Abstract
Macrocyclic chelators have been widely employed in the realm of nanoparticle-based positron emission tomography (PET) imaging, whereas its accuracy remains questionable. Here, we found that 64Cu can be intrinsically labeled onto nanographene based on interactions between Cu and the π electrons of graphene without the need of chelator conjugation, providing a promising alternative radiolabeling approach that maintains the native in vivo pharmacokinetics of the nanoparticles. Due to abundant π bonds, reduced graphene oxide (RGO) exhibited significantly higher labeling efficiency in comparison with graphene oxide (GO) and exhibited excellent radiostability in vivo. More importantly, nonspecific attachment of 1,4,7-triazacyclononane-1,4,7-triacetic acid (NOTA) on nanographene was observed, which revealed that chelator-mediated nanoparticle-based PET imaging has its inherent drawbacks and can possibly lead to erroneous imaging results in vivo.
Keywords: graphene, imaging agents, nanoparticles, positron emission tomography, radiolabeling
Graphical abstract
Chelator-free radiolabeling based on interactions between Cu and the π electrons of nanographene was shown, which can potentially substitute chelator-assisted labeling. The reliability of in vivo positron emission tomography imaging is enhanced while bypassing the inherent drawbacks of conventional chelator-based labeling.
Macrocyclic chelators, such as DOTA (1,4,7,10-tetraazacy-clododecane-1,4,7,10-tetraacetic acid) or NOTA (1,4,7-triazacyclononane-1,4,7-triacetic acid), have been routinely employed for small molecule and antibody-based positron emission tomography (PET), offering sensitive, quantitative and non-invasive functional detection of diseases at cellular or molecular levels.[1] In the past decade, with the explosive advances in nanotheranostic research, chelator conjugation has been expanded to the realm of nanoparticle-based PET imaging,[2] which has assisted in the evaluation of new nanoparticles by better understanding their in vivo biodistribution. Owing to the numerous reports of successful nanoparticle-based PET imaging employing macrocyclics-aided radiometal chelation,[3] it is now considered as the gold standard in PET-based evaluation of nanoparticle kinetics. However, chelator-based radiolabeling and PET imaging have their inevitable challenges. The incorporation of chelators might alter the size, surface charge, and hydrophilicity of the tracers, which may eventually lead to completely different imaging consequences.[4] In addition, due to their macrocyclic structure and relatively hydrophobic nature,[5] chelators might also interact with nanoparticles, influencing the overall imaging results. Therefore, although chelators have been widely employed in PET imaging of nanomaterials in the past decade, it is still questionable whether such a practice is precise enough to depict their real biodistribution.
To better understand the in vivo behavior of the nanoparticles and avoid the influence of the chelators, a novel chelator-free radiolabeling approach can be employed, whereby radiometals can directly label onto the nanoparticles through certain surface interactions.[6] Intrinsically radiolabeled nanoparticles would potentially maintain their native biodistribution and pharmacokinetics, thereby accurately reflecting the real in vivo behavior of the nanoparticles.[7]
In this study, nanographene, one of the most attractive nanomaterials in the research community, was employed for chelator-free 64Cu-radiolabeling and PET imaging. Two subtypes of graphene nanomaterials, reduced graphene oxide (RGO) and graphene oxide (GO), were labeled with 64Cu by transition metal–π electron interaction based on the electron transfer between 64Cu2+ cation and π bond on the surface of graphene nanosheets. The chelator-free radiolabeling was achieved by simply mixing RGO and GO with 64Cu in 0.1M sodium acetate buffer (Figure 1a). Theoretically, 64Cu2+ ions (3d9) need one electron to form a stable electronic configuration; π bonds of nanographene are able to provide the additional electron to stably incorporate the 64Cu2+ acceptor ions onto the surface of graphene.[8] Therefore, the amount of π bonds on the graphene nanomaterials becomes an essential factor influencing the labeling efficiency and radiostability.
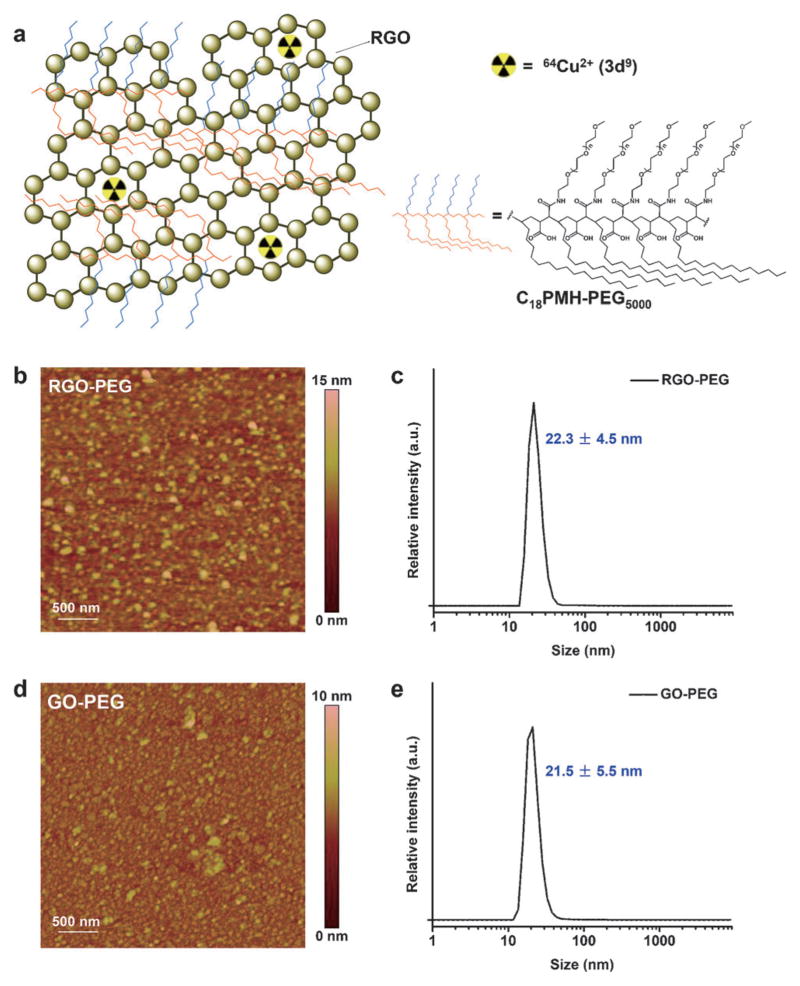
Figure 1.
a) Schematic illustration of 64Cu chelator-free radiolabeling on RGO nanoparticles. The blue and orange chains represent the hydrophilic and hydrophobic parts of the PEG. The sizes of RGO-PEG and GO-PEG were characterized by AFM (b and d) and DLS (c and e).
To validate our hypothesis regarding the mechanism of chelator-free labeling, we performed 64Cu labeling on both PEG-modified RGO (RGO-PEG, 22.3 ± 4.5 nm, zeta-potential −9.68 ± 0.86 mV; Figure 1b,c) and GO-PEG (21.5 ± 5.5 nm, zeta-potential −0.85 ± 0.27 mV; Figure 1d,e) at different concentrations (0.05, 0.2 and 0.5 mgmL−1) and temperatures (37 and 75°C). Due to more abundant π bonds on RGO-PEG, we observed that it has a significantly higher labeling yield than that of GO-PEG under each labeling condition (Figure 2, Figures S1–S6, and Table S1 in the Supporting Information), confirming our hypothesis that the chelator-free labeling is highly π bond-dependent. The chelator-free radiolabeling is also influenced by temperature and concentration (Figure 2g, and Table S1). With 0.5 mgmL−1 at 75°C, the labeling yield of RGO-PEG reached as high as 75.5 ± 1.7% after 60 minutes incubation. Such rapid and high radiolabeling yields are comparable to that achieved with NOTA, the most efficient commercially available 64Cu chelator,[9] suggesting the promising potential of chelator-free 64Cu radiolabeling of RGO-PEG for in vivo PET imaging applications. To confirm that chelator-free radiolabeling indeed took place on RGO and GO rather than PEG chains, RGO and GO without PEGylation were labeled under the same condition (Figure S7). Higher labeling yields were achieved, since more π bonds are accessible to 64Cu without PEG coating.
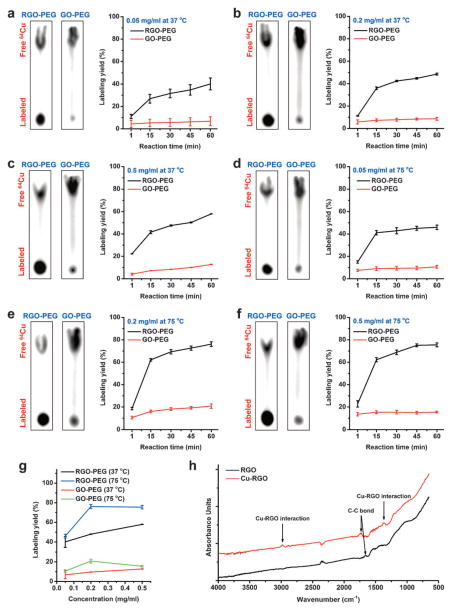
Figure 2.
a) Autoradiographic images of TLC plates and labeling yields of RGO-PEG and GO-PEG at different time points were acquired at 37°C with concentrations of a) 0.05 mgmL−1, b) 0.2 mgmL−1, and c) 0.5 mgmL−1. The same reactions were conducted at 75°C, with concentrations of d) 0.05 mgmL−1, e) 0.2 mgmL−1, and f) 0.5 mgmL−1. g) Comparison of the labeling yields of RGO-PEG and GO-PEG at different concentrations after 60 minutes incubation at 37 and 75°C. h) FTIR spectra of RGO and Cu chelator-free labeled RGO.
To further confirm the mechanism behind the chelator-free radiolabeling of RGO, Fourier transform infrared spectroscopy (FTIR) was conducted on both RGO and Cu-RGO. As shown in Figure 2h, two new peaks were observed at 1350 cm−1 and 3000 cm−1 from Cu-RGO, which resembled the peak of C–H bonds, representing the newly formed electron interactions between Cu and C. In addition, a red shift was observed for Cu-RGO bond from 1600 cm−1 to 1750 cm−1, indicating that C–C bond is lengthened by incorporation of Cu into the graphene carbon structures. The radiolabeling specificity by transition metal–π electron interactions was subsequently examined by directly mixing the nanosheets with 89Zr4+ (4p6), which does not have π electrons in the outermost atomic orbital. With 0.2 mgmL−1 at 37°C, both RGO-PEG and GO-PEG exhibited minimal labeling yields (Figure S8), demonstrating the important role of transition metal–π electron interactions in intrinsic radiolabeling.
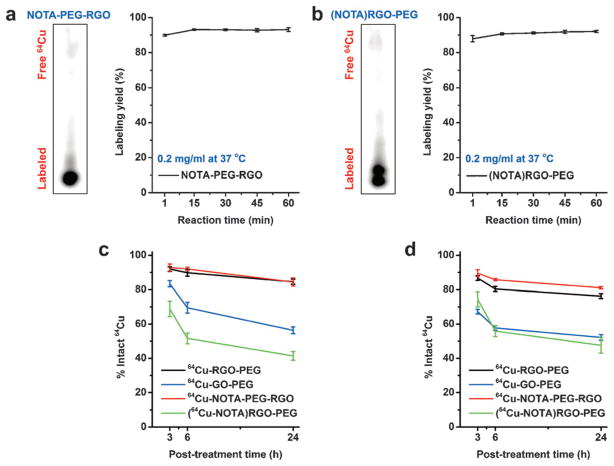
Conventional chelator-based radiolabeling was also conducted after NOTA conjugation to RGO by reaction with amino groups on PEG, as a comparison to chelator-free radiolabeling. As expected, excellent labeling yields were achieved even at low nanoparticle concentration (Figure 3a, Figure S9a, and Table S2). Interestingly we found that NOTA itself could also be nonspecifically loaded onto the RGO nanosheets (loading efficiency =28.9%) without the need of covalent linkage, when we accidentally used RGO-PEG without amino groups (unable to react with NOTA). The loading may be attributed to the hydrophobic interactions or the interaction between benzene structures in p-SCN-Bn-NOTA and π bonds of RGO, which is relatively weak as almost half of NOTA released from RGO within one hour (Figure S10). Surprisingly NOTA-loaded RGO-PEG (denoted as (NOTA)RGO-PEG) also exhibited excellent labeling yield (Figure 3b, Figure S9b, Table S2), almost similar to that with NOTA-conjugated RGO (NOTA-PEG-RGO).
Figure 3.
Autoradiographic images of TLC plates and labeling yields at different time points were acquired after incubation 64Cu with a) NOTA-conjugated RGO-PEG and b) NOTA-loaded RGO-PEG at concentrations of 0.2 mgmL−1 at 37°C. The labeling stability of them was tested in both c) PBS and d) complete mouse serum during 24 h incubation (n =3).
In vitro labeling stability was investigated before in vivo applications, since PET imaging can only detect the signal from the isotopes regardless the real biodistribution of the nanoparticles.[10] After incubation in PBS and complete mouse serum for 24 h, both chelator-free labeled and chelator-based labeled RGO showed excellent in vitro stability (Figure 3c,d), however the radiostability of chelator-free labeled GO was relatively low, indicating that the amount of π bonds not only affects the labeling efficiency but also the stability of the incorporated isotope. In addition, although (64Cu-NOTA)RGO-PEG had excellent labeling efficiency, the labeling stability was relatively low, suggesting that possible NOTA loading may lead to decreased radiostability and therefore reduced imaging accuracy by conventional chelator-based imaging with graphene nanoparticles.
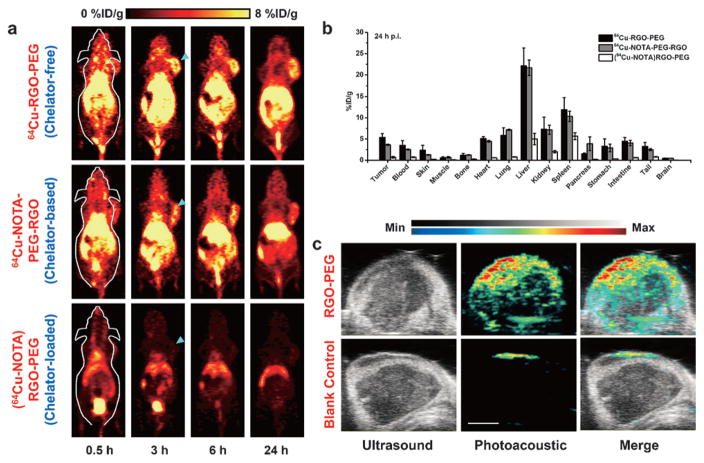
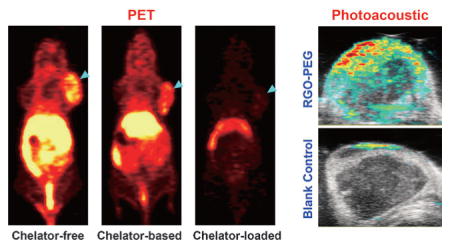
Serial PET imaging was performed with 64Cu-RGO-PEG after tail vein injection in 4T1 tumor-bearing mice. Due to their suitable size and optimized PEGylation, prolonged blood circulation of 64Cu-RGO-PEG was observed, which induced a prompt and persistent tumor uptake (n =6; Figure 4a, Figure S11, and Table S3) by an enhanced permeation and retention (EPR) effect. The prolonged blood circulation and superb tumor uptake also showed the excellent in vivo radiostability of 64Cu-RGO-PEG with minimal 64Cu detachment. The accuracy of PET imaging was validated by ex vivo biodistribution studies (Figure 4b, Table S4), which corroborated well with the region-of-interest (ROI) data from the PET images. As a control 64Cu-NOTA-PEG-RGO was also tested, showing slightly shorter blood circulation and lower tumor uptake (n =3; Figure 4a, Figure S11, and Table S3). Slight bladder uptake could be observed in mice injected with 64Cu-NOTA-PEG-RGO, stemming from the detachment of adsorbed 64Cu-NOTA from RGO nanosheets.
Figure 4.
a) Serial coronal PET images at different time points post injection of 64Cu-RGO-PEG, 64Cu-NOTA-PEG-RGO and (64Cu-NOTA)-RGO-PEG were acquired in 4T1 tumor-bearing mice. b) Ex vivo biodistribution of them at 24 h post injection. c) Ultrasound and photo-acoustic imaging upon intravenous injection of RGO-PEG (200 μL, 0.2 mgmL−1) and the same volume of PBS in a 4T1 tumor-bearing mouse.
Accordingly, although (64Cu-NOTA)RGO-PEG had the same labeling efficiency as that of 64Cu-NOTA-PEG-RGO, the in vivo stability results were completely different, as evidenced by the PET imaging of mice injected with (64Cu-NOTA)RGO-PEG under similar conditions. The radioactive signal from (64Cu-NOTA)RGO-PEG was strongly depressed as early as 0.5 h post injection and most activity could only be detected in the bladder by 3 h post injection (n =3; Figure 4a, Figure S11, and Table S3), indicating that 64Cu-NOTA detached from RGO-PEG immediately after entering the blood circulation and was excreted in the urine using the renal clearance pathway. These interesting results indicate that the decade-old gold-standard NOTA conjugated nanoparticles might not be the most accurate approach for PET imaging, since we cannot assure the purity of NOTA-conjugated RGO without any nonspecific loading. Taken together, PET imaging clearly illustrated that intrinsically radiolabeled RGO possesses higher in vivo radiostability and hence allows for more reliable evaluation of graphene biodistribution.
Taking advantage of the strong light absorbance of graphene nanomaterials,[11] photoacoustic imaging was performed in RGO-PEG injected mice to further confirm successful tumor retention, and test the multimodality imaging ability of our nanoconstructs. As expected, significantly enhanced signal was observed in tumors injected with RGO-PEG than that from the blank control (Figure 4c), further corroborating the accuracy of the PET imaging results.
In conclusion, we report successful intrinsic radiolabeling of nanographene with 64Cu based on transition metal–π electron interactions without the need of chelator conjugation. This post-synthesis chelator-free radiolabeling can be conducted very simply under mild conditions and achieve excellent labeling efficiency and in vivo radiostability, thereby exhibiting great clinical translation potential. By investigating the mechanism of chelator-free radiolabeling of graphene, we broke the stereotype that NOTA or DOTA conjugation is a necessary for 64Cu-based in vivo PET imaging. Our study provides important guidelines for future research on radio-chemistry and in vivo applications of nanomaterials: 1) Interaction between Cu and π electrons are widely applicable to 64Cu labeling with graphene, but not limited to it. Other π-bond-rich nanomaterials can be also used as promising candidates for chelator-free radiolabeling of 64Cu. 2) Scientists have frequently overlooked the possible nonspecific interactions between the chelators and the nanoparticles, which may significantly influence the PET results. For example, NOTA/DOTA conjugation may not be suitable for hydrophobic and aromatic nanoparticles. As such, thorough investigation of the chelator-conjugated nanoparticles is warranted, both in terms of their physicochemical properties as well as radiostability. 3) It is beneficial to try chelator-free labeling before conventional chelator-based labeling, which may uncover a novel labeling mechanism providing better labeling efficiency and stability.
Supplementary Material
Acknowledgments
This work is supported, in part, by the University of Wisconsin–Madison, the National Institutes of Health (NIBIB/NCI 1R01CA169365, 1R01EB021336, P30CA014520, T32CA009206 and T32GM008505), the American Cancer Society (125246-RSG-13-099-01-CCE), and Grainger Foundation (Wisconsin Distinguished Graduate Fellowship). We also gratefully acknowledge the Analytical Instrumentation Center of the School of Pharmacy at University of Wisconsin–Madison for obtaining FTIR spectra.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supporting information for this article can be found under: http://dx.doi.org/10.1002/anie.201610649.
Contributor Information
Sixiang Shi, Materials Science Program, University of Wisconsin-Madison 1509 University Ave, Madison, WI 53706 (USA).
Cheng Xu, Department of Radiology, University of Wisconsin-Madison 600 Highland Ave, Madison, WI 53792 (USA).
Dr. Kai Yang, Jiangsu Key Laboratory for Carbon-Based Functional Materials and Devices Functional Nano and Soft Materials Laboratory (FUNSOM) Soochow University, 199 Renai Rd, Suzhou, Jiangsu 215123 (China)
Shreya Goel, Materials Science Program, University of Wisconsin-Madison 1509 University Ave, Madison, WI 53706 (USA).
Hector F. Valdovinos, Department of Medical Physics University of Wisconsin-Madison 1111 Highland Ave, Madison, WI 53705 (USA)
Dr. Haiming Luo, Department of Radiology, University of Wisconsin-Madison 600 Highland Ave, Madison, WI 53792 (USA)
Emily B. Ehlerding, Department of Medical Physics University of Wisconsin-Madison 1111 Highland Ave, Madison, WI 53705 (USA)
Dr. Christopher G. England, Department of Medical Physics University of Wisconsin-Madison 1111 Highland Ave, Madison, WI 53705 (USA)
Dr. Liang Cheng, Jiangsu Key Laboratory for Carbon-Based Functional Materials and Devices Functional Nano and Soft Materials Laboratory (FUNSOM) Soochow University, 199 Renai Rd, Suzhou, Jiangsu 215123 (China)
Dr. Feng Chen, Department of Radiology, University of Wisconsin-Madison 600 Highland Ave, Madison, WI 53792 (USA)
Prof. Robert J. Nickles, Department of Medical Physics University of Wisconsin-Madison 1111 Highland Ave, Madison, WI 53705 (USA)
Prof. Zhuang Liu, Jiangsu Key Laboratory for Carbon-Based Functional Materials and Devices Functional Nano and Soft Materials Laboratory (FUNSOM) Soochow University, 199 Renai Rd, Suzhou, Jiangsu 215123 (China)
Prof. Weibo Cai, Materials Science Program, University of Wisconsin-Madison 1509 University Ave, Madison, WI 53706 (USA). Department of Radiology, University of Wisconsin-Madison 600 Highland Ave, Madison, WI 53792 (USA). Department of Medical Physics University of Wisconsin-Madison 1111 Highland Ave, Madison, WI 53705 (USA). University of Wisconsin Carbone Cancer Center 600 Highland Ave, Madison, WI 53792 (USA)
References
- 1.a) Gambhir SS. Nat Rev Cancer. 2002;2:683–693. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]; b) Price EW, Orvig C. Chem Soc Rev. 2014;43:260–290. doi: 10.1039/c3cs60304k. [DOI] [PubMed] [Google Scholar]; c) Cai W, Wu Y, Chen K, Cao Q, Tice DA, Chen X. Cancer Res. 2006;66:9673–9681. doi: 10.1158/0008-5472.CAN-06-1480. [DOI] [PubMed] [Google Scholar]; d) Luo H, Hernandez R, Hong H, Graves SA, Yang Y, England CG, Theuer CP, Nickles RJ, Cai W. Proc Natl Acad Sci USA. 2015;112:12806–12811. doi: 10.1073/pnas.1509667112. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Yang Y, Hernandez R, Rao J, Yin L, Qu Y, Wu J, England CG, Graves SA, Lewis CM, Wang P, Meyerand ME, Nickles RJ, Bian XW, Cai W. Proc Natl Acad Sci USA. 2015;112:E6525–E6534. doi: 10.1073/pnas.1502648112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Goel S, England CG, Chen F, Cai W. Adv Drug Deliver Rev. 2016 doi: 10.1016/j.addr.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Aryal S, Key J, Stigliano C, Landis MD, Lee DY, Decuzzi P. Small. 2014;10:2688–2696. doi: 10.1002/smll.201303933. [DOI] [PubMed] [Google Scholar]
- 3.a) Cai W, Chen K, Li ZB, Gambhir SS, Chen X. J Nucl Med. 2007;48:1862–1870. doi: 10.2967/jnumed.107.043216. [DOI] [PubMed] [Google Scholar]; b) Chen F, Hong H, Zhang Y, Valdovinos HF, Shi S, Kwon GS, Theuer CP, Barnhart TE, Cai W. ACS Nano. 2013;7:9027–9039. doi: 10.1021/nn403617j. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chen F, Goel S, Hernandez R, Graves SA, Shi S, Nickles RJ, Cai W. Small. 2016;12:2775–2782. doi: 10.1002/smll.201600194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Albanese A, Tang PS, Chan WC. Annu Rev Biomed Eng. 2012;14:1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]; b) Blanco E, Shen H, Ferrari M. Nat Biotechnol. 2015;33:941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF, Chan WCW. Nat Rev Mater. 2016;1:16014. [Google Scholar]; d) Stasiuk GJ, Long NJ. Chem Commun. 2013;49:2732–2746. doi: 10.1039/c3cc38507h. [DOI] [PubMed] [Google Scholar]
- 5.Cox JPL, Craig AS, Helps IM, Jankowski KJ, Parker D, Eaton MAW, Millican AT, Millar K, Beeley NRA, Boyce BA. J Chem Soc Perkin Trans 1. 1990:2567–2576. [Google Scholar]
- 6.a) Zhang Y, Jeon M, Rich LJ, Hong H, Geng J, Shi S, Barnhart TE, Alexandridis P, Huizinga JD, Seshadri M, Cai W, Kim C, Lovell JF. Nat Nanotechnol. 2014;9:631–638. doi: 10.1038/nnano.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang Y, Wang D, Goel S, Sun B, Chitgupi U, Geng J, Sun H, Barnhart TE, Cai W, Xia J, Lovell JF. Adv Mater. 2016;28:8524–8530. doi: 10.1002/adma.201602373. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chen F, Ellison PA, Lewis CM, Hong H, Zhang Y, Shi S, Hernandez R, Meyerand ME, Barnhart TE, Cai W. Angew Chem Int Ed. 2013;52:13319–13323. doi: 10.1002/anie.201306306. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2013;125:13561–13565. [Google Scholar]
- 7.a) Goel S, Chen F, Ehlerding EB, Cai W. Small. 2014;10:3825–3830. doi: 10.1002/smll.201401048. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sun X, Cai W, Chen X. Acc Chem Res. 2015;48:286–294. doi: 10.1021/ar500362y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Shekar SC, Swathi RS. J Phys Chem C. 2015;119:8912–8923. [Google Scholar]; b) Ma L, Koka J, Stace AJ, Cox H. J Phys Chem A. 2014;118:10730–10737. doi: 10.1021/jp506530g. [DOI] [PubMed] [Google Scholar]; c) Yi HB, Lee HM, Kim KS. J Chem Theory Comput. 2009;5:1709–1717. doi: 10.1021/ct900154x. [DOI] [PubMed] [Google Scholar]
- 9.a) Cooper MS, Ma MT, Sunassee K, Shaw KP, Williams JD, Paul RL, Donnelly PS, Blower PJ. Bioconjugate Chem. 2012;23:1029–1039. doi: 10.1021/bc300037w. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang Y, Hong H, Engle JW, Bean J, Yang Y, Leigh BR, Barnhart TE, Cai W. PLoS One. 2011;6:e28005. doi: 10.1371/journal.pone.0028005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong H, Yang K, Zhang Y, Engle JW, Feng L, Yang Y, Nayak TR, Goel S, Bean J, Theuer CP, Barnhart TE, Liu Z, Cai W. ACS Nano. 2012;6:2361–2370. doi: 10.1021/nn204625e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Weber J, Beard PC, Bohndiek SE. Nat Methods. 2016;13:639–650. doi: 10.1038/nmeth.3929. [DOI] [PubMed] [Google Scholar]; b) Yang K, Hu L, Ma X, Ye S, Cheng L, Shi X, Li C, Li Y, Liu Z. Adv Mater. 2012;24:1868–1872. doi: 10.1002/adma.201104964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.