Abstract
Parkinson's disease is associated with the deposition and accumulation of α-synuclein fibrils in the brain. A30P and A53T mutations have been linked to the early-onset familial disease state. Time-resolved tryptophan fluorescence energy-transfer measurements have been used to probe the structures of pseudo-wild-type and mutant (A30P) α-synucleins at physiological pH (7.4), in acidic pH (4.4) solutions, and in the presence of SDS micelles, a membrane mimic. Fluorescent donor–energy acceptor (DA) distance distributions for six different tryptophan/3-nitro-tyrosine pairs reveal the presence of compact, intermediate, and extended conformations of the protein. CD spectra indicate that the protein develops substantial helical structure in the presence of SDS micelles. DA distributions show that micelles induce compaction in the N-terminal region and expansion of the acidic C terminus. In acidic solutions, there is an increased population of collapsed structures in the C-terminal region. Energy-transfer measurements demonstrate that the average DA distances for the W4–Y19 and Y19–W39 pairs are longer in one of the two disease-related mutants (A30P).
Keywords: tryptophan, 3-nitrotyrosine
Parkinson's disease (PD) is the most common age-related neurodegenerative movement disorder (1). The primary symptoms of PD are caused by the loss of dopaminergic neurons in the substantia nigra region of the brainstem (2). A diagnostic hallmark of PD is the presence in the cerebral cortex of intracellular inclusions (Lewy bodies and neurites) (3, 4), but the role of these proteinacious materials in the pathogenic process has not been established. α-Synuclein (α-syn), an abundant 140-residue neuronal protein of unknown function (Fig. 1) (5), is the primary component of the fibrillar inclusions. Autosomal dominant early-onset PD has been linked to two point mutations (A53T and A30P) in the gene encoding α-syn (6, 7). Introduction of human wild-type, A30P-mutant, or A53T-mutant α-syn into transgenic animal models produces age-dependent motor dysfunction, neuronal deposits of fibrillar protein, and loss of dopaminergic neurons, consistent with suggestions that PD may arise from these protein aggregates (8, 9). In vitro fibrillogenesis experiments also have shown that the two mutants aggregate faster than the wild-type protein (10, 11), consistent with the proposal that the disease is caused by fibril formation. Fibrillar α-syn, however, may be a symptom rather than a cause of the disease. Recent reports suggest that the formation of soluble oligomeric intermediates is accelerated in the A30P mutant (12). Although there are contradictory results, the A30P mutant has been reported to bind more weakly to lipid vesicles than does the wild-type protein (13–16). These observations point to a role for prefibrillar species in PD (12, 17) and, further, suggest that α-syn interaction with synaptic-vesicle membranes may be involved in the pathogenic mechanism (18, 19). Indeed, it has been proposed that fibrillar aggregates are a byproduct of neuronal death and that the formation of Lewy bodies could be a protective mechanism to sequester neurotoxic species (20–23).
Fig. 1.
Primary amino acid sequence of human α-syn. The seven imperfect repeats are underlined. Two point mutations linked to early-onset PD (A30P and A53T) are colored gray. Red, acidic; blue, basic; green, aromatic. Residues in boldface have been either mutated (W, F, and Y) or chemically modified [Y(NO2)] for FET studies.
α-syn is localized in the cytosol and presynaptic terminals of neurons, with some fractions associated with synaptic-vesicle membranes (24–27). Although its function has not been fully determined, suggestions include a role in neuronal plasticity and synaptogenesis (24, 28) and as a protein-folding chaperone (29). The amino acid sequence includes seven imperfect repeats (consensus XKTKEGVXXXX) in the N-terminal portion that are similar to an 11-residue repeating motif in exchangeable apolipoproteins (Fig. 1) (24). This similarity suggests that the protein may be capable of reversibly binding to the surfaces of lipid membranes (14, 15, 18, 30). A distinguishing characteristic of the amino acid sequence is the highly acidic C-terminal region. In vitro, α-syn has been characterized as a monomeric, intrinsically unstructured (natively unfolded) protein (31). Traditional physical techniques such as x-ray diffraction and NMR spectroscopy provide little insight into the properties of dynamic and heterogeneous polypeptide ensembles. Small-angle x-ray scattering studies indicate that the protein has a radius of gyration (Rg) of ≈40 Å, larger than expected for a folded globular (15 Å) polypeptide (32). However, in the presence of acidic phospholipid vesicles, the protein undergoes a conformational change, forming some α-helical structure (estimated to be as high as 71% helical content) observable by CD (30) and NMR spectroscopies (33–35), as well as by EPR studies (36, 37). Interestingly, in contrast to the monomeric protein, larger oligomeric intermediates form β-sheet structures that cause membrane leakage in vitro (19, 38), providing further support for the hypothesis that prefibrillar structures may be cytotoxic. It also has been shown that, at higher protein concentrations (>100 μM), α-syn exhibits β-structure before fibril formation (10). Large environmentally induced conformational changes of this type are particularly interesting. Toxic conformers of otherwise benign proteins have been invoked as pathogens in a number of neurodegenerative diseases (39, 40).
We have used time-resolved fluorescence energy-transfer (FET) measurements to probe the structure and dynamics of pseudo-wild-type and disease-related (A30P) α-syn. FET rates depend on the inverse sixth power of the fluorescent donor–energy acceptor (DA) distance (41); analysis of fluorescence decay kinetics can provide probability distributions of DA distances (42–45). A fluorescent amino acid [tryptophan (Trp)] and a chemically modified tyrosine (Tyr) {3-nitrotyrosine [Y(NO2)]} were chosen as donor and acceptor, respectively; this pair has a critical Förster distance (ro) of 26 Å (46). Trp fluorescence-decay and energy-transfer kinetics were used to characterize the conformational heterogeneity of the protein at physiological pH (7.4), in acidic (pH 4.4) solutions, and in the presence of SDS micelles, a membrane mimic. Distance distributions have been determined for six DA pairs [F4W/A19Y(NO2), A19Y(NO2)/Y39W, F4W/Y39(NO2), V74Y(NO2)/F94W, F94W/Y136(NO2), and F4W/Y136(NO2)]. Also, FET kinetics were measured for two DA pairs in the vicinity of Pro-30 [F4W/A19Y(NO2) and A19Y(NO2)/Y39W] to probe the effects of this disease-related mutation on the structural and dynamical properties of the protein.
Materials and Methods
Materials. N-acteyl-tryptophanamide (NATA), tetranitromethane, and SDS were used as received from Sigma.
Protein Preparation, Modification, and Characterization. The wild-type human α-syn expression plasmid (pRK172) was provided by M. Goedert (Medical Council Research Laboratory of Molecular Biology, Cambridge, U.K.) (47). Single Trp residues were introduced at three different aromatic-residue positions (F4, Y39, and F94) by site-directed mutagenesis. Because there are four native Tyr residues (Y39, Y125, Y133, and Y136) in α-syn, all but one Tyr were mutated to phenylalanine (Phe). In two of the mutant proteins, all native Tyr residues were mutated to Phe, and additional Tyr residues were introduced (A19 and V74). All site-directed mutagenesis reactions were performed by using a QuikChange kit (Stratagene). All mutations were confirmed by DNA sequencing at the California Institute of Technology DNA Sequencing Core Facility. Recombinant α-syn was expressed and purified by using minor modifications of procedures described in ref. 37. After boiling and acid-precipitation, α-syn was chromatographed on a Q-Sepharose Fast Flow 16/10 column (Amersham Biosciences) equilibrated with 20 mM Tris buffer (pH 8.0) and eluted with a linear gradient from 0 to 0.5 M NaCl. Fractions containing α-syn were pooled and further purified on a Mono-Q 10/10 column (Amersham Biosciences). Protein concentrations were determined by using a molar extinction coefficient estimated on the basis of amino acid content: ε280 = 6,970 M–1·cm–1 (48).
Nitrated proteins were generated according to procedures described in ref. 49, with minor modifications. Purified α-syn was concentrated by ultrafiltration to 70–100 μM in 20 mM Tris buffer with 200–300 mM NaCl at pH 8.0. Tetranitromethane in ethanol [1% (vol/vol), 75 μl] was added dropwise to a stirred protein solution (850 μl) in the dark. After 10 min, a second aliquot of tetranitromethane was added. After another 10 min, the reaction was terminated by gel filtration chromatography with a HiPrep Desalting 26/10 column (Amersham Biosciences) and finally purified on a Mono Q 10/10 column. Nitrated-protein concentrations were determined by using the published molar extinction coefficient: ε381 = 2,200 M–1·cm–1 (50). The purity of all protein samples was assessed by SDS/PAGE on a Pharmacia Phastsystem (Amersham Biosciences). The molecular weights of recombinant and chemically modified proteins were confirmed by electrospray ionization–MS at the California Institute of Technology Protein/Peptide Microanalytical Laboratory. Absorption, CD, and luminescence spectra were measured on a Hewlett–Packard 8452 diode array spectrophotometer, a 62ADS spectropolarimeter (Aviv Associates, Lakewood, NJ), and a Fluorolog2 spectrof luorimeter (Jobin Yvon, Longjumeau, France), respectively. All purified proteins were concentrated by using Centriprep YM-3 (molecular weight cutoff of 3,000; Millipore) and were stored at –80°C.
Time-Resolved Fluorescence Measurements. Trp fluorescence-decay kinetics measurements were carried out as described in ref. 46. Protein samples (3–5 μM, unless otherwise stated) were deoxygenated by repeated evacuation/Ar-fill cycles on a Schlenk line. A polarized laser pulse (35° from vertical) from the third harmonic (290 nm) of a regeneratively amplified femtosecond Ti:sapphire laser (Spectra-Physics) was used as an excitation source, and a picosecond streak camera (C5680, Hamamatsu Photonics, Hamamatsu City, Japan) was used in the photon-counting mode for detection. Trp emission was selected by an interference filter (λ = 355 ± 5 nm). To determine the fluorescence anisotropy decay {r(t) = [I∥(t) – I⊥(t)]/[I∥(t) + 2I⊥(t)]} (51), polarized luminescence decays [I⊥(t) and I∥(t)] were collected simultaneously with an optical fiber array. All protein samples were filtered through Microcon YM-100 spin filter units (molecular weight cutoff of 100,000; Millipore) to remove oligomeric material before the experiments.
Data Analysis. Trp luminescence decay kinetics were modeled with the function I(t) = Io(t)∫P(r)exp(–ket(r)t)dr, where Io(t) is the Trp fluorescence decay in the absence of energy transfer to Tyr(NO2), P(r) is the probability of observing a conformation with DA distance r, and ket(r) is the rate of energy transfer at distance r. Two fitting protocols were used to analyze the kinetics. In one case, a Levenberg–Marquardt least-squares minimization algorithm (52) was used to fit to the distance distribution of an ideal freely jointed polymer (Gaussian chain) {P(r)dr = [3/(2π 〈r2〉)]3/2[exp(–3r2/2〈r2〉)]4π r2dr} (53). This simplest polymer chain consists of N bonds, each of fixed length and freely rotating. A single adjustable parameter, the mean-squared DA distance (〈r2〉) was extracted from these fits. Within the Gaussian chain model, the mean DA distance is proportional to the rms distance [〈r〉= (8〈r2〉/3π)1/2], and the effective length of the chain segment (l′) is related to the mean squared distance and contour (maximum) length, L, of the polymer (l′ = 〈r2〉/L) (54). In the second approach, we extracted distance distributions directly from the decay kinetics by numerical inversion of the Laplace transform describing I(t). The critical constraint for this inversion is the requirement that P(r) ≥ 0. A linear least-squares (LLS) matlab (Mathworks, Natick, MA) algorithm with a nonnegativity constraint (LSQNONNEG) was used to project the narrowest P(k) distributions from the fluorescence kinetics. A transformation using the Förster equation [r = Do(kR/ket)1/6, where Do = RoΦD1/6 and kR is the measured radiative decay rate constant of excited NATA] recasts the P(k) distributions obtained by LSQNONNEG into distributions over r (41). The experimental limit for distances extracted from this analysis is ≈40 Å.
Computational Methods. Canonical α-helices and β-strands were constructed with side chains for the following DA pairs: F4W/A19Y(NO2), A19Y(NO2)/Y39W, F4W/Y39(NO2), V74Y(NO2)/F94W, and F94W/Y136(NO2). Molecular-dynamics simulations were carried out with the mpsim program (54) to optimize the terminal W and Y side-chain positions while holding the coordinates of all other atoms fixed. In this optimization, a DREIDING force field (55) with CHARMM22 (56) charges assigned to all atoms was used. Center-to-center distances were obtained for an α-helix and a β-strand for each DA pair.
Results and Discussion
Trp Fluorescence and Anisotropy Decay. Trp residues incorporated at the sites of three different aromatic amino acids (F4, Y39, and F94) in α-syn exhibit fluorescence maxima at 348 nm, indicating that the indole side chains are largely water-exposed at pH 7.4 and 4.4 (emission maximum of NATA in water is 352 nm). In the presence of SDS micelles (40 mM in sodium phosphate buffer at pH 7.4), Trp fluorescence exhibits a pronounced blue shift: W4, 328 nm; W39 and W94, 338 nm (the fluorescence maximum of NATA is 347 nm under comparable conditions). The observation that the N terminus binds to the micelle is in good agreement with NMR studies (33–35).
The Trp fluorescence decay in the protein is not well modeled by a single exponential function. This behavior is typical of Trp-containing peptides and proteins; it has been attributed to multiple, slowly interconverting (>10 ns) side-chain conformations. Under all solution conditions examined (pH 7.4 sodium phosphate buffer, pH 4.4 sodium acetate buffer, and in the presence of SDS micelles at pH 7.4), there are at least three distinct conformations for all three Trp residues: τ1 ≈ 4 ns (35–50%), τ2 ≈ 1.6 ns (35–50%), and τ3 ≈ 0.5 ns (<10%). We have used time-resolved fluorescence polarization measurements to characterize Trp mobility in the entire protein (57): ≈90% of Trp fluorescence depolarizes within ≈150 ps, and the residual anisotropy (ro = 0.06) decays with a 1-ns time constant in pH 7.4 solution [for l-Trp in water, the anisotropy in the absence of rotational diffusion is 0.4 (58)]. In the presence of SDS micelles, the residual anisotropy for all Trp sites increases (0.09), and its decay slows to 3–5 ns, revealing an increase in the microenvironment viscosity and rigidity of the polypeptide structure. In comparison to other lipid-binding proteins, the micelle-induced changes in α-syn Trp anisotropy decay are rather modest. Melittin binding to liposomes, for example, produces a larger ro value (0.185) and longer rotational correlation time (10 ns) (59); Trp rotation in human apolipoprotein C-I slows dramatically (44 ns) in the presence of micelles (60). Although the blue shift of the Trp fluorescence indicates that the indole is in a strongly hydrophobic micellar environment, the rapid anisotropy decay reveals that the side chain retains a considerable degree of rotational freedom.
State of Aggregation. To probe the equilibrium state of aggregation in solution, Trp fluorescence decay kinetics were examined at different α-syn concentrations. For W39 and W94, the observed kinetics were independent of protein concentration (1–30 μM). However, W4 fluorescence decay kinetics exhibited a modest dependence on [α-syn], suggesting some type of interprotein interaction. At elevated concentrations, the fast-decay (0.3 ns) component disappears, producing an increase in the intermediate-decay (1 ns) population, as well as a slight increase (10%) in the amplitude of the slow-phase rate constant. Interprotein FET kinetics measurements with mixtures of fluorescent donor-only and energy acceptor-only proteins [F4W/Y125F/Y133F/Y136F and Y39(NO2)/Y125F/Y133F/Y136F; Y39W/Y125F/Y133F/Y136F and Y39(NO2)/Y125F/Y133F/Y136F], however, provided no evidence for short interprotein contacts in the 3–15 μM concentration range. We conclude that the state of protein aggregation within the low-micromolar concentration range is constant and that, most likely, only monomeric species are present. Moreover, Trp fluorescence decay provides no evidence to suggest that the A30P mutation induces a change in the aggregation state of the protein at concentrations <30 μM. To explore the effects of Tyr nitration, we examined the dependence of intraprotein FET kinetics on protein concentration by using the A19Y(NO2)/Y39W pair (300 nM to 30 μM). The kinetics varied only slightly as protein concentration increased: three distinct rates were needed to fit the data, and the amplitude for the intermediate population increased 15%.
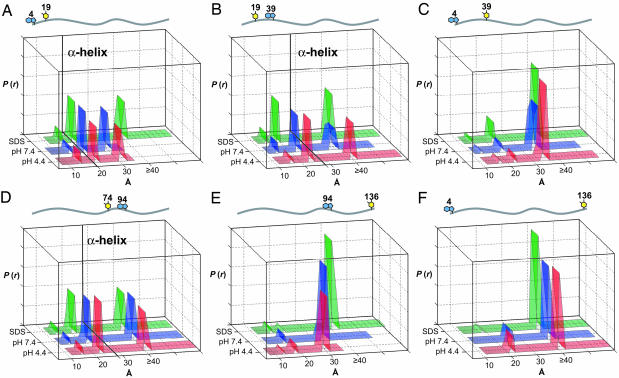
Conformational Mapping. As a first approximation, we fit the α-syn FET kinetics to DA distance distributions for freely jointed polymer chains (Fig. 2 and Table 1) (53). The nonrandom residuals and large reduced χ2 values indicate that this model, with a single adjustable parameter, does not capture all of the features of W–Y(NO2) energy transfer in the protein. Nevertheless, the results provide an approximate description of the conformational heterogeneity in terms of a well defined polymer model (Gaussian chain) (53). As expected, the mean DA distances scale with the number of residues separating D and A (Table 1). The W4–Y136 pair exhibits very little quenching, so that the distances extracted from these fits will have larger errors. The mean squared distance between two points in a Gaussian chain varies as Ll′, where L is the contour (maximum) length of the polymer and l′ is the effective length of the chain segment (53). For a freely jointed polypeptide, the length of the chain segment is generally taken to be 3.8 Å (the length of a residue). Stiffer polypeptide chains will have larger values of l′. At pH 7.4, five of the DA pairs (except W4–Y136) have chain segments in the 13- to 16-Å range, corresponding in each case to three to four amino acid residues. Under acidic conditions (pH 4.4), in which α-syn aggregation is accelerated (32), the chain segments in the N-terminal region of the protein lengthen slightly. The C-terminal DA pair (W94–Y136) exhibits a dramatic decrease in chain-segment length (about two amino acid residues) at pH 4.4. Apparently, the neutralization of the negative charges in the C terminus produces a relatively flexible polymer that behaves like a Gaussian coil. Upon addition of SDS {[SDS] = 40 mM; ≈650 μM micelles (61)} above the critical micelle concentration of ≈4 mM (62), CD spectra indicate that our mutant α-syns adopt ≈65% helical secondary structure (CD = 222 nm). The Gaussian fits to the FET data, however, do not reveal a consistent trend in segment lengths in SDS. The greatest effect appears as a substantial increase in segment length (about five residues) in the C-terminal region of the protein.
Fig. 2.
Trp-to-Y(NO2) distance distributions for freely jointed polymer chain models of α-syns under a variety of solution conditions. Green, 40 mM SDS in 20 mM NaPi buffer, pH 7.4; blue, 20 mM NaPi buffer, pH 7.4; red, 20 mM NaOAc buffer, pH 4.4.
Table 1. Trp/Y(NO2) sites and extracted mean DA distances for α-syn.
| Solution | 〈r〉LLS,* Å | χr2† | 〈r〉gc,‡ Å | I′,§ Å | χr2† | |
|---|---|---|---|---|---|---|
| W4-Y(NO2)19 (n = 15) | pH 7.4¶ | 25 | 1.2 | 23 | 13 | 43 |
| SDS∥ | 26 | 1.2 | 26 | 14 | 48 | |
| pH 4.4** | — | 1.3 | 26 | 14 | 32 | |
| Y(NO2)19-W39 (n = 20) | pH 7.4 | 30 | 1.1 | 29 | 13 | 7.6 |
| SDS | — | 1.4 | 26 | 10 | 12 | |
| pH 4.4 | — | 1.1 | 31 | 15 | 9.7 | |
| W4-Y(NO2)39 (n = 35) | pH 7.4 | 34 | 1.4 | 43 | 16 | 1.4 |
| SDS | 34 | 1.2 | 43 | 17 | 2.9 | |
| pH 4.4 | — | 1.3 | 45 | 18 | 1.4 | |
| Y(NO2)74-W94 (n = 20) | pH 7.4 | — | 1.2 | 31 | 15 | 28 |
| SDS | — | 1.4 | 26 | 11 | 39 | |
| pH 4.4 | — | 1.3 | 31 | 15 | 40 | |
| W94-Y(NO2)136 (n = 42) | pH 7.4 | 33 | 1.2 | 43 | 14 | 1.7 |
| SDS | — | 1.1 | 52 | 20 | 5.4 | |
| pH 4.4 | 28 | 1.1 | 34 | 8.5 | 1.3 | |
| W4-Y(NO2)136 (n = 132) | pH 7.4 | — | 1.6 | 62 | 8.9 | 3.8 |
| SDS | — | 2.1 | 72 | 12 | 4.5 | |
| pH 4.4 | — | 1.5 | 47 | 5.2 | 1.3 |
n, number of chain elements (residues); —, no entry.
Mean DA distance obtained from LLS fits.
Reduced χ2.
Mean DA distance obtained from fits to a Gaussian chain model.
Effective length for a chain element.
20mM NaPi buffer.
40 mM SDS in 20 mM NaPi buffer, pH 7.4.
20 mM NaOAc buffer, pH 4.4.
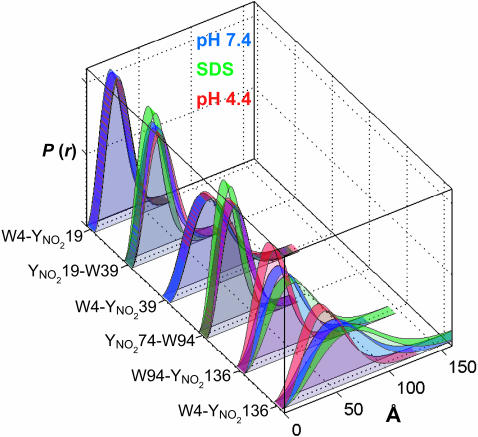
Fits to the FET data reveal that the Gaussian chain-distance distribution is a rough approximation of the conformational heterogeneity of α-syn. Consequently, we elected to extract DA distances directly from the Trp fluorescence decay kinetics by using a LLS procedure without recourse to a specific polymer model. This fitting procedure produces the narrowest distance distributions required to fit the data (using χ2 as a quality-of-fit criterion). In all cases, the LLS method gave better fits to the data than did the Gaussian chain model, but the general trends in the fitting results were comparable (Fig. 3 and Table 1). For comparison with the Gaussian chain results, we calculated mean DA distances from the LLS fits for the pairs that do not contain populations with r ≥ 40 Å (Table 1). The distance distributions projected from Trp decay kinetics in pH 7.4 solutions reveal both compact and highly extended conformations. For proteins with the DA pair separated by 15 and 20 residues, regardless of location in the sequence, the protein ensemble includes short (15 Å; ≤10%), intermediate (≈20 Å; ≈45%), and extended (≥30 Å; ≈45%) polypeptides (Fig. 3 A, B, and D). The short and intermediate populations are consistent with α-helical structures (average center-to-center distance calculated for an ideal helix: W4–Y19, 20 Å; Y19–W39, 28 Å; and Y74–W94, 28 Å; see Computational Methods). By contrast, if the polypeptide chains were entirely extended (as in a β-strand), we would expect distances ≥60 Å (W4–Y19, 59 Å; Y19–W39, 65 Å; and Y74–W94, 69 Å). For the Y74–W94 pair, about half of the polypeptide ensemble exhibits distances ≥40 Å, consistent with highly extended conformations. As expected, increasing the number of residues between DA pairs shifts the majority of the population to distances beyond our experimental limit (Fig. 3 C, E, and F).
Fig. 3.
Distributions of Trp-to-Y(NO2) distances in α-syns extracted from FET kinetics. Results are shown for W4–Y(NO2)19 (A), Y(NO2)19–W39 (B), W4–Y(NO2)39 (C), Y(NO2)74–W94 (D), W94–Y(NO2)136 (E), and W4–Y(NO2)136 (F) under a variety of solution conditions. Green, 40 mM SDS in 20 mM NaPi buffer, pH 7.4; blue, 20 mM NaPi buffer, pH 7.4; red, 20 mM NaOAc buffer, pH 4.4. Center-to-center distances calculated for an ideal helix are shown as black lines in A–D.
The LLS fits to the FET data collected in the presence of SDS provide more insight into the micelle-induced conformational changes (Fig. 3). For internal pairs, average DA distances decrease in the presence of SDS, particularly for the Y19–W39 and W4–Y39 pairs (Fig. 3 B and C). Based on chemical shifts in Cα NMR spectra, residues 18–31 are expected to have the highest degree of α-helical structure (33). However, the 19-Å DA distance found for Y19–W39 (≈40% amplitude) is substantially less than that of an ideal α-helix (28 Å). Moreover, this pair also exhibits a large population with distances >40 Å. Similarly, for the Y74–W94 pair, the intermediate DA distance shortens slightly (23–20 Å) and is well below that expected for a single α-helical segment. Interestingly, the distance distribution obtained for the W4–Y19 pair remains relatively unchanged despite the fact that the W4 fluorescence spectrum undergoes the largest blue shift in SDS. As found in the Gaussian chain fits, the distance between the W94–Y136 pair increases significantly in SDS, presumably owing to repulsive electrostatic interactions with the negative micelle surface. The most striking feature of the α-syn structures in the presence of SDS micelles is the lack of tertiary contacts; there is no evidence for globular structure despite the development of α-helical secondary structure. Although α-syn has substantial helical content in the presence of micelles, the FET data reveal that the protein remains highly disordered and largely extended, in accord with previous NMR work that indicated a lengthened helix (three turns per 11 residues) for the first 100 residues, with a break around residues 42–44 (34). Site-directed spin-labeling experiments also are consistent with an extended helical structure for α-syn in the presence of phospholipid vesicles (37).
Acidic conditions do not change the general character of the six DA distance distributions, but there is a slight increase in the amplitudes of the shorter-distance populations. Our results are consistent with small-angle x-ray scattering studies that indicate that the average radius of gyration shrinks from 40 Å at pH 7 to 30 Å at pH 3.0 (32). Interestingly, the distances separating the C-terminal and N- to C-terminal pairs shorten at lower pH (Fig. 3 E and F). In the W94-Y136 pair, a 12-Å population appears, and the amplitude of the extended population decreases and shifts to a shorter distance (30 Å). As found in the Gaussian chain fits, the greatest structural change induced by acidic conditions is the contraction in the C-terminal region. The correlation with accelerated aggregation under acidic conditions (32) raises the possibility that the high charge and extended conformation in the C terminus serve to inhibit aggregation. When the acidic side chains become neutralized, reduced electrostatic repulsion induces shortening of the C terminus and overall collapse of the polypeptide chain. Notably, C-terminally truncated α-syn (1–110) aggregates faster than the full-length protein (63, 64).
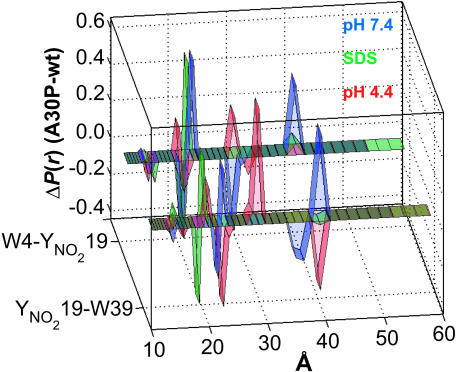
Structural Effects of the A30P Mutation. The finding that single α-syn mutations (A30P and A53T) are associated with familial early-onset PD opens the way for understanding the pathogenic role of this protein. Consequently, FET kinetics of two W/Y(NO2) pairs in the vicinity of A30P were examined as functions of solution conditions (pH 7.4, pH 4.4, and with SDS micelles at pH 7.4). Interestingly, the presence of this mutation leads to an increase in the average DA distance under all solution conditions except acidic pH, in which some of the polypeptide chains become shorter (Fig. 4). In pH 7.4 solutions with and without SDS micelles, the N-terminal (W4–Y19) pair is more extended than the internal (Y19–W39) pair. With the Pro-30 mutation, both pairs lose populations with DA distances <20 Å. The populations with intermediate DA distances (20–22 Å) shift to slightly greater separations upon introduction of the A30P mutation. The extended population in W4–Y19 moves from a DA distance of 30 to ≥40 Å. In the Y19–W39 pair, the extended population also moves to ≥40 Å. The expansion of the polypeptide in the vicinity of the Pro-30 mutation may be attributable to the increased stiffness of the polypeptide backbone (segment length of about five residues) and to the helix-disrupting property of Pro. From Cα chemical shift analysis, the slight bias (10%) for residual helical conformation in the N-terminal region is abolished in the A30P protein in solution (65). However, in the presence of SDS micelles, the labeled A30P derivatives exhibit nearly identical CD spectra with helical content comparable to the corresponding labeled pseudo-wild-type proteins. Although there are few acidic residues in the N-terminal region (one between W4 and Y19; three between Y19 and W39), the labeled A30P derivatives undergo a sizeable compaction in acidic solutions, possibly because of the presence of a turn in that region.
Fig. 4.
Difference distance distributions of disease-related A30P α-syns extracted from FET kinetics under a variety of solution conditions. Results are shown for W4/Y(NO2)19/A30P and Y(NO2)19/A30P/W39. Green, 40 mM SDS in 20 mM NaPi buffer, pH 7.4; blue, 20 mM NaPi buffer, pH 7.4; red, 20 mM NaOAc buffer, pH 4.4.
α-syn and PD. The finding that single mutations in α-syn are linked to familial early-onset forms of PD points to a central role for the protein in the etiology of the disease. A crucial question is whether a conformationally altered (misfolded) protein is directly involved in the pathogenic mechanism. Our FET kinetics measurements emphasize that α-syn is a highly disordered polymer at pH 7.4 and 4.4 and in the presence of micelles. On average, the polypeptide is more extended than expected for a freely jointed polymer, and under some conditions the protein is substantially less flexible than a random coil. Nevertheless, it is likely that the protein is highly dynamic and that conformers interchange on very short (approximately microsecond) time scales. We find that modifications of solution conditions and amino acid sequence do not produce unique conformational changes; rather, these perturbations result in subtle redistributions of the structures comprising the protein ensemble. The likelihood of identifying a unique α-syn toxic conformer seems rather remote.
It has been suggested that the death of dopaminergic neurons in PD is the ultimate result of a cascade of events involving inhibition of mitochondrial complex I, α-syn aggregation, and proteosome dysfunction (39, 40). The role of α-syn in this complex disease progression is likely to involve its interactions with other biomolecules (e.g., vesicles, enzymes, chaperones, and proteosomes) (39, 40). We have demonstrated that FET kinetics measurements can provide unique insights into the conformational heterogeneity of α-syn that are not apparent from other spectroscopic measurements. The greatest power of our approach may lie in its ability to determine how different α-syn subpopulations interact with neuronal compounds and structures that are implicated in the pathogenic process.
Acknowledgments
This work was supported by the Parkinson's Disease Foundation (J.R.W.), the National Parkinson Foundation (J.R.W.), the Beckman Macular Research Center (H.B.G. and R.L.), and the Arnold and Mabel Beckman Foundation (H.B.G. and J.R.W.), National Institutes of Health Grants GM068461 (to J.R.W.) and P50 AG05142 (to R.L.), and Department of Energy Grant DE-FG02-02ER15359 (to J.R.W.). J.C.L. thanks the Arnold and Mabel Beckman Foundation for a Beckman Senior Research Fellowship.
Author contributions: J.C.L., H.B.G., and J.R.W. designed research; J.C.L. and P.A.H. performed research; R.L. contributed new reagents/analytic tools; J.C.L. and J.R.W. analyzed data; and J.C.L., H.B.G., and J.R.W. wrote the paper.
Abbreviations: α-syn, α-synuclein; DA, fluorescent donor–energy acceptor; FET, fluorescence energy-transfer; LLS, linear least-squares; NATA, N-acteyl-tryptophanamide; PD, Parkinson's disease; Trp, tryptophan; Tyr, tyrosine; Y(NO2), 3-nitrotyrosine.
References
- 1.Parkinson, J. (1817) An Essay on the Shaking Palsy (Sherwood, Neely & Jones, London).
- 2.Dunnett, S. B. & Bjorklund, A. (1999) Nature 399, A32–A39. [DOI] [PubMed] [Google Scholar]
- 3.Baba, M., Nakajo, S., Tu, P. H., Tomita, T., Nakaya, K., Lee, V. M. Y., Trojanowski, J. Q. & Iwatsubo, T. (1998) Am. J. Pathol. 152, 879–884. [PMC free article] [PubMed] [Google Scholar]
- 4.Spillantini, M. G., Schmidt, M. L., Lee, V. M. Y., Trojanowski, J. Q., Jakes, R. & Goedert, M. (1997) Nature 388, 839–840. [DOI] [PubMed] [Google Scholar]
- 5.Clayton, D. F. & George, J. M. (1998) Trends Neurosci. 21, 249–254. [DOI] [PubMed] [Google Scholar]
- 6.Polymeropoulos, M. H., Lavedan, C., Leroy, E., Ide, S., Dehejia, A., Dutra, A., Pike, B., Root, H., Rubenstein, J., Boyer, R., et al. (1997) Science 276, 2045–2047. [DOI] [PubMed] [Google Scholar]
- 7.Kruger, R., Kuhn, W., Muller, T., Woitalla, D., Graeber, M., Kosel, S., Przuntek, H., Epplen, J. T., Schols, L. & Riess, O. (1998) Nat. Genet. 18, 106–108. [DOI] [PubMed] [Google Scholar]
- 8.Beal, M. F. (2001) Nat. Rev. Neurosci. 2, 325–332. [DOI] [PubMed] [Google Scholar]
- 9.Dawson, T. M. (2000) Cell 101, 115–118. [DOI] [PubMed] [Google Scholar]
- 10.Conway, K. A., Harper, J. D. & Lansbury, P. T. (1998) Nat. Med. 4, 1318–1320. [DOI] [PubMed] [Google Scholar]
- 11.Narhi, L., Wood, S. J., Steavenson, S., Jiang, Y., Wu, G. M., Anafi, D., Kaufman, S. A., Martin, F., Sitney, K., Denis, P., et al. (1999) J. Biol. Chem. 274, 9843–9846. [DOI] [PubMed] [Google Scholar]
- 12.Conway, K. A., Lee, S.-J., Rochet, J.-C., Ding, T. T., Williamson., R. E. & Lansbury, P. T., Jr. (2000) Proc. Natl. Acad. Sci. USA 97, 571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jo, E., Fuller, N., Rand, R. P., George-Hyslop, P. S. & Fraser, P. E. (2002) J. Mol. Biol. 315, 799–807. [DOI] [PubMed] [Google Scholar]
- 14.McLean, P. J., Kawamata, H., Ribich, S. & Hyman, B. T. (2000) J. Biol. Chem. 275, 8812–8816. [DOI] [PubMed] [Google Scholar]
- 15.Perrin, R. J., Woods, W. S., Clayton, D. F. & George, J. M. (2000) J. Biol. Chem. 275, 34393–34398. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, P. H., Nielsen, M. S., Jakes, R., Dotti, C. G. & Goedert, M. (1998) J. Biol. Chem. 273, 26292–26294. [DOI] [PubMed] [Google Scholar]
- 17.Ding, T. T., Lee, S.-J., Rochet, J.-C. & Lansbury, P. T., Jr. (2002) Biochemistry 41, 10209–10217. [DOI] [PubMed] [Google Scholar]
- 18.Jo, E., McLaurin, J., Yip, C. M., George-Hyslop, P. S. & Fraser, P. E. (2000) J. Biol. Chem. 275, 34328–34334. [DOI] [PubMed] [Google Scholar]
- 19.Volles, M. J. & Lansbury, P. T., Jr. (2002) Biochemistry 41, 4595–4602. [DOI] [PubMed] [Google Scholar]
- 20.Forno, L. S. (1996) J. Neuropathol. Exp. Neurol. 55, 259–272. [DOI] [PubMed] [Google Scholar]
- 21.Tompkins, M. M. (1997) Brain Res. 775, 24–29. [DOI] [PubMed] [Google Scholar]
- 22.Tompkins, M. M., Basgall, E. J., Zamrini, E. & Hill, W. D. (1997) Am. J. Pathol. 150, 119–131. [PMC free article] [PubMed] [Google Scholar]
- 23.Forno, L. S. & Langston, J. W. (1993) Neurodegeneration 2, 19–24. [Google Scholar]
- 24.George, J. M., Jin, H., Woods, W. S. & Clayton, D. F. (1995) Neuron 15, 361–372. [DOI] [PubMed] [Google Scholar]
- 25.Withers, G. S., George, J. M., Banker, G. A. & Clayton, D. F. (1997) Dev. Brain Res. 99, 87–94. [DOI] [PubMed] [Google Scholar]
- 26.Iwai, A., Masliah, E., Yoshimoto, M., Ge, N. F., Flanagan, L., Desilva, H. A. R., Kittel, A. & Saitoh, T. (1995) Neuron 14, 467–475. [DOI] [PubMed] [Google Scholar]
- 27.Maroteaux, L., Campanelli, J. T. & Scheller, R. H. (1988) J. Neurosci. 8, 2804–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu, L. J., Mallory, M., Xia, Y., Veinbergs, I., Hashimoto, M., Yoshimoto, M., Thal, L. J., Saitoh, T. & Masliah, E. (1998) J. Neurochem. 71, 338–344. [DOI] [PubMed] [Google Scholar]
- 29.Rekas, A., Adda, C. G., Aquilina, J. A., Barnham, K. J., Sunde, M., Galatis, D., Williamson, N. A., Masters, C. L., Anders, R. F., Robinson, C. V., et al. (2004) J. Mol. Biol. 340, 1167–1183. [DOI] [PubMed] [Google Scholar]
- 30.Davidson, W. S., Jonas, A., Clayton, D. F. & Georges, J. M. (1998) J. Biol. Chem. 273, 9443–9449. [DOI] [PubMed] [Google Scholar]
- 31.Weinreb, P. H., Zhen, W. G., Poon, A. W., Conway, K. A. & Lansbury, P. T. (1996) Biochemistry 35, 13709–13715. [DOI] [PubMed] [Google Scholar]
- 32.Uversky, V. N., Li, J. & Fink, A. L. (2001) J. Biol. Chem. 276, 10737–10744. [DOI] [PubMed] [Google Scholar]
- 33.Eliezer, D., Kutluay, E., Bussell, R., Jr., & Browne, G. (2001) J. Mol. Biol. 307, 1061–1073. [DOI] [PubMed] [Google Scholar]
- 34.Bussell, R., Jr., & Eliezer, D. (2003) J. Mol. Biol. 329, 763–778. [DOI] [PubMed] [Google Scholar]
- 35.Chandra, S., Chen, X., Rizo, J., Jahn, R. & Sudhof, T. C. (2003) J. Biol. Chem. 278, 15313–15318. [DOI] [PubMed] [Google Scholar]
- 36.Ramakrishnan, M. & Marsh, D. (2003) Biochemistry 42, 12919–12926. [DOI] [PubMed] [Google Scholar]
- 37.Jao, C. C., Der-Sarkissian, A., Chen, J. & Langen, R. (2004) Proc. Natl. Acad. Sci. USA 101, 8331–8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, M., Li, J. & Fink, A. L. (2003) J. Biol. Chem. 278, 40186–40197. [DOI] [PubMed] [Google Scholar]
- 39.Dawson, T. M. & Dawson, V. L. (2003) Science 302, 819–822. [DOI] [PubMed] [Google Scholar]
- 40.von Bohlen und Halbach, O., Schober, A. & Krieglstein, K. (2004) Prog. Neurobiol. 73, 151–177. [DOI] [PubMed] [Google Scholar]
- 41.Förster, T. (1948) Ann. Phys. (Leipzig) 2, 55–75. [Google Scholar]
- 42.Beechem, J. M. & Hass, E. (1989) Biophys. J. 55, 1225–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Navon, A., Ittah, V., Landsman, P., Scheraga, H. A. & Hass, E. (2001) Biochemistry 40, 105–118. [DOI] [PubMed] [Google Scholar]
- 44.Lyubovitsky, J. G., Gray, H. B. & Winkler, J. R. (2002) J. Am. Chem. Soc. 124, 14840–14841. [DOI] [PubMed] [Google Scholar]
- 45.Lee, J. C., Engman, K. C., Tezcan, F. A., Gray, H. B. & Winkler, J. R. (2002) Proc. Natl. Acad. Sci. USA 99, 14778–14782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, P. G. & Brand, L. (1994) Anal. Biochem. 218, 1–13. [DOI] [PubMed] [Google Scholar]
- 47.Jakes, R., Spillantini, M. G. & Goedert, M. (1994) FEBS Lett. 345, 27–32. [DOI] [PubMed] [Google Scholar]
- 48.Gill, S. C. & von Hippel, P. H. (1989) Anal. Biochem. 182, 319–326. [DOI] [PubMed] [Google Scholar]
- 49.Rischel, C., Tyberg, P., Riglere, R. & Poulsen, F. M. (1996) J. Mol. Biol. 257, 877–885. [DOI] [PubMed] [Google Scholar]
- 50.Riordan, J. F. & Vallee, B. L. (1972) Methods Enzymol. 25, 515–521. [DOI] [PubMed] [Google Scholar]
- 51.Lakowicz, J. R. (1983) in Principles of Fluorescence Spectroscopy (Plenum, New York), pp. 155–181.
- 52.Press, W. H., Flannery, B. P., Teukolsky, S. A. & Vetterling, W. T. (1989) Numerical Recipes (Cambridge Univ. Press, Cambridge, U.K.).
- 53.Flory, P. J. (1969) Statistical Mechanics of Chain Molecules (Interscience, New York).
- 54.Lim, K. T., Brunett, S., Iotov, M., McClurg, R. B., Vaidehi, N., Dasgupta, S., Taylor, S. & Goddard, W. A. (1997) J. Comput. Chem. 18, 501–521. [Google Scholar]
- 55.Mayo, S. L., Olafson, B. D. & Goddard, W. A. (1990) J. Phys. Chem. 94, 8897–8909. [Google Scholar]
- 56.MacKerell, A. D., Bashford, D., Bellot, M., Dunbrack, R. L., Evanseck, J. D., Field, M. J., Fischer, S., Gao, J., Guo, H., Ha, S., et al. (1998) J. Phys. Chem. 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- 57.Beechem, J. M. & Brand, L. (1985) Annu. Rev. Biochem. 54, 43–71. [DOI] [PubMed] [Google Scholar]
- 58.Ruggiero, A. J., Todd, D. C. & Fleming, G. R. (1990) J. Am. Chem. Soc. 112, 1003–1014. [Google Scholar]
- 59.Georghiou, S., Thompson, M. & Mukhopadhyay, A. K. (1982) Biochim. Biophys. Acta 688, 441–452. [DOI] [PubMed] [Google Scholar]
- 60.Jonas, A., Privat, J. P., Wahl, P. & Osborne, J. C. (1982) Biochemistry 21, 6205–6211. [DOI] [PubMed] [Google Scholar]
- 61.Helenius, A. & Simons, K. (1975) Biochim. Biophys. Acta 415, 29–79. [DOI] [PubMed] [Google Scholar]
- 62.Chatterjee, A., Moulik, S. P., Sanyal, S. K., Mishra, B. K. & Puri, P. M. (2001) J. Phys. Chem. B 105, 12823–12831. [Google Scholar]
- 63.Crowther, R. A., Jakes, R., Spillantini, M. G. & Goedert, M. (1998) FEBS Lett. 436, 309–312. [DOI] [PubMed] [Google Scholar]
- 64.Kim, T. D., Paik, S. R. & Yang, C.-H. (2003) Biochemistry 41, 13782–13790. [DOI] [PubMed] [Google Scholar]
- 65.Bussell, R., Jr., & Eliezer, D. (2001) J. Biol. Chem. 276, 45996–46003. [DOI] [PubMed] [Google Scholar]