Abstract
A number of transcription factor proteins contain domains that are fully or partially unstructured. The means by which such proteins acquire naturally folded conformations are not well understood. When they encounter their proper binding partner(s), several of these proteins adopt a folded conformation through an induced-fit mechanism. The glucocorticoid receptor (GR) is a ligand-activated transcription factor. Expressed independently as a recombinant peptide, the N-terminal transactivation domain (AF1) of the GR shows little structure and appears to exist as a collection of random coil configurations. The GR AF1 is known to interact with other transcription factors, including a critical component of the general transcription machinery proteins, the TATA box binding protein (TBP). We tested whether this interaction can lead to acquisition of structure in the GR AF1. Our results show that recombinant GR AF1 acquires a significant amount of helical content when it interacts with TBP. These structural changes were monitored by Fourier transform infrared and NMR spectroscopies, and by proteolytic digestions. Our results support a model in which TBP binding interaction with the GR AF1 induces significantly greater helical structure in the AF1 domain. This increased helical content in the GR AF1 appears to come mostly at the expense of random coil conformation. These results are in accordance with the hypothesis that an induced-fit mechanism gives structure to the GR AF1 when it encounters TBP.
Keywords: glucocorticoid receptor, N-terminal activation function, coregulatory protein, protein folding
The glucocorticoid receptor (GR) is a ligand-activated transcription factor with the domain structural arrangement typical of the nuclear hormone receptors superfamily (1–4). The GR regulates transcription of target genes by binding DNA at specific hormone response elements and/or by interacting with other transcription factors (5–8). Although the structural organization of steroid receptors into N-terminal domain (NTD), DNA binding domain (DBD), and ligand binding domain is well characterized (9–14), precisely how transcription is regulated by them is largely unknown. For all steroid receptors, this is in part due to the lack of information about their transcription activating domain AF1, located in the NTD (9, 10). The GR AF1 sequence resembles those of acidic transactivation domains of several transcription factors (15, 16). When expressed independently, the GR AF1 shows little structure and seems to exist as an ensemble of largely unstructured conformers (17, 18). The GR AF1 is known to interact with other transcription factors, and conditional folding has been suggested to be the key for these interactions (19–22).
It has been reported that in the presence of trifluoroethanol, the “core” of AF1 (AF1c, amino acids 187–242), located toward the C-terminal end of AF1, adopts three helical segments (17). Independent experiments have shown that substitution of the α-helix-breaking amino acid proline for natural residues at critical positions in these putative helices significantly reduces the transcriptional potency of the GR (17). Other experiments have shown that mutations of the hydrophobic amino acids in AF1 reduce both its interactions with other coregulatory proteins and its transactivation function (21). We recently demonstrated that the recombinant GR AF1 adopts a native-like folded conformation when incubated in the presence of the naturally occurring osmolyte, trimethylamine N-oxide, and in this folded conformation it strongly interacts with certain specific coregulatory proteins. These include the TATA box binding protein (TBP), the CREB binding protein, and steroid receptor coactivator 1 (22). Taken together, the data strongly suggest that proper folding of AF1 is an important requirement for its actions in regulating transcription. How AF1 acquires this structure under physiological conditions is still an unanswered question. Three major hypotheses have been proposed: (i) that in AF1, an acidic cluster of amino acids without highly defined structure interacts with proteins of the transcription machinery; (ii) that an induced fit process structures AF1 when it encounters specific binding partners; and (iii) that intramolecular signals, such as those coming from posttranscriptional modifications, interdomain interactions, or binding to its DNA, glucocorticoid response element, induce structure in AF1. Mutational analysis does not support the role of acidic clusters (23). Models ii and iii may not be mutually exclusive, and we have shown previously that binding to a glucocorticoid response element induces a folded configuration in the GR NTD (24). Recent studies have shown that several transactivation domains undergo a transition to a folded state upon interaction with coregulatory proteins, which included some steroid receptors (25–28). The question remaining is, does induced fit occur when the AF1 domain encounters a proper binding partner protein?
It is interesting that the GR AF1 can interact directly with TBP (22, 25), the critical protein that forms the basis for the multiprotein transcription initiation complex. This direct interaction between AF1 and TBP is not part of currently popular models for transcriptional regulation by the GR (29). These models instead focus on indirect actions of the GR, through intermediary proteins, to affect the accumulation of the proteins of the primary transcription complex (30). Direct GR AF1:TBP interaction raises the possibility that the GR AF1 domain somehow directly influences the primary transcription machinery. In vitro transcription studies indicated that the holo-GR acts to stabilize the preinitiation complex (31, 32). One possibility is that a structured conformation of AF1, induced or stabilized by its interaction with TBP, is involved in the platform for TBP-associated factors. TBP:steroid receptor interaction is not limited to the GR. A recent study has shown that the intrinsically unstructured NTD of the ERα interacts with the TBP and that this interaction leads to stabilization of structure in the estrogen receptor α (ER-α) NTD (28), which contains an AF1 region. However, it is not clear whether these structural changes specifically involve the mapped AF1 region in the ER-α NTD. Data are not available on TBP:NTD interaction for other steroid receptors. In terms of primary sequence homology, the steroid receptors' NTDs are poorly conserved, yet they all possess a potent transactivation region, and where studied they all show little or no structure in aqueous solutions. It is therefore important to find out whether TBP binding can induce structure specifically in an AF1. Because TBP is a specific binding partner for the GR AF1, in this study we tested whether a direct TBP binding to the otherwise mostly unstructured recombinant GR AF1 domain induces more compact structure in the GR AF1. Structural changes were monitored by Fourier transform infrared (FTIR) and NMR spectroscopies, and by proteolytic digestions.
Materials and Methods
Protein Expression and Purification. The GR AF1 domain was constructed from human GR cDNA digested with BglII and inserted into an expression vector pGEX-4T-1 (Amersham Pharmacia Biotech). The expression and purification of AF1 protein is described in refs. 18 and 22. The pET11d.TBP expression vector encoding His-TBP was kindly provided by Robert Roeder (The Rockefeller University, New York). The recombinant expression plasmid pET11d.TBP was selected and transformed into Escherichia coli BL21(DE3). The bacteria containing the recombinant vector for His-TBP were induced with isopropyl-β-d-thiogalactopyranoside at 1 mM concentration for 3 h, lysed, and extracted. Recombinant His-TBP was purified on a Ni-NTA column. The purity of protein was analyzed on SDS/PAGE gel by Coomassie blue staining. Centriprep (Amicon) filters were used to concentrate protein samples.
FTIR Spectroscopy. FTIR spectra were recorded with an ABB Bomem (Quebec City, QC, Canada) MB Series FTIR spectrometer equipped with a dTGS detector and purged constantly with dry air generated by a Balston (Haverhill, MA) air dryer. Protein samples (100 μM) were loaded in a liquid IR cell (ABB Bomem) with CaF2 windows and a 6-μm path length. For each spectrum, a 200-scan interferogram was collected at single-beam mode with a 4-cm–1 resolution. Reference spectra were recorded under identical scanning conditions with only the corresponding buffer in the cell. Protein spectra were obtained according to previously established criteria and a subtraction procedure (33). The residual water vapor signals, if present, in the spectrum of protein were removed by subtracting the spectrum of gaseous water. This removal was confirmed by examination of the spectra between 1,700 and 1,800 cm–1. Second-derivative spectra were obtained with a seven-point Savitsky–Golay derivative function. All second-derivative spectra were baseline-corrected and area-normalized as described in ref. 34. Good signal-to-noise ratios were obtained. The secondary-structure content of the protein was determined by curve-fitting analysis of the inverted second-derivative spectrum from infrared second-derivative amide 1 spectra as described in ref. 35.
Expression and Purification of 15N-AF1 for NMR Studies. An overnight culture of E. coli strain BL21 containing the pGX-4T-AF1 construct was grown in LB broth supplemented with 5 g/liter glucose and 0.10 g/liter ampicillin. The overnight culture was diluted 1:100 in a previously described minimal media containing 15NH4Cl (36) and grown at 33°C with vigorous shaking to an A600 of 0.35–0.45. Isopropyl β-d-thiogalactoside added to a final concentration of 1 mM, and the culture was shaken at 33°C for 3 h. The cells were collected by centrifugation at 4°C and frozen at –80°C. 15N-labeled AF1 and unlabeled TBP proteins were purified as described above.
NMR Spectroscopy. All NMR spectra were collected at 25°C on Varian Unity Plus 750- and 600-MHz instruments with 5 mM triple-resonance probes equipped with actively shielded pulsed field gradients. Quadrature detection in the indirectly detected dimensions was achieved by using the States-TPPI schemes (37). The sensitivity-enhanced pulsed field gradient approach was used for the collection of the 1H-15N heteronuclear sequential quantum correlation (HSQC) spectra (38) and all HSQC-based triple-resonance experiments (39).
CD Spectroscopy. The CD spectra of the purified recombinant AF1, TBPC, and AF1:TBPC mixtures were recorded at 22°C on an Aviv 62 spectropolarimeter by using a 1.0-cm quartz cell, with a bandwidth of 1.0 nm and a scan step of 0.5 nm. The spectra were recorded at a protein concentration of 45 μM and were corrected for the contribution of solute concentrations. Each spectrum is a result of five spectra accumulated, averaged, and smoothed.
Limited Proteolytic Digestion. Three sets of purified proteins (AF1, TBP, and AF1:TBP mixture) were digested by using trypsin or chymotrypsin (Promega). The ratios of AF1:TBP complex are indicated in the figure legend. In another set, a nucleotide containing TATA box 5′-gctataaaagggca-3′ and 5′-tgcccttttatagc-3′ was added in the mixture of AF1:TBP to form a AF1:TBP:TATA complex (1:1:1). Digestions were carried out at 4°C by using a protein:enzyme mass ratio of 100:1. Reactions were terminated by adding SDS loading buffer and placing the sample tubes in boiling water. The proteolytic digestion products were resolved on an SDS/PAGE gel followed by either Coomassie blue R-250 staining or immunoblotting with an antibody for AF1.
Results
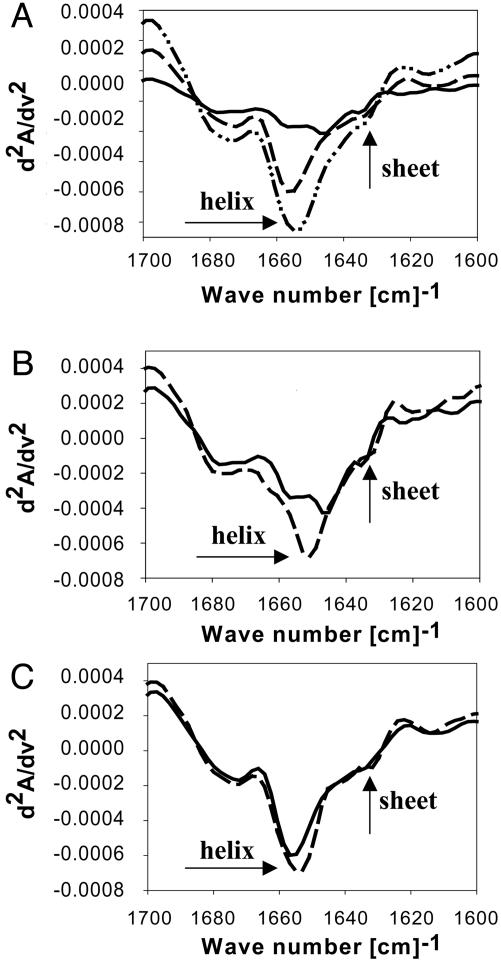
FTIR Spectra Indicate Formation of Structure in the GR AF1 When It Encounters TBP. A topological diagram of the GR showing its major functional domains is presented in Fig. 1A. Three sets of purified recombinant protein samples (Fig. 1B), AF1, TBP, and a 1:1 mixture of AF1:TBP were compared in FTIR experiments. To obtain information regarding the nature of the secondary structural elements in the proteins, we plotted the data as second-derivative amide I spectra (Fig. 2A). All spectra exhibited four basic band components that on the basis of previous infrared studies of proteins in aqueous solution (40) can be assigned to α-helix (≈1,656 cm–1), β-sheet (≈1,635 cm–1), and β-turns (1,685 and 1,675 cm–1). An additional band component at 1,647 cm–1, ascribable to random structure, also was revealed by the curve-fitting procedure. Compared with the individual proteins' spectra, the spectrum of the AF1:TBP mixture (Fig. 2 A) shows that the peak around 1,656 cm–1 is significantly increased, suggesting that the complex has more helical content than either AF1 or TBP alone (Fig. 2 A). Consistent with this interpretation, simple summation of the second-derivative spectrum of AF1 and TBP showed that the AF1:TBP mixture has a slightly higher peak around 1,656 cm–1 (data not shown). A more careful comparison was made by mathematically extracting the contribution of each individual protein from the mixture spectrum. A comparison of the second-derivative spectrum of AF1 with that of the mixture after the contribution of TBP had been removed indicated a significant increase in the helical content in AF1 when mixed with TBP, as shown by the ≈1,656-cm–1 peak (Fig. 2B), suggesting that TBP interaction with AF1 gives more helical structure to the AF1 domain. Similar plots of TBP spectra with and without AF1 complexed showed that only a slight difference is found in the secondary structural elements of TBP, whether or not it is complexed with AF1 (Fig. 2C), suggesting that most of the structural changes taking place are, in fact, happening in the AF1 domain. Quantitative estimates of the secondary structural elements calculated from these spectra are summarized in Table 1. It is evident from Table 1 that the helical content in AF1 after binding to TBP is increased by >50%. This increased helical content comes at the expense of random coil conformation.
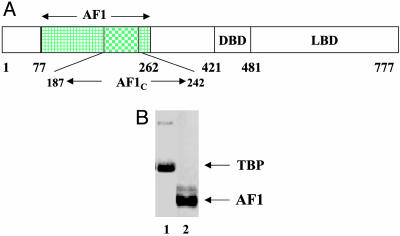
Fig. 1.
Proteins studied in this work. (A) Topological diagram of the human GR showing its major functional domains. The highlighted region represents the N-terminal activation domain AF1. LBD, ligand binding domain. (B) Coomassie blue-stained SDS/PAGE of 5 μg each of purified TBP (lane 1) and GR AF1 (lane 2) polypeptides.
Fig. 2.
Second derivative FTIR spectra showing that binding of TBP to AF1 enhances helical content in AF1. (A)—,AF1;–––, TBP; – ·· –, AF1:TBP complex. (B)—,AF1;–––, AF1:TBP complex after subtracting the contribution of TBP. (C)—,TBP;–––, AF1:TBP complex after subtracting the contribution of AF1. The spectra shown are one of the two independent experiments each carried out on an independent preparation of TBP and AF1.
Table 1. A summary of quantitative estimation of secondary structural elements of the GR AF1 and TBP before and after complex formation.
| Spectrum | Helix, % | Sheet, % | Bend, % | Turn, % | Coil, % |
|---|---|---|---|---|---|
| AF1 | 26.7 | 11.6 | 10.0 | 12.3 | 39.4 |
| AF1:TBP-TBP | 42.1 | 11.8 | 9.3 | 12.4 | 24.4 |
| TBP | 41.3 | 16.1 | 9.4 | 12.4 | 20.8 |
| AF1:TBP-AF1 | 41.0 | 16.4 | 9.6 | 12.4 | 20.6 |
The data are calculated from second-derivative FTIR spectra. AF1:TBP-TBP indicates the secondary structures attributable to AF1 in the AF1:TBP complex, after removal of the contributions of TBP; and AF1:TBP-AF1 indicates the structures of TBP in the complex after removal of the contribution of AF1.
The data in Fig. 2 also show that binding of TBP to AF1 shifts the α-helix and β-sheet bands to lower wave numbers (compare the minima around 1,656 cm–1 in Fig. 2B). These TBP-induced changes in the peak wave number of the α-helix and β-sheet bands in AF1 (Fig. 2B) indicate conformational changes at tertiary structural levels, consequent on an environmental alteration for the α-helix and β-sheet structures. Because the vibrational energy of carbonyl stretching is inversely related to the strength of hydrogen bonding, the downshift in the α-helix and β-sheet band wave numbers induced by binding TBP implies a strengthening in hydrogen bonding within α-helices and between β-strands (41). These data are further evidence for a more compact structure being formed in the AF1 domain due to AF1:TBP interaction.
To test for nonspecific protein interactions, in another set of experiments, we recorded the FTIR spectra of AF1, a non-AF1-interacting protein, glutathione S-transferase (GST), and a mixture of AF1:GST. The results demonstrate that structural changes observed in AF1 are due to specific binding with TBP. As expected, there were no significant conformational changes observed in AF1 with GST present (data not shown). These results confirm that the helical increase observed in AF1 in the presence of TBP is not due to the presence of another protein in the mixture but is a result of TBP binding.
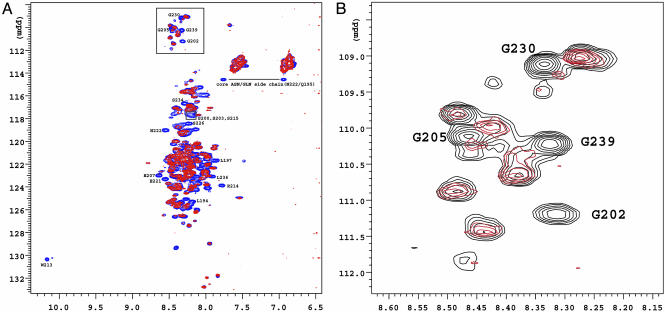
NMR HSQC Spectra Show Structural Alterations in AF1 Conformation After TBP Binding. We carried out NMR experiments to determine by a different method the effect of TBP interaction on the structure of the AF1 domain. NMR studies were performed on both full-length AF1 and the 60-aa AF1c. This “core” of AF1 contains the regions of key putative helices and is capable of mediating transcriptional activation to transfected promoter–reporter constructs (17). The lack of resonance dispersion in the 1H-15N HSQC spectra of both AF1 (Fig. 3A) and AF1c (data not shown) unbound to TBP suggest that both of these proteins are mostly unstructured. This conclusion is supported by the random coil-like chemical shifts observed for all protons and carbons of the AF1c domain (data not shown). We have assigned nearly every amino acid in the AF1c, and the corresponding assigned peaks are also present in the 1H-15N HSQC spectra of the AF1, shown in Fig. 3. For example, in the box near the top of Fig. 3A, Gly-202, -205, -230, and -239 assigned in the AF1 are based on the amino acid assignments from AF1c. We further recorded 1H-15N HSQC spectra of the GR AF1 bound to unlabeled TBP to test whether TBP binding can bring structural changes in AF1, as was seen by FTIR experiments. Addition of TBP creates significant chemical shift perturbations for certain residues in AF1 (Fig. 3A, compare peaks in blue and red), and this is easily observed for three (Gly-202, -230, and -239) of the four Gly residues within AF1c (Fig. 3B). Interestingly, these Gly residues are present in the region of AF1 that is predicted to have propensity to form helical segments. Helicity appears to be important for the GR AF1's transcriptional activation activity (17). The chemical-shift changes in AF1 that we document in the AF1:TBP mixture are consistent with the formation of helical regions in AF1 after the AF1:TBP complex is formed. Notable changes are observed for several correlations in the 1H-15N HSQC spectra of the GR AF1, when compared with and without TBP bound (Fig. 3A). Again, these changes are readily seen in the area containing the random coil Gly residues of unbound AF1. Although most of the AF1 Gly residues are unaffected by TBP, several Gly peaks are not present in the HSQC spectrum taken in the presence of TBP (Fig. 3B). These peaks may have disappeared because of exchange-broadening, or they may have moved into the large clump of resonances in the center of the spectrum. In either event, several of the Gly residues are differentially affected by the addition of TBP. This is also true of the lone Trp residue (W213) in AF1, which disappears upon addition of TBP (Fig. 3A). Like the Gly residues, the lone Trp affected by TBP binding is also found near a putative helix in the AF1c region (17). These NMR results are in agreement with the FTIR data (Fig. 2 and Table 1), indicating that TBP binding brings structural changes in AF1.
Fig. 3.
1H-15N HSQC NMR spectra. (A) 15N-labeled GR AF1 with (red) and without (blue) TBP bound. The numbering of amino acid residues indicates the positions of amino acids in the AF1c.(B) Expanded view of Gly residues (boxed in A) of 15N-labeled GR AF1 in the absence (black) or presence (red) of TBP.
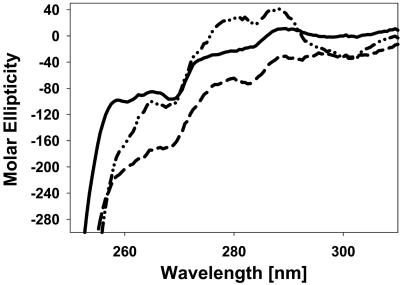
Near-UV CD Spectra Support the Conformational Changes in the AF1C Region After AF1:TBP Interaction. To validate NMR results showing conformational changes in the AF1C region after AF1:TBP binding, we recorded near-UV CD spectra of AF1 with and without TBP bound. In the AF1 there is only one Trp residue, located in the AF1C region. Approximately 180 aa toward the C terminus of the TBP (TBPC) are mapped to be interacting with the AF1 (21). The amino acid sequence of this TBPC peptide does not possess any Trp residues. Therefore, we used this peptide to follow whether Trp residue W213 in AF1, which we found in our HSQC NMR spectra to be moving to more helical conformation (Fig. 3A), shows perturbation due to AF1:TBP interaction. A comparison of near-UV CD spectra of AF1, TBPC, and AF1:TBPC complex shows that W213 in AF1 indeed moves to a more compact structure in AF1, as assessed by enhanced peak intensity around a 290-nm wavelength in the complex compared with the spectra of either AF1 or TBPC alone (Fig. 4). Because there is no Trp residue in TBPC, this conformational change is arising from the lone Trp residue in AF1C. These results further support our NMR data.
Fig. 4.
Near-UV CD spectra showing perturbation of W213 in the AF1 domain after AF1:TBP interaction. —, AF1;–––,TBPC;– ·· –, AF1:TBPC.
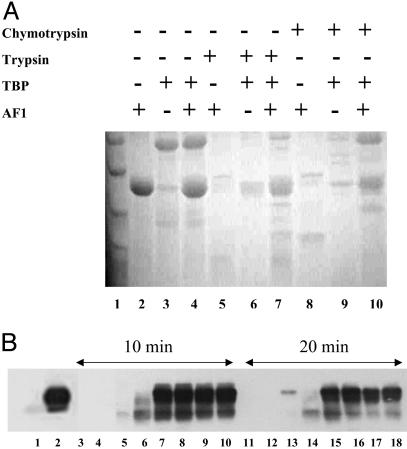
The Structure Induced by AF1:TBP Interaction Resists Proteolysis. To evaluate the changes in the tertiary structure of the AF1 region brought about by TBP binding, we carried out limited proteolytic digestions. The patterns of proteolytic products of AF1, TBP, and AF1:TBP mixture after digestion by trypsin or chymotrypsin are shown in Fig. 5. It is evident from the Coomassie blue-stained gel (Fig. 5A) that, alone, AF1 and TBP are mostly digested under these conditions of proteolysis (compare lanes 2 and 3 with lanes 5 and 6), whereas they are partially protected in the mixture (compare lane 4 with lane 7), suggesting that a more compact tertiary structure is formed in the complex, such that the residues attacked by trypsin are moved to positions not easily reached by them. Similar protection from chymotrypsin is shown in lanes 8–10.
Fig. 5.
Limited proteolysis of AF1, TBP, and the AF1:TBP complex. (A) Coomassie blue-stained gel showing product of proteolytic digestion after 15 min. Lanes: 1, molecular weight markers; 2–4, undigested AF1, TBP, and AF1:TBP mixture, respectively; 5–7, trypsin-digested AF1, TBP, and AF1:TBP mixture, respectively; 8–10, chymotrypsin-digested AF1, TBP, and AF1:TBP complex, respectively. (B) Immunoreactions with an antibody raised against amino acids 150–175 in the human GR AF1 showing products of trypsin digestion. Lanes: 1, undigested TBP; 2, undigested AF1; 3–10, digested with trypsin for 10 min; 11–18, digested for 20 min at 4°C; 3, TBP; 4, AF1; 5–9 and 13–17, AF1:TBP mixture at a ratio of 1:0.25, 1:0.5, 1:1, 1:1.5, and 1:2, respectively; 10 and 18, AF1:TBP:TATA (1:1:1) mixture. TATA represents a double-stranded oligonucleotide containing TATA box sequences.
To further confirm these proteolytic data, we followed tryptic digests immunochemically. Tryptic digestions were carried out by using AF1:TBP mixture at various ratios of AF1:TBP in the presence or absence of an oligonucleotide containing a TATA box sequence. We fixed the AF1 protein concentration and added various amounts of TBP to make AF1:TBP ratios of 1:0.25, 1:0.5, 1:1, 1:1.5, and 1:2. These samples were then digested with trypsin for 10 or 20 min at 4°C. After the digestion, products of proteolysis were resolved on an SDS/PAGE gel and examined by immunoreaction with an anti-AF1 antibody. It is evident from Fig. 5B that when digested, AF1 alone (lanes 4 and 12) does not show any protected band that can be recognized with AF1 antibody, whereas when AF1 is complexed with TBP, protected AF1 bands can be seen, and this pattern of protection depends on the ratio of AF1:TBP until it plateaus at 1:1 (lanes 5–10 and 13–18). There is no cross-reaction with the antibody used in the lanes (1, 3, and 11) where there is only TBP present. Also, no changes are observed in the intensity of bands between AF1:TBP and AF1:TBP:TATA (compare lane 7 with lane 10 and lane 15 with lane 18), suggesting that binding of TBP to a TATA box affects neither binding of TBP to AF1 nor TBP binding-induced AF1 folding. A comparison of band intensities at 10 and 20 min of tryptic digestion showed a decrease in the intensity of the bands after 20 min compared with those after 10 min, suggesting that, over time, the enzyme is able to reach some of the protected sites in AF1.
These observations suggest that AF1 folds to a protease-resistant shape and that it remains in this conformation as long as TBP is present. It is evident from these results that the TBP-induced structure in AF1 is not confined to secondary structural elements but that tertiary structural changes are also taking place in the protein. The proteolytic protection observed for TBP suggests as well that AF1:TBP interaction protects tryptic sites in TBP.
Discussion
Significant regions of many proteins contain sequences that do not automatically fold into their fully condensed, functional structures, even in the presence of chaperone proteins. The high proportion of these sequences in the proteomes of all organisms argues for important, as-yet-unknown functions. Characterization of the conformational propensities and function of such non-globular protein sequences represents a major challenge. Strikingly, among proteins with such “natively” unfolded regions are many transcription factors, including steroid receptors (42). The primary amino acid sequences of the NTDs of the steroid receptors, which contain transactivation function region AF1, are much less conserved than are DBD and ligand binding domain regions, and several studies have shown that the isolated NTDs are not fully structured in aqueous solution (17, 18, 27, 28). Although the isolated NTDs of steroid receptors have poor sequence homology, in solution they share the characteristic of disordered structure. By investigating one such segment in detail, i.e., the GR AF1, we hope to gain insights relevant to a broad range of proteins engaged in transcription regulation.
Such transcription factors almost always work in conjunction with other proteins, and by multiple mechanisms, coregulatory proteins influence or modulate the transcriptional activity of the GR. Coregulators may link the GR with the basal transcriptional machinery, may link the GR with intervening regulators, and/or may covalently modify chromatin. Coregulatory proteins known to interact with the GR are steroid receptor coactivator 1, TIF2, CREB binding protein/p300, and RIP140. The GR also interacts with TBP, part of the basal transcriptional machinery (30). The GR AF1 domain is known to play an important role in many of these interactions. The ability of GR AF1 to interact with components of the general transcriptional machinery or with coregulator complexes provides a broad insight into the process of transcriptional initiation. However, it remains to be determined exactly how these proteins interact with the GR AF1.
In many cases, the unfolded or partially folded regions of proteins take full shape when the protein interacts with its proper binding partner(s), the molecules to which it must bind to carry out its function (42). Applied to the GR, this induced fit model of folding hypothesizes that AF1 is not fully structured in vivo until it binds one or another key partner molecule (17, 43). We hypothesize that an induced conformation or limited set of conformations occurs in AF1 in order for it to carry out its transcription function. Formally, induced fit could occur by initial nonspecific interactions between the unfolded AF1 domain and the binding partner. In this version of the model, when such interactions occur, the proximity of the two proteins leads to rapid acquisition of proper structure in AF1. Alternatively, initially there could be more specific interactions of binding partner with a partially folded AF1, or even with a tiny pool of fully folded AF1 molecules creating a kinetic “sink” into which the general population falls (discussed in ref. 43).
TBP has a central role in the basal transcription machinery and is involved in binding to a number of transcriptional activators (21, 22, 25, 28). These multiprotein interactions may help to efficiently recruit TBP to the TATA box. We and others have previously shown that the GR AF1 is able to directly interact with TBP in vitro (19, 22). TBP has been shown to interact also with the ERα NTD, which contains several AF regions (28), but the precise effects in any of these AFs were not determined. In this paper we show that complex formation between the GR AF1, specifically, and TBP is accompanied by a change in protein conformation. The most likely cause of this observed effect is that the highly structured TBP induces a folding event in otherwise mostly unstructured AF1. We note that this effect does not require DNA. We have shown earlier that GR DBD:glucocorticoid response element binding causes structure to form in AF1 (24). We propose that this conformation and that resulting from AF1:TBP binding are similar. Thus, the approximation of GR to TBP after glucocorticoid response element binding may facilitate AF1:TBP interaction, depending on the geometry of the promoter involved. GR tethered to DNA via a heterologous transcription factor could also approximate GR AF1 and TBP to enhance their interaction.
Our structural data from FTIR, NMR, and proteolytic digestion experiments clearly reconfirm that the GR AF1 has but little structure in aqueous solution (Table 1). Binding with TBP leads to imposition of greater structure in the AF1 domain, an example of an induced fit mechanism. Thus, the binding of TBP to an otherwise not fully structured AF1 domain is not a simple tethering of the two molecules, but rather an important step toward giving a folded functional structure to the GR AF1 domain. An induced fit mechanism has been reported also for the activation domain of c-Myc specifically through interaction with TBP (21). Similar results have been reported for an interaction between a steroid receptor AF domain (AR AF1) and a coregulatory protein (27, 44). Although limited, these data indicate that the activation domains of these transcription factors may be adopting a folded functional conformation under physiological conditions through interaction with coregulatory proteins of the transcriptional machinery complex. In the context of full-length steroid receptor, AF1 structure may be influenced as well by interdomain signaling. For the glucocorticoid and progesterone receptors, data support induced conformational stabilization of their AF1 domains through inherent interdomain influences (24, 43). Beyond this inherent structural effect, we have shown also that binding the DBD of the GR to its cognate DNA response element causes structure to form in the GR NTD (24). Others have shown DNA binding effects on NTD/AF1 structure of the progesterone receptor (45). It has been proposed that DNA sequence of the various response elements found in specific genes may influence the fold and therefore the specific actions of AF1 (46, 47). We hypothesize that in the holoreceptor, under physiological conditions, AF1 exists as a partially folded structure due to interdomain influences. Each receptor's DNA binding site interaction leads to further acquisition of AF1 structure, with specific variations in DNA sequence providing different signals. The resulting structurally modified forms of AF1 are well suited for its varied interactions with other critical coregulatory proteins, essential for gene regulation by the receptor. These interactions give the final folded structure to AF1 and the basis for the multiprotein assemblies involved in steroid receptor-mediated regulation of transcription.
Our data suggest that the assembly of GR AF1:binding partner complexes are an essential step in realizing AF1's properly folded, functioning structure. It remains to be found what kind of functional structure(s) AF1 adopts. If we assume that the folded form of AF1 caused by osmolyte, DNA binding, and TBP binding are similar, it is possible to begin to predict the general structure of this form. From the location of the sites available to proteolytic enzymes (22), one sees that amino acids 77 to ≈215 are folded. The infolded W213 and the S211 are just within the fold. Several enzymes demonstrate cuts just beyond it. NMR results herein show that G202 is well within this folded region. NMR also suggests that G230 and G234 are in more a ordered region. Finally, we repeatedly have demonstrated increased helical content upon folding. The predicted helices in AF1 all fall within the subdomain protected from proteases. The overall picture, thus, is one of a helix-based folded AF1 with areas exposed to water around amino acids 130 and 215–220. The TBP binding surface is formed by the fold of amino acids 187–244 (unpublished results). Our studies may provide a basis for future structural studies to determine the three-dimensional structure of the AF1 domain.
Acknowledgments
We thank Dr. Bo Xu and the staff of the University of Texas Medical Branch Protein Core Laboratory for help in overexpressing and purifying the AF1 protein, and Emily F. Welch and Justin Serrette for excellent technical assistance. The TBP construct was kindly provided by Dr. Robert Roeder, and the TBPC construct was a kind gift from Dr. Z. Sean Jou (Yale University, New Haven, CT). This work was supported by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grant 1RO1 DK58829 (to R.K. and E.B.T.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DBD, DNA binding domain; FTIR, Fourier transform infrared; GR, glucocorticoid receptor; NTD, N-terminal domain; TBP, TATA box binding protein.
References
- 1.Evans, R. M. (1988) Science 240, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beato, M. (1989) Cell 56, 335–344. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto, K. R. (1985) Annu. Rev. Genet. 19, 209–252. [DOI] [PubMed] [Google Scholar]
- 4.Simons, S. S., Jr. (1994) in Vitamins and Hormones, ed. Litwack, G. (Academic, London), pp. 49–130.7810076
- 5.Freedman, L. P., Luisi, B. F., Korszun, Z. R., Basavappa, R., Sigler, P. B. & Yamamoto, K. R. (1988) Nature 334, 543–546. [DOI] [PubMed] [Google Scholar]
- 6.Kumar, R. & Thompson, E. B. (1999) Steroids 64, 310–319. [DOI] [PubMed] [Google Scholar]
- 7.Freedman, L. P., Yamamoto, K. R., Luisi, B. F. & Sigler, P. B. (1988) Cell 54, 41–44. [DOI] [PubMed] [Google Scholar]
- 8.Glass, C. K., Rose, D. W. & Rosenfeld, M. G. (1997) Curr. Opin. Cell Biol. 9, 222–232. [DOI] [PubMed] [Google Scholar]
- 9.Giguere, V., Hollenberg, S. M., Rosenfeld, M. G. & Evans, R. M. (1986) Cell 46, 645–652. [DOI] [PubMed] [Google Scholar]
- 10.Danielian, P. S., White, R., Lees, J. A. & Parker, M. G. (1992) EMBO J. 11, 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luisi, B. F., Xu, W. X., Otwinowski, Z., Freedman, L. P., Yamamoto, K. R. & Sigler, P. B. (1991) Nature 352, 497–505. [DOI] [PubMed] [Google Scholar]
- 12.Bledsoe, R. K., Montana, V. G., Stanley, T. B., Delves, C. J., Apolito, C. J., McKee, D. D., Consler, T. G., Parks, D. J., Stewart, E. L., Willson, T. M., et al. (2002) Cell 110, 93–105. [DOI] [PubMed] [Google Scholar]
- 13.Dieken, E. S. & Miesfeld, R. L. (1992) Mol. Cell. Biol. 12, 5895–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godowski, P. J., Rusconi, S., Miesfeld, R. & Yamamoto, K. R. (1987) Nature 325, 365–368. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, S. (1993) Cell 72, 481–483. [DOI] [PubMed] [Google Scholar]
- 16.Sigler, P. B. (1988) Nature 333, 210–212. [DOI] [PubMed] [Google Scholar]
- 17.Dahlman-Wright, K., Baumann, H., McEwan, I. J., Almlof, T., Wright, A. P. H., Gustafsson, J. A. & Hard, T. (1995) Proc. Natl. Acad. Sci. USA 92, 1699–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baskakov, I. V., Kumar, R., Srinivasan, G., Ji, Y., Bolen, D. W. & Thompson, E. B. (1999) J. Biol. Chem. 274, 10693–10696. [DOI] [PubMed] [Google Scholar]
- 19.Henriksson, A., Almlof, T., Ford, J., McEwan, I. J., Gustafsson, J. A. & Wright, A. P. H. (1997) Mol. Cell. Biol. 17, 3065–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ford, J., McEwan, I. J., Wright, A. P. H. & Gustafsson, J. A. (1997) Mol. Endocrinol. 11, 1467–1475. [DOI] [PubMed] [Google Scholar]
- 21.Almlof, T., Wallberg, A. E., Gustafsson, J. A. & Wright, A. P. H. (1998) Biochemistry 37, 9586–9594. [DOI] [PubMed] [Google Scholar]
- 22.Kumar, R., Lee, J. C., Bolen, D. W. & Thompson, E. B. (2001) J. Biol. Chem. 276, 18146–18152. [DOI] [PubMed] [Google Scholar]
- 23.Almlof, T., Wright, A. P. H. & Gustafsson, J. A. (1995) J. Biol. Chem. 270, 17535–17540. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, R., Baskakov, I. V., Srinivasan, G., Bolen, D. W., Lee, J. C. & Thompson, E. B. (1999) J. Biol. Chem. 274, 24737–24741. [DOI] [PubMed] [Google Scholar]
- 25.McEwan, I. J., Dahlman-Wright, K., Ford, J. & Wright, A. P. H. (1996) Biochemistry 35, 9584–9593. [DOI] [PubMed] [Google Scholar]
- 26.Shen, F., Triezenberg, S. J., Hensley, P., Porter, D. & Knutson, J. R. (1996) J. Biol. Chem. 271, 4827–4837. [DOI] [PubMed] [Google Scholar]
- 27.Reid, J., Kelly, S. M., Watt, K., Price, N. C. & McEwan, I. J. (2002) J. Biol. Chem. 277, 20079–20086. [DOI] [PubMed] [Google Scholar]
- 28.Warnmark, A., Wikstrom, A., Wright, A. P. H., Gustafsson, J. A. & Hard, T. (2001) J. Biol. Chem. 276, 45939–45944. [DOI] [PubMed] [Google Scholar]
- 29.Ito, M. & Roeder, R. G. (2001) Trends Endocrinol. Metabol. 12, 127–134. [DOI] [PubMed] [Google Scholar]
- 30.McKenna, N. J., Lanz, R. B. & O'Malley, B. W. (1999) Endocr. Rev. 20, 321–344. [DOI] [PubMed] [Google Scholar]
- 31.Deroo, B. J. & Archer, T. K. (2001) Oncogene 20, 3039–3046. [DOI] [PubMed] [Google Scholar]
- 32.Fryer, C. J., Lee, H. L., Zaniewski, E., Liang, T. & Mymryk, J. S. (1995) J. Steroid Biochem. Mol. Biol. 5, 421–429. [DOI] [PubMed] [Google Scholar]
- 33.Dong, A. & Caughey, W. S. (1994) Methods Enzymol. 232, 139–175. [DOI] [PubMed] [Google Scholar]
- 34.Dong, A., Prestrelski, S. J. & Carpenter, J. C. (1995) J. Pharm. Sci. 84, 415–424. [DOI] [PubMed] [Google Scholar]
- 35.Dong, A., Caughey, B., Caughey, W. S., Bhat, K. S. & Coe, J. E. (1992) Biochemistry 31, 9364–9370. [DOI] [PubMed] [Google Scholar]
- 36.Volk, D. E., House, P. G., Thiviyanathan, V., Luxon, B. A. Zhang, S., Lloyd, R. S. & Gorenstein, D. G. (2000) Biochemistry 39, 7331–7336. [DOI] [PubMed] [Google Scholar]
- 37.Marion, D., Ikura, M., Tschudin, R. & Bax, A. (1989) J. Magn. Reson. 85, 393–399. [Google Scholar]
- 38.Muhandiram, D. R. & Kay, L. E. (1994) J. Magn. Reson. B103, 203–216. [Google Scholar]
- 39.Wittekind, M. & Mueller, L. (1993) J. Magn. Reson. B101, 201–205. [Google Scholar]
- 40.Susi, H. & Byler, D. M. (1986) Methods Enzymol. 130, 290–311. [DOI] [PubMed] [Google Scholar]
- 41.Jackson, M. & Mantsch H. H. (1995) Crit. Rev. Biochem. Mol. Biol. 30, 95–120. [DOI] [PubMed] [Google Scholar]
- 42.Uversky, V. N., Gillespie J. R. & Fink, A. L. (2000) Proteins 41, 415–427. [DOI] [PubMed] [Google Scholar]
- 43.Kumar, R. & Thompson, E. B. (2003) Mol. Endocrinol. 17, 1–10. [DOI] [PubMed] [Google Scholar]
- 44.Kumar, R., Betney, R., Li, J., Thompson, E. B. & McEwan, I. J. (2004) Biochemistry 43, 3008–3013. [DOI] [PubMed] [Google Scholar]
- 45.Bain, D. L., Franden, M. A., McMananam, J. L., Takimoto, G. S. & Horwitz, K. B. (2000) J. Biol. Chem. 275, 7313–7320. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, E. B. & Kumar, R. (2003) Biochem. Biophys. Res. Commun. 306, 1–4. [DOI] [PubMed] [Google Scholar]
- 47.Leftsin, J. A. & Yamamoto, K. R. (1998) Nature 392, 885–888. [DOI] [PubMed] [Google Scholar]