (38.91%) were positive for scrub typhus by recombinant IgM ELISA.
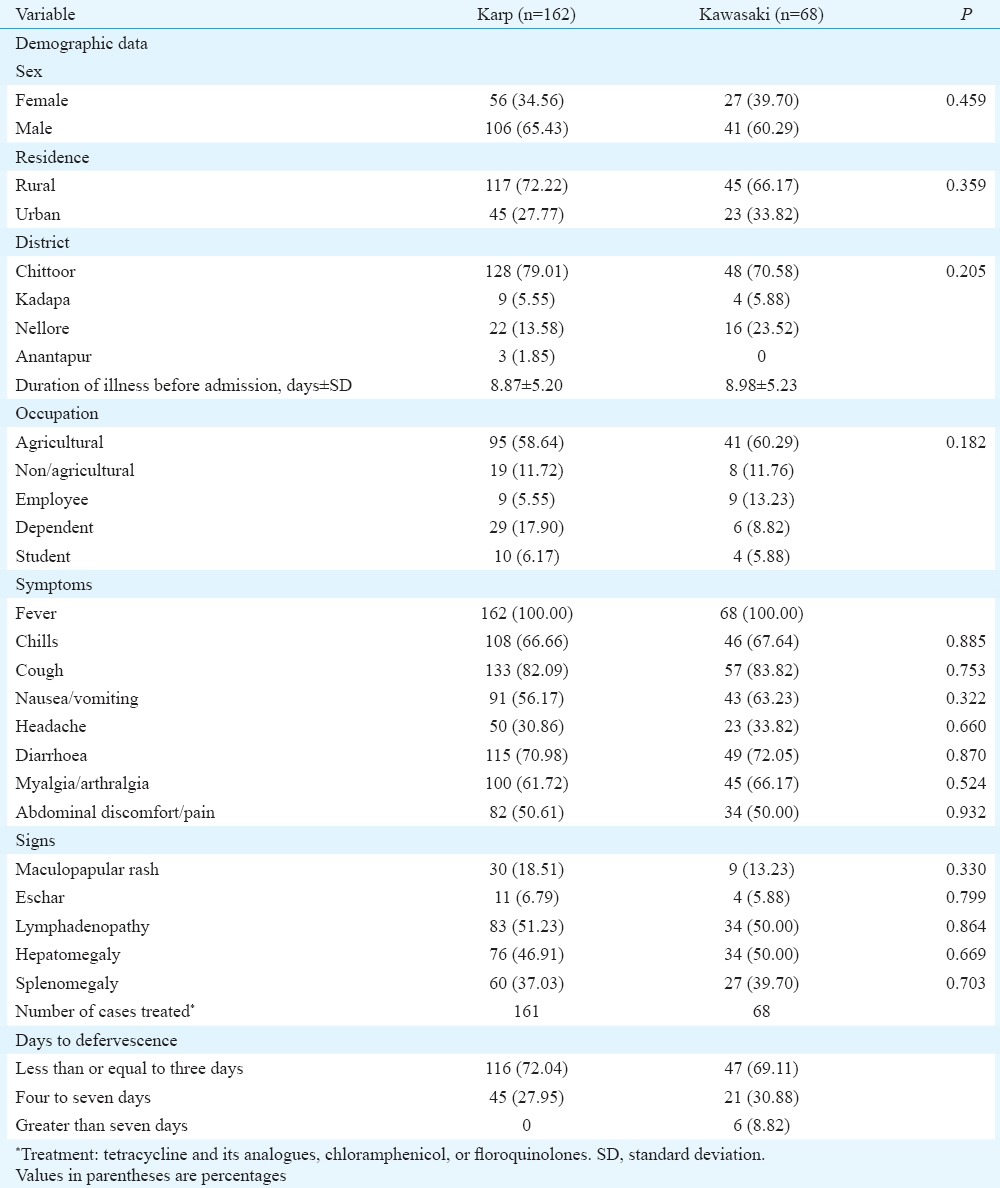
The species-specific serotypes were detected in 230 (34.69%) samples only. The serotypes, Karp (230 bp) and Kawasaki (523 bp) were detected among the examined specimens (Figure), whereas other serotypes, Gilliam (407 bp), Kuroki (220 bp) and Kato (242 bp) were not identified. It was observed that Karp (n=162) was the predominant serotype followed by Kawasaki (n=68) (Table II). Seven patients had dual infection with both Karp and Kawasaki serotypes. No significant difference was observed in the demographics or duration of fever between the Karp and Kawasaki serotype-positive patients. Cough was a prominent symptom in both Karp and Kawasaki serotype-affected patients (Table III). Only 6.70 per cent of Karp serotype-positive patients and 5.88 per cent of Kawasaki serotype-positive patients had eschar. All patients were treated with one of the tetracycline group of agents with defervescence of fever (in days) as shown in (Table III).
Figure.
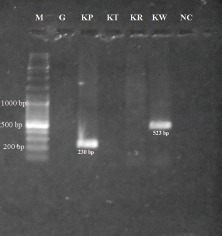
Agarose gel electrophoresis of amplified DNAs by nested polymerase chain reaction with serotype-specific primers. M: 1kb DNA ladder (Fermentas, code no. #SM0331); G, Gilliam; Kp, Karp; Kt, Kato; Kr, Kuroki; Kw, Kawasaki; NC, Negative control.
Table II.
Presence of different serotypes of Orientia tsutsugamushi in southern Andhra Pradesh region, India
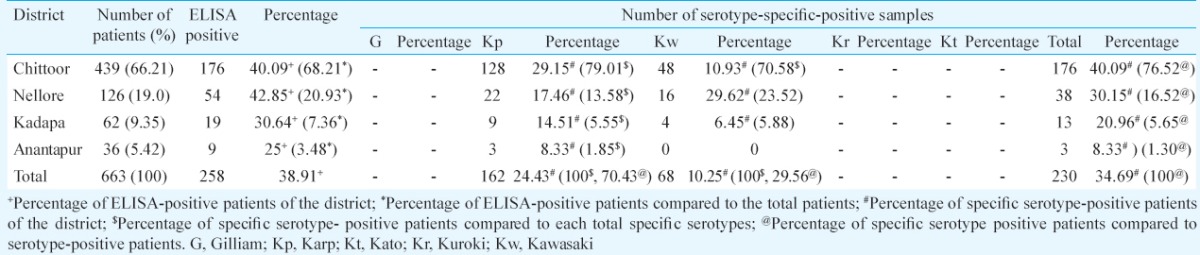
Table III.
Demographic and clinical characteristics of confirmed patients of scrub typhus (n=258)
Discussion
In the present study, circulating serotypes of O. tsutsugamushi were studied in southern Andhra Pradesh region. The public health importance of O. tsutsugamushi with the geographical differences in pathogenicity18 and the emergence of antibiotic-resistant strains19 have stimulated numerous studies on this organism. Moreover, there is significant antigenic and genotypic diversity of this organism from the endemic areas. In this study, only those cases that occurred during September to December were selected as the incidence of scrub typhus peaks during these months. The diagnosis of scrub typhus is often missed due to non-specific signs and symptoms and non-availability of the relevant laboratory tests. Kim et al20 found significant differences in frequencies of eschars, rashes, general weakness and conjunctival injection between Boryoung and Karp clusters. They also suggested that frequency of eschars and rashes in scrub typhus patients may depend on the genotypes of O. tsutsugamushi. However, in our study, no significant difference in clinical features was found between the Karp and Kawasaki serotypes found in this area. The presence of the pathognomonic eschar is an important diagnostic clue for diagnosis of scrub typhus21, but this definition is not suitable for the Indian subcontinent as eschar and rash are seen in <10 per cent of cases12. There is less accessibility to the gold standard test of indirect immunofluorescence antibody (IFA) or indirect immunoperoxidase because of the non-availability of the epidemic or endemic serotypes of whole O. tsutsugamushi bacteria from the particular geographic region added to the cost-effectiveness of these tests22. The scrub typhus ELISA is a flexible alternative to the IFA technique23 and has specificities and sensitivities of >90 per cent for detecting specific antibodies12. In moderately equipped laboratories in endemic regions, recombinant antigen-based ELISA is a useful alternative technique for diagnosis24.
Of the 258 IgM ELISA-positive patients’ samples, 230 (89.14%) were amplified by Karp-specific and Kawasaki-specific primers and the other 28 (10.85%) samples were not amplified by other sets of specific primers. This may be due to the absence of Orientia-specific DNA in the blood samples due to institution of specific treatment before collecting the samples while the IgM was still present in the blood. Another reason may be the non-availability of the primers needed to identify the diverse serotypes of this organism which are present in nature. The PCR technique has great potential for the diagnosis of infections caused by organisms which are difficult to cultivate, and hence, it can be employed for detecting O. tsutsugamushi in clinical samples25. The 56-kDa protein gene and 16s rRNA gene have been used to differentiate between Orientia and other genera/species26. We targeted 56-kDa-type-specific antigen gene as it contains four variable domains (domains I to IV) that differ between the strains27. It has been reported that O. tsutsugamushi has a variety of serotypes, the prevalence of which vary between different endemic areas28. In our study, Karp serotype was predominant followed by Kawasaki. Other serotypes, namely, Gilliam, Kato and Kuroki, were not detected in our study. A study from Himachal Pradeshl13 reported Kuroki serotype from this region by serotype-specific PCR, but they studied only ten randomly selected samples, of which eight were positive to Kuroki type and other two samples were non-responsive to the serotype-specific primers. A study from Korea reported the Boryong serotype being distributed throughout the country except for Cheju Island29. In another study Karp serotype was found to be distributed in all regions of Thailand whereas the Kawasaki serotype was found in the southern region of Thailand30.
One of the major limitations of this study was that all the ELISA-positive samples could not be serotyped using the primers selected for the study. This signifies that other serotypes may be in circulation in this region and additional primers need to be used to detect all existing serotypes.
In conclusion, the results of the present study revealed that two serotypes of O. tsutsugamushi were circulating in this region. The presence of dual infection with dual serotypes was also observed, and this might lead to emergence of newer strains with genetic variations that could alter the disease profile in future outbreaks of scrub typhus in this part of India. This information would be useful in understanding the O. tsutsugamushi evolution and may help in correlating disease severity to serotypes or genotypes during future outbreaks.
Acknowledgment
The authors thank all the government and private health centres in and around Chittoor and other neighbouring districts of southern Andhra Pradesh, India, for sending the clinically suspected scrub typhus patients’ blood samples to SVIMS. The authors also thank Prof. K.V. S. Sharma, Department of Statistics, SVU for performing statistical analysis for the data.
Footnotes
Conflicts of Interest: None.
References
- 1.Kelly DJ, Fuerst PA, Ching WM, Richards AL. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin Infect Dis. 2009;48(Suppl 3):S203–30. doi: 10.1086/596576. [DOI] [PubMed] [Google Scholar]
- 2.Tantibhedhyangkul W, Prachason T, Waywa D, El Filali A, Ghigo E, Thongnoppakhun W, et al. Orientia tsutsugamushi stimulates an original gene expression program in monocytes: relationship with gene expression in patients with scrub typhus. PLoS Negl Trop Dis. 2011;5:e1028. doi: 10.1371/journal.pntd.0001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxena A, Khiangte B, Tiewsoh I. Scrub typhus complicated by acute respiratory distress syndrome and multiorgan failure; an unrecognized alarming entity in central India: a report of two cases. J Family Med Prim Care. 2014;3:80–3. doi: 10.4103/2249-4863.130334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebisawa II. Current epidemiology and treatment of tsutsugamushi disease in Japan. J Travel Med. 1995;2:218–20. doi: 10.1111/j.1708-8305.1995.tb00662.x. [DOI] [PubMed] [Google Scholar]
- 5.Shishido A. Identification and serological classification of the causative agent of scrub typhus in Japan. Jpn J Sci Biol. 1962;15:308–21. [Google Scholar]
- 6.Yamamoto S, Kawabata N, Tamura A, Urakami H, Ohashi N, Murata M, et al. Immunological properties of Rickettsia tsutsugamushi, Kawasaki strain, isolated from a patient in Kyushu. Microbiol Immunol. 1986;30:611–20. doi: 10.1111/j.1348-0421.1986.tb02988.x. [DOI] [PubMed] [Google Scholar]
- 7.Ohashi N, Tamura A, Sakurai H, Yamamoto S. Characterization of a new antigenic type, Kuroki, of Rickettsia tsutsugamushi isolated from a patient in Japan. J Clin Microbiol. 1990;28:2111–3. doi: 10.1128/jcm.28.9.2111-2113.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura A, Takahashi K, Tsuruhara T, Urakami H, Miyamura S, Sekikawa H, et al. Isolation of Rickettsia tsutsugamushi antigenically different from Kato, Karp, and Gilliam strains from patients. Microbiol Immunol. 1984;28:873–82. doi: 10.1111/j.1348-0421.1984.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 9.Ohashi N, Koyama Y, Urakami H, Fukuhara M, Tamura A, Kawamori F, et al. Demonstration of antigenic and genotypic variation in Orientia tsutsugamushi which were isolated in Japan, and their classification into type and subtype. Microbiol Immunol. 1996;40:627–38. doi: 10.1111/j.1348-0421.1996.tb01120.x. [DOI] [PubMed] [Google Scholar]
- 10.Chang WH. Current status of tsutsugamushi disease in Korea. J Korean Med Sci. 1995;10:227–38. doi: 10.3346/jkms.1995.10.4.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurung S, Pradhan J, Bhutia PY. Outbreak of scrub typhus in the North East Himalayan region-Sikkim: an emerging threat. Indian J Med Microbiol. 2013;31:72–4. doi: 10.4103/0255-0857.108729. [DOI] [PubMed] [Google Scholar]
- 12.Varghese GM, Abraham OC, Mathai D, Thomas K, Aaron R, Kavitha ML, et al. Scrub typhus among hospitalised patients with febrile illness in South India: magnitude and clinical predictors. J Infect. 2006;52:56–60. doi: 10.1016/j.jinf.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Bakshi D, Singhal P, Mahajan SK, Subramaniam P, Tuteja U, Batra HV. Development of a real-time PCR assay for the diagnosis of scrub typhus cases in India and evidence of the prevalence of new genotype of O. tsutsugamushi. Acta Trop. 2007;104:63–71. doi: 10.1016/j.actatropica.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 14.Kumar V, Kumar V, Yadav AK, Iyengar S, Bhalla A, Sharma N, et al. Scrub typhus is an under-recognized cause of acute febrile illness with acute kidney injury in India. PLoS Negl Trop Dis. 2014;8:e2605. doi: 10.1371/journal.pntd.0002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Premaratna R, Loftis AD, Chandrasena TGAN, Dasch GA, de Silva HJ. Rickettsial infections and their clinical presentations in the Western Province of Sri Lanka: a hospital-based study. Int J Infect Dis. 2008;12:198–202. doi: 10.1016/j.ijid.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Furuya Y, Yoshida Y, Katayama T, Yamamoto S, Kawamura A., Jr Serotype-specific amplification of Rickettsia tsutsugamushi DNA by nested polymerase chain reaction. J Clin Microbiol. 1993;31:1637–40. doi: 10.1128/jcm.31.6.1637-1640.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1997;74:5463–7. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawamura A, Tanaka H, Tamura A. R. Tsutsugamushi infection and pregnancy. In: Kawamura A, Tanaka H, Tamura A, editors. Tsutsugamushi disease. Tokyo, Japan: University of Tokyo Press; 1995. [Google Scholar]
- 19.Watt G, Chouriyagune C, Ruangweerayud R, Watcharapichat P, Phulsuksombati D, Jongsakul K, et al. Scrub typhus infections poorly responsive to antibiotics in Northern Thailand. Lancet. 1996;348:86–9. doi: 10.1016/s0140-6736(96)02501-9. [DOI] [PubMed] [Google Scholar]
- 20.Kim DM, Yun NR, Neupane GP, Shin SH, Ryu SY, Yoon HJ, et al. Differences in clinical features according to Boryoung and Karp genotypes of Orientia tsutsugamushi. PLoS One. 2011;6:e22731. doi: 10.1371/journal.pone.0022731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayen JJ, Pond HS, Forrester JS, Wood FC. Scrub typhus in Assam and Burma; a clinical study of 616 cases. Medicine (Baltimore) 1946;25:155–214. doi: 10.1097/00005792-194605000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Kim YJ, Yeo SJ, Park SJ, Woo YJ, Kim MW, Kim SH, et al. Improvement of the diagnostic sensitivity of scrub typhus using a mixture of recombinant antigens derived from Orientia tsutsugamushi serotypes. J Korean Med Sci. 2013;28:672–9. doi: 10.3346/jkms.2013.28.5.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dasch GA, Halle S, Bourgeois AL. Sensitive microplate enzyme-linked immunosorbent assay for detection of antibodies against the scrub typhus rickettsia, Rickettsia tsutsugamushi. J Clin Microbiol. 1979;9:38–48. doi: 10.1128/jcm.9.1.38-48.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Land MV, Ching WM, Dasch GA, Zhang Z, Kelly DJ, Graves SR, et al. Evaluation of a commercially available recombinant-protein enzyme-linked immunosorbent assay for detection of antibodies produced in scrub typhus rickettsial infections. J Clin Microbiol. 2000;38:2701–5. doi: 10.1128/jcm.38.7.2701-2705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furuya Y, Yoshida Y, Katayama T, Kawamori F, Yamamoto S, Ohashi N, et al. Specific amplification of Rickettsia tsutsugamushi DNA from clinical specimens by polymerase chain reaction. J Clin Microbiol. 1991;29:2628–30. doi: 10.1128/jcm.29.11.2628-2630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enatsu T, Urakami H, Tamura A. Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologies of 56-kDa type-specific antigen genes. FEMS Microbiol Lett. 1999;180:163–9. doi: 10.1111/j.1574-6968.1999.tb08791.x. [DOI] [PubMed] [Google Scholar]
- 27.Ohashi N, Nashimoto H, Ikeda H, Tamura A. Diversity of immunodominant 56-kDa type-specific antigen (TSA) of Rickettsia tsutsugamushi. Sequence and comparative analyses of the genes encoding TSA homologues from four antigenic variants. J Biol Chem. 1992;267:12728–35. [PubMed] [Google Scholar]
- 28.Shirai A, Coolbaugh JC, Gan E, Chan TC, Huxsoll DL, Groves MG. Serologic analysis of scrub typhus isolates from the Pescadores and Philippine Islands. Jpn J Med Sci Biol. 1982;35:255–9. doi: 10.7883/yoken1952.35.255. [DOI] [PubMed] [Google Scholar]
- 29.Choi MS, Chang WJ, Park SK, Huh MS, Kim HR, Han TH, et al. Seroepidemiological survey of scrub typhus in Korea, 1995-1996. J Korean Soc Microbiol. 1997;32:219–26. [Google Scholar]
- 30.Manosroi J, Chutipongvivate S, Auwanit W, Manosroi A. Determination and geographic distribution of Orientia tsutsugamushi serotypes in Thailand by nested polymerase chain reaction. Diagn Microbiol Infect Dis. 2006;55:185–90. doi: 10.1016/j.diagmicrobio.2006.01.014. [DOI] [PubMed] [Google Scholar]


