Abstract
Phenazines produced by Pseudomonas and Streptomyces spp. are heterocyclic nitrogen-containing metabolites with antibiotic, antitumor, and antiparasitic activity. The antibiotic properties of pyocyanin, produced by Pseudomonas aeruginosa, were recognized in the 1890s, although this blue phenazine is now known to be a virulence factor in human disease. Despite their biological significance, the biosynthesis of phenazines is not fully understood. Here we present structural and functional studies of PhzF, an enzyme essential for phenazine synthesis in Pseudomonas spp. PhzF shares topology with diaminopimelate epimerase DapF but lacks the same catalytic residues. The structure of PhzF in complex with its substrate, trans-2,3-dihydro-3-hydroxyanthranilic acid, suggests that it is an isomerase using the conserved glutamate E45 to abstract a proton from C3 of the substrate. The proton is returned to C1 of the substrate after rearrangement of the double-bond system, yielding an enol that converts to the corresponding ketone. PhzF is a dimer that may be bifunctional, providing a shielded cavity for ketone dimerization via double Schiff-base formation to produce the phenazine scaffold. Our proposed mechanism is supported by mass and NMR spectroscopy. The results are discussed in the context of related structures and protein sequences of unknown biochemical function.
Keywords: active site residues, catalytic activity, x-ray structure
The phenazines include >80 heterocyclic nitrogen-containing natural products synthesized by fluorescent Pseudomonas spp., members of a few other bacterial genera, and thousands of chemically synthesized derivatives (1). Most naturally derived phenazines are pigments and many exhibit broad-spectrum antibiotic activity against bacteria, fungi, and parasites. Structurally simple phenazines such a phenazine-1-carboxylic acid (PCA) and its hydroxy and carboxamide derivatives are produced by beneficial strains of Pseudomonas on the roots of plants, where they have a critical role in suppressing fungal pathogens (2, 3) and can contribute to the ecological competence of the strains that produce them (4). The antibiotic properties of pyocyanin, the blue phenazine synthesized in human tissues by the opportunistic pathogen Pseudomonas aeruginosa, were first reported in the late 1890s, although interest now focuses on its role as a virulence determinant in chronic and acute lung disease (5, 6). The antibiotic, antiparasitic, and antitumor activities of naturally occurring phenazine compounds also have stimulated interest in the development of synthetic analogues tailored to particular therapeutic applications, but progress has been limited by the lack of efficient and generally applicable methods for the synthesis of substituted phenazines (1).
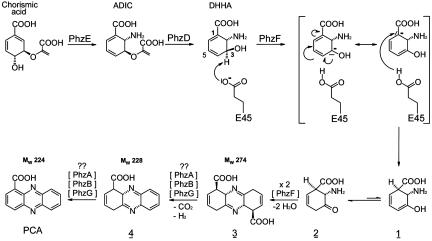
The conserved seven-gene operon phzABCDEFG is responsible for the synthesis of PCA by Pseudomonas fluorescens 2-79 and other fluorescent Pseudomonas spp. (7–10). PhzC, PhzD, and PhzE are similar to enzymes of the shikimic acid pathway and, together with PhzF, are absolutely required for phenazine synthesis. Using transformants of Escherichia coli expressing all or different subsets of these genes, McDonald et al. (11) demonstrated that 2-amino-2-deoxyisochorismic acid (ADIC), formed by PhzE from chorismate, is converted by PhzD, an isochorismatase, to trans-2,3-dihydro-3-hydroxyanthranilic acid (DHHA) (Fig. 1). Subsequently, poorly characterized steps likely involving the condensation of two identical DHHA molecules are then required to form the phenazine ring system. These reactions are carried out by PhzF, which lacks sequence similarity to proteins of known function. Here we provide detailed structural and biochemical analyses of PhzF, its substrate, and its products, and we propose a catalytic mechanism for the condensation of DHHA to PCA. PhzF exhibits significant structural similarity to members of the diaminopimelate epimerase (DapF) fold family of proteins, but differences in key active site residues suggest greater structural and functional diversity within this family than previously was appreciated.
Fig. 1.
Biosynthesis of PCA from chorismic acid via DHHA. Also shown is the proposed mechanism of action of PhzF.
Materials and Methods
Protein Purification and Crystallization. The Phz enzymes of P. fluorescens 2-79 and YddE of E. coli were cloned into pET15b (Novagen) and expressed in E. coli. Proteins were purified by standard procedures, including Ni2+-affinity and size-exclusion chromatography (12). Seleno-l-methionine labeling was achieved by suppression of methionine biosynthesis. The QuikChange II XL system (Stratagene) was used for site-directed mutagenesis.
All crystallization experiments were performed at room temperature with the hanging-drop vapor-diffusion method. PhzF in the unliganded form was crystallized as described in ref. 12 or as a complex with 3-hydroxyanthranilic acid (3OHAA) by incubating the protein (10 mg·ml–1) with 10 mM ligand in 24–30% (wt/vol) polyethylene glycol 4000/0.1 M sodium citrate (pH 4.4–5.0)/0.2 M NH4OAc. The latter crystals were soaked in mother liquor with 10 mM DHHA for 10 days to obtain the DHHA complex. Cryoprotection was achieved by washing crystals in mother liquor amended with 30% (vol/vol) glycerol (PhzF) or 10% (vol/vol) glycerol (complexes). Data were collected at 100 K at the European Synchrotron Radiation Facility (Grenoble, France) or on a rotating anode (PhzF–DHHA complex). Crystals of unliganded PhzF belong to space group P3221 with a = b = 56 Å, and c = 156 Å and hold one monomer per asymmetric unit. Complexes of PhzF crystallized in space group P21212 with cell parameters of a = 93 Å, b = 100 Å, and c = 57 Å and contained one dimer in the asymmetric unit. Data were reduced in xds (13). YddE was crystallized with 50 mM sodium citrate (pH 5.3)/2 M (NH4)2SO4/5% (vol/vol) 2-propanol. Crystals were cryoprotected with 2.4 M (NH4)2SO4/8% (vol/vol) 2-propanol/15% (vol/vol) ethylene glycol for data collection at 100 K on beamline X4A of the National Synchrotron Light Source (Brookhaven, NY). The crystals belong to space group C2221 with a = 125 Å, b = 131 Å, and c = 49 Å and hold one monomer per asymmetric unit. Data were reduced in denzo/scalepack (14).
The structure of PhzF was determined from selenium single-wavelength anomalous dispersion (SAD) data collected at 1.7-Å resolution on ID14-EH1 (fixed wavelength, 0.934 Å) of the European Synchrotron Radiation Facility (12). Selenium atoms were located by shelxd97 (15) and refined in sharp (16). The model was automatically built in arp/warp 6.0 (17). The second crystal form was phased by molecular replacement by using molrep (18). Models were manually corrected in o (19) and refined in refmac5 (20).
The structure of YddE was determined from selenium multi-wavelength anomalous dispersion data. The structure was solved with solve (21) and manually traced in o. Refinement was carried out in cns (22). Data collection and refinement statistics are given in Table 1. Coordinates have been deposited in the Protein Data Bank (23).
Table 1. Data collection and refinement statistics.
| Se-SAD/sulfate | 3OHAA | DHHA | YddE (E. coli) | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength,* Å | 0.934/E-ID14/1 | 1.001/E-ID14/4 | 1.542/Rotating anode | 0.9793/N-X4A |
| Resolution, Å | 30.0-1.7 (1.8-1.7)† | 20.0-1.35 (1.45-1.35)† | 20.0-1.88 (1.98-1.88)† | 20.0-2.3 (2.38-2.3)† |
| Space group | P3221 | P21212 | P21212 | C2221 |
| Cell constants, Å | a = b = 56.3, c = 156.4 | a = 92.9, b = 100.1, c = 57.1 | a = 92.8, b = 99.8, c = 57.4 | a = 125.0, b = 130.6, c = 48.6 |
| Cell angles, ° | α = β = 90, γ = 120 | α = β = γ = 90 | α = β = γ = 90 | α = β = γ = 90 |
| Average redundancy | 20.6 (15.3)† | 4.0 (3.7)† | 4.9 (4.9)† | 4.3 (27)† |
| I/σ | 27.4 (9.8)† | 11.3 (3.1)† | 14.0 (5.4)† | 12.6 (3.0)† |
| Completeness, % | 98.2 (91.1)† | 99.7 (99.3)† | 97.8 (95.7)† | 89.5 (85.6)† |
| Anom. completeness‡ % | 98.3 (90.9)† | — | — | 96 (86)† |
| Rsym§ | 7.5 (26.8)† | 6.8 (41.5)† | 7.5 (28.4)† | 7.5 (24.7)† |
| Wilson B factor, Å2 | 22 | 20 | 24 | 38 |
| Refinement | ||||
| Resolution, Å | 20.0-1.35 (1.74-1.70)† | 20.0-1.35 (1.39-1.35)† | 20.0-1.88 (1.93-1.88)† | 20.0-2.3 (2.44-2.30) |
| R¶ | 13.0 (14.9)† | 11.8 (22.2)† | 15.0 (20.8)† | 21.8 (27.1)† |
| Rfree∥ | 15.3 (17.3)† | 15.0 (24.2)† | 19.7 (24.1)† | 26.0 (31.7)† |
| Rmsd bonds, Å/angles, ° | 0.025/1.562 | 0.026/2.058 | 0.025/1.885 | 0.007/1.4 |
| B-factor deviation bonds/angles, Å2 | ||||
| Main chain | 1.0/1.8 | 2.3/3.0 | 1.2/2.0 | 1.3/2.1 |
| Side chain | 3.2/5.0 | 4.2/5.9 | 3.2/5.0 | 1.9/2.5 |
| Residues in Ramachandran core,** % | 93 | 92 | 91 | 87 |
| Protein atoms | 2,156 | 4,468 | 4,443 | 2,379 |
| Solvent atoms | 331 | 679 | 373 | 25 |
| Ligand atoms | 16 | 36 | 22 | 144 |
| Average B factor, Å2 | 14 | 12 | 17 | 34 |
| PDB accession code | 1U1V | 1U1W | 1U1X | 1SDJ |
Radiation source: E, European Synchrotron Radiation Facility (Grenoble, France); N, National Synchrotron Light Source (Brookhaven, NY).
Values for the highest shell are shown in parentheses.
Completeness calculations treat Friedel-pairs as separate observations.
Rsym = ΣΣ I (h)j — 〈I(h)〉/ΣΣ I(h)j where I(h)j is the measured diffraction intensity and the summation includes all observations.
R is the R factor = (Σ|FO| — Σ|FC|)/Σ|FO|.
Rfree is the R factor calculated by using 5% of the data that were excluded from the refinement.
Ramachandran core refers to the most favored regions in the ϕ/ψ Ramachandran plot.
Enzymatic Assays. Substrate turnover and product formation by Phz enzymes was followed by HPLC and either A at 270 and 370 nm or electrospray ionization-MS and electrospray ionization-MS/MS mass detection. DHHA was either supplied or generated in situ from chorismic acid by recombinant PhzE and PhzD. Binding of 3OHAA and DHHA to PhzF was assessed fluorometrically.
For NMR spectroscopy, 10 mM DHHA was prepared in sodium phosphate buffer (pH 7.5) in 2H2O or a 9:1 H2O/2H2O mixture. Solutions were degassed, and samples were prepared under argon for oxygen-free experiments. PhzF was added to 3 μM immediately before data collection. For quenching experiments, the reaction was allowed to develop for 10 min before NaBH4 was added to 25 mM. NMR spectra were acquired at 15°C on a 600-MHz spectrometer equipped with shielded Z gradients. 2,2-Dimethyl-2-silapentane-5-sulfonate sodium salt was used as an internal standard to calibrate proton resonances. Water resonance was suppressed with a watergate sequence (24) or by presaturation. 1D 1H NMR spectra were recorded in 5-min intervals with water suppression. Two-dimensional gradient-enhanced COSY and heteronuclear single quantum coherence spectra were collected for 2 h after enzyme addition.
Supporting Information. For full experimental details, refer to Supporting Methods, Table 2, and Figs. 4–11, which are published as supporting information on the PNAS web site.
Results and Discussion
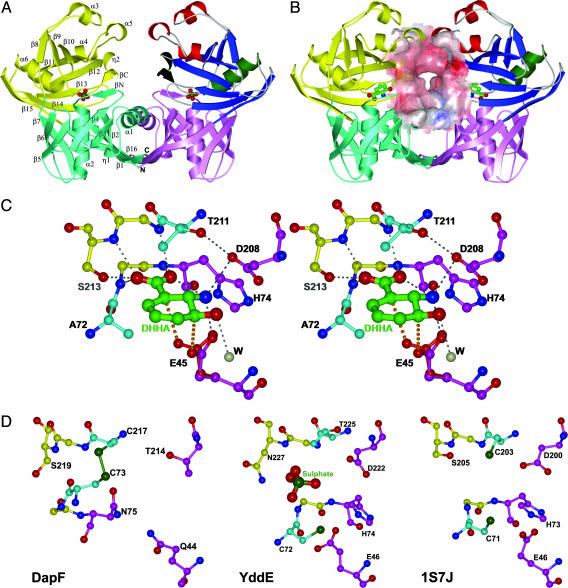
Overall Structure. The PhzF structure, as predicted by 3d-pssm (25), is related to DapF (26). A striking feature of the monomer is the presence of two similar ≈130-residue domains with topology best described as a central α-helix (α2 and α6) surrounded by an eight-stranded mixed β-sheet and decorated with one α-helix in the N-terminal domain (α1) or three α-helices in the C-terminal domain (α3, α4 and α5) (Fig. 2A). Each domain also holds a short β-strand (βN and βC) that is not part of the β-barrel and a short 310-helix (η1 and η2). The order and orientation of the core secondary structural elements are the same in both domains if the second β-strand of the N-terminal domain (β2) is superimposed on the first β-strand of the C-terminal domain (β8). Residues 258–278 form a long β-strand that spans both domains (β15 and β16). Together with β6 and β14, it connects the two domains by a three-stranded antiparallel β-sheet. At the same time, it aligns with and runs antiparallel to the N terminus, bringing the termini close together. Consequently, the entire monomer is potentially a building block for larger proteins. Main differences from DapF exist in two additional decorating α-helices in the C-terminal domain (α3 and α4) and the presence or absence of small secondary structure elements that are not part of the fold core (Fig. 5).
Fig. 2.
Ribbon diagrams of PhzF in the open and closed forms, substrate binding to the active site, and comparison to the active sites of related proteins. Overall structure of PhzF in the open (A) and closed (B) forms. Binding partners are sulfate (A) and 3OHAA (B). Key building blocks of the C-terminal domain in one monomer are color-coded: green, central α-helix; blue, eight-stranded β-barrel; and red, decorating α-helices. Secondary structure is labeled in the other monomer. The surface in B demonstrates the size of the intermonomer cavity in the closed form. (C) Stereoview of DHHA binding to the active site of PhzF. The proposed position of E45 in reprotonation is shown in red. Conserved residues are shown in magenta, and the positions of the catalytic cysteines in DapF are shown in cyan. (D) Active sites of DapF (Left), YddE (Center), and phenazine-biosynthesis protein from Enterococcus faecalis V583 (Right). All structures were prepared with bobscript (32).
The sequence identity between N- and C-terminal domains in PhzF is only 15%, indicating an ancient gene duplication event. Eighty-five residues of both domains can be superimposed within 1.9 Å. This similarity is lower than in DapF (99 residues in 1.8 Å) (26) and reflects the presence of the additional decorating helices in PhzF.
PhzF eluted at an apparent molecular mass of 42 kDa in gel filtration (calculated monomer molecular mass: 32 kDa). The crystal structure clarifies that the enzyme is a dimer in an up/up configuration with the two active sites facing each other (Fig. 2A). The dimer in the “open” form, generated by crystal symmetry (sulfate complex), interacts mainly through the α1-helices and the β16-strands in the N-terminal domains. The C-terminal domains are not in direct contact but are relatively close. The contact area is 1,316 Å2, corresponding to 10.9% of the monomer surface.
In the “closed” conformation generated during ligand binding (Fig. 2B), the secluded surface area is increased to 12.2% because of newly established direct and water-mediated hydrogen bonds as well as hydrophobic contacts between residues 170–174 in the α5-helices of the C-terminal domains. These interactions result from a twist rotation along the interconnecting β15/β16 strands that leads to a maximum displacement of ≈5 Å for residues of the C-terminal domain when the N-terminal domains of the open and closed forms are superimposed. This movement, which brings the C-terminal domains into contact and closes a channel leading from the active sites into the void between the two monomers, makes the active site inaccessible to the solvent and is achieved by shifting residues 202–208 closer to residues of the N-terminal domain and establishing a strong hydrogen bond (2.6 Å) between S44 and D208. These two residues are conserved in the related proteins YddE from E. coli (this study and PDB entries 1QY9 and 1QYA, ref. 27) and a “phenazine biosynthesis protein” from Enterococcus faecalis V583 (PDB entry 1S7J) but not in DapF. The active sites remain separated by ≈30 Å, as in the open form, indicating functional independence. Opening and closing of the active sites, however, is likely to be coordinated through the contacts between the C-terminal domains: when one monomer opens or closes, it will support the same conformational transition in the second chain because of the symmetrical nature of the interactions. The new cavity formed during dimer closure (Fig. 2B) is large enough to hold the tricyclic phenazine scaffold and clearly is the location through which products of the active sites will detach from the protein. This finding is highly significant in view of the fact (11) that phenazines are synthesized by dimerization of a precursor molecule.
Interestingly, the deposited crystal form of DapF generates crystallographic dimers with interactions between the N-terminal domains of neighboring monomers similar to those in PhzF. However, the contact area is only 5.5% of the accessible monomer surface area, and the lack of interaction between the C-terminal domains leaves them extremely open. The single epimerization catalyzed by DapF does not suggest a catalytic advantage for the dimeric form, and the significance of these dimers therefore is uncertain. In contrast, YddE from E. coli and the phenazine biosynthesis protein from Enterococcus faecalis form dimers like those of PhzF. However, except for PhzF, these proteins were crystallized in the absence of natural ligands, and it is not clear if they would close during ligand binding.
Active Site. Sequence conservation and comparison with DapF suggest that the active center of PhzF lies in the cleft between the N- and C-terminal domains. However, whereas the fold shows close relationship to DapF, key residues from the active site differ between the two enzymes. For DapF, which catalyzes epimerization of l,l-diaminopimelate to l,d-meso-diaminopimelate, acid/base catalytic activity has been attributed to two conserved cysteines, neither of which is present in PhzF. Superimposition shows that C73 of DapF is replaced by A72 and C217 is replaced by T211 in PhzF. In contrast, the active center residues E45, H74, and D208 are conserved among PhzF and more distantly related members of this enzyme family identified by blast (28) database searches. These amino acids, in turn, are not found in DapF, indicating functional divergence between the two proteins. Their importance is further highlighted by their interactions with the inhibitor 3OHAA and the substrate DHHA in crystal complexes with PhzF.
The most striking feature of these complexes is the short contact between E45's Oε2 carboxylate atom and carbon atom C3 of the ligands (Fig. 2C). For 3OHAA, this distance is 3.1 Å, and in the WT/DHHA structure, the two atoms are only 2.7 Å apart. This finding indicates the presence of a CH···O hydrogen bond that could develop into a transition state leading to proton abstraction from C3 of DHHA, the substrate of PhzF. E45 also interacts with the amino group of the ligand. Mutation to alanine or glutamine leads to a complete loss of activity, whereas fluorescence spectroscopy suggests that the affinity of 3OHAA is not altered significantly [Kd(WT), 1.4 μM; Kd(E45A), 0.11 μM; and Kd(E45Q), 4.0 μM]. E45 is solvent accessible in the open conformation but becomes secluded in the ligand-bound form.
The side chain of D208 is part of an extensive hydrogen bonding network including N18, S44, A210, T211, and the amino group of the ligand. The angle of approach, however, suggests that it is not directly involved in the chemical conversion of the substrate. Its function likely resides in positioning a negative charge in the vicinity of the amino group. This positioning ensures that the amino group remains positively charged and, thereby, decreases the pKa value of the proton bound to C3. The importance of D208 in closing the active site has been mentioned above. Mutation of D208 to alanine abolished activity. This mutant crystallizes under conditions similar to the WT but shows no affinity toward 3OHAA, in agreement with the idea that D208 has an important function in ligand binding.
An ambiguity exists for the χ2 torsion angle of H74. This side chain can form hydrogen bonds either with the carbonyl of P19 and the amino group of DHHA (although neither bond would have optimal geometry) or with Oε1 of the carboxylate group of E45, which could activate this residue in catalysis. Surprisingly, mutation of H74 to alanine decreased enzymatic activity only 4-fold. Affinity for 3OHAA, on the other hand, was increased [Kd(H74A), 69 nM]. The crystal structure shows that additional water molecules bind to the active site and that N18 adopts a different conformation in the open form of this mutant (data not shown). This result suggests that the function of H74 lies in providing hydrogen bonds that can, to some extent, be emulated by water molecules.
The carboxylate group of the ligand is entrenched in a dense hydrogen bonding network with backbone and side-chain atoms from the N termini of the central α-helices of both domains (G73, H74, G212, and S213). Taking the hydrogen bond to the ligand's own amino group into account, both oxygen atoms of the carboxylate group form three hydrogen bonds, leading to neutralization of the negative charge. This neutralization will be important for activation of the ligand by the mechanism proposed below because it also will also decrease the pKa of the proton bound to C3. The open form of PhzF could be crystallized only with anions such as sulfate or tartrate bound to this position, presumably because of an unfavorable interaction between the positive poles of the central α-helices resulting in increased flexibility of the monomer.
In the complexed form, the active site of PhzF contains a single water molecule that forms a hydrogen bond with the 3-hydroxy group of the ligand. Other residues lining the active site include L69, A72, H155, A183, M198, Y203, V205, and T211. They are mostly in hydrophobic contact with the ligand, and their degree of conservation is rather low when distant relatives of PhzF are taken into account. The active center in the closed conformation is clearly too small to hold a second ligand like DHHA, indicating that it is not the site of precursor dimerization required to generate the phenazine scaffold.
Catalytic Activity. HPLC–spectroscopy experiments with various combinations of recombinant Phz enzymes and DHHA generated in situ or supplied as a pure compound show clearly that DHHA is the substrate of PhzF and not of PhzG, as had been suggested previously (11). PhzG, like PhzA or PhzB, did not alter DHHA (Supporting Methods) and, surprisingly, PhzF alone converted DHHA to PCA, albeit with <10% of the yield obtained when all of the enzymes were present. The close proximity of E45's side chain to C3 of the ligands in our complexes suggests that PhzF is an acid/base catalyst that isomerizes DHHA. The product can then undergo PCA formation through several conversions, some of which likely do not require enzymatic catalysis. The proposed mechanism involves proton abstraction from C3 of DHHA by E45, followed by rearrangement of the double bonds and reprotonation at C1 (Fig. 1). The structure suggests that E45 carries out this reprotonation itself, effectively recycling the proton of the substrate. No other residue in the active center is close enough to C1 or C5, the alternative position for reprotonation. The carboxylate of E45, in contrast, can easily be brought close to C1 by making small adjustments of the side-chain torsion angles and, taking into account that in the course of the reaction, C1 of DHHA will change its hybridization state from sp2 to sp3, causing the ring to pucker toward E45. The hydrogen bond between the second oxygen atom of E45's carboxylate and the amino group of the ligand acts as a pivot point in the reorientation of the side chain. The function of E45 as a proton shuttle is supported by results from NMR spectroscopy (see below).
After reprotonation the resulting enol 1 (Fig. 1) will tautomerize to yield the reactive ketone 2. Condensation of two such ketone molecules by double Schiff-base formation gives phenazine precursor 3. The yield of this dimerization will be increased if release of ketone 2 into the solvent, where it can undergo side reactions, is avoided and the probability of encounter with a second molecule of 2 is increased. The size of the cavity and the relative orientation of the monomers in the PhzF dimer indeed suggest that condensation occurs before the ligands are released from the enzyme. As pointed out above, the products of the two active sites can leave the enzyme only through the intermonomer cavity. In the complexes, the hydroxyl and amino groups of the ligands face each other, favoring the proposed condensation reaction.
Several lines of experimental evidence support this mechanism. HPLC–electrospray ionization/MS analysis identifies a species of 274 m/z that appears soon after PhzF is added to DHHA. This molecular mass is consistent with the proposed phenazine precursor 3 and, as expected, this species does not absorb UV at 280 nm because of the absence of a conjugated double-bond system. However, the compound is not stable and rapidly loses 46 mass units, corresponding to an oxidative decarboxylation or loss of formic acid to form intermediate 4, which absorbs UV at 280 nm. The driving force for the reaction is the higher stability of the conjugated double-bond system, which also may explain the observation (11) that the phenazine biosynthetic pathway in Pseudomonas yields PCA and not phenazine-1,6-dicarboxylic acid. PCA is detected later in the electrospray ionization/MS experiments. The time course with which it and the second intermediate 4 build up suggests uncatalyzed spontaneous formation from phenazine precursor 3.
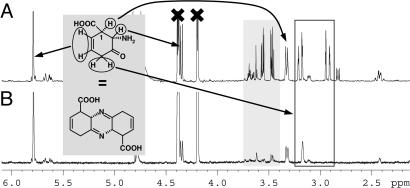
To further characterize the first product of PhzF, we followed turnover of DHHA by NMR spectroscopy in H2O and 2H2O. Five minutes after addition of the enzyme, ≈25% of the substrate was consumed and new signals emerged that can be assigned to a single cyclic species based on coupling constants and size and time-dependent change of their integrals (Figs. 3 and 6–8). The resonances are characteristic of one methylene group, two tertiary protons, and two double-bond protons of an almost identical chemical shift. Homonuclear double-quantum filtered-COSY spectra show that the double-bond protons couple weakly with the methylene protons and the tertiary proton at 3.3 ppm. The spectrum agrees with ketone 2 or the symmetrical phenazine precursor 3. Further evidence for this assignment is derived from an experiment aimed at trapping reactive compounds 2 or 3 by reduction with NaBH4. This trial yielded a single species with one additional proton at position 3, in agreement with the corresponding alcohol or diamine.
Fig. 3.
1H-NMR spectra of DHHA and PhzF in H2O(A) or 2H2O(B) 5 min after addition of the enzyme. Resonances of DHHA have been crossed. The assignment of the first product of PhzF is indicated. Strong peaks not belonging to this product are highlighted in gray, and methylene resonances are shown in a black box. Note that the proton at C1 is present in both solvents.
Only a single methylene proton is visible when DHHA and PhzF react in 2H2O. The observation that only one resonance is depleted indicates stereospecific tautomerization. At present we cannot distinguish between a thermodynamically/kinetically controlled event involving water or specific catalysis by a solvent-accessible functional group of the protein.
The tertiary proton at 3.3 ppm is assigned to C1 of the product. The fact that it is not depleted when the experiment is conducted in 2H2O strongly supports the role of E45 in shuttling a proton from C3 to C1, because reprotonation by a different residue would lead to incorporation of deuterium at C1 in 2H2O.
A complex mixture of compounds accumulates in reactions of DHHA with PhzF followed over time by NMR or MS. These likely arise because of instability of some of the intermediates, e.g., by hydrolysis or double-bond rearrangement, and account in part for the low yield of PCA produced from DHHA by PhzF in the absence of PhzA, PhzB, and PhzG. The solution in the NMR tube turns yellow with a brown layer on top, indicating that oxygen is involved in some of the conversions, but carrying out the reaction under argon does not reduce the complexity of the mixture (Fig. 6). Efficient conversion of DHHA to PCA also requires PhzA, PhzB, and PhzG, suggesting that these proteins act downstream of PhzF, perhaps in a multienzyme complex, catalyzing specific reactions and/or preventing the release of unstable intermediates into the solvent.
Comparison with Related Proteins. Results of database searches indicate that PhzF-like proteins are distributed widely among microorganisms, often with multiple paralogues present in a single bacterial species (Fig. 11). Except for PhzF, however, all of these proteins have been deduced from recently sequenced microbial genomes and are functionally and biochemically uncharacterized. Until recently, DapF-like proteins also were underrepresented in the Protein Data Bank, with only two diaminopimelate structures deposited and no protein–ligand complexes known. The situation is now changing because several entries resulting from structural genomics initiatives have emerged in the last year. Together with the insight gained from PhzF and comparison with sequences of more distantly related proteins, a broader picture of this enzyme family can now be drawn (Fig. 2D and Supporting Methods).
It is obvious that the DapF topology provides a scaffold for enzymatic acid/base catalysis. Although the substrates of the other DapF homologues are not known, they likely contain a carboxylate group because the residues in its binding site at the N termini of the central α-helices of both domains are highly conserved (Fig. 2D). Tight binding of the carboxylate is important because it will decrease the pKa of these ligands and activate them for proton abstraction.
The catalytic side chains that act as the acid or base are less conserved than might be expected, with DapF and PhzF apparently representing two extremes within this family. Whereas DapF requires two cysteines for activity, PhzF relies on glutamic acid E45 and aspartate D208, the latter not directly involved in turnover but in substrate binding and activation. The other two known family members, YddE from E. coli and phenazine-biosynthesis protein from Enterococcus faecalis V583, are of mixed type: they contain at least the basic cysteine of DapF as well as E45, H74, and D208, the three conserved PhzF residues (Fig. 2D). This finding suggests that their substrates contain an amino group in a β-position to the carboxylate. However, their substrates may not be as reactive as DHHA, making the presence of the more active cysteine necessary for proton abstraction. The function of E45 in this case might be either to reprotonate the substrate or to activate the base because both residues are close to each other in the active site (Fig. 2D). Interestingly, C72 was found covalently modified in one structure of YddE, indicating its high reactivity (29). YddE lacks the acidic cysteine of DapF, and the conserved glutamate presumably fulfills the role of the acid by forming a proton transfer system with cysteine. Despite having all of the catalytic residues of PhzF, YddE does not use DHHA as a substrate, probably because the space requirements of the additional thiol group interfere with binding. Indeed, binding of 3OHAA or DHHA to YddE could not be detected by fluorescence measurements.
The Enterococcus faecalis V583 structure 1S7J contains both cysteines in addition to the PhzF active-site residues. The functional assignment of 1S7J as a phenazine biosynthesis protein seems unlikely because the gene is not part of a phenazine biosynthesis operon and DHHA isomerization does not require the two cysteine residues. This enzyme more likely is an epimerase that utilizes the PhzF residues for binding of an amino group.
Two PhzF-related proteins of eukaryotic origin also have been identified. Aes1 from Schizosaccharomyces pombe has 28% sequence identity with PhzF and was found in a screen for factors that enhance antisense RNA-mediated gene silencing (29). In addition to E45, H74, and D208 from PhzF, this protein contains the acidic cysteine residue C217 of DapF (Fig. 2D), suggesting that the unknown substrate is deprotonated by glutamic acid and reprotonated by cysteine. A second related sequence, MAWD-binding protein (MAWDBP) (30), is of human origin and was identified in a two-hybrid screen for binding partners of the WD-40 domains of MAWD (putative human mitogen-activated protein kinase activator with WD repeats), a protein that may be involved in breast cancer (31). MAWDBP shares 22% identity with PhzF, and its active site contains the basic cysteine residue, making it similar to YddE from E. coli. It will be interesting to see how the enzymatic activity of MAWDBP is associated with physiological function. This relationship ultimately may lead to the discovery of new signaling molecules.
Supplementary Material
Acknowledgments
We thank Petra Herde for excellent technical assistance; Bernhard Griewel for help with NMR data collection; Roger Goody and Ilme Schlichting for support; Heinz Floss for helpful discussions; Volker Lorbach and Michael Müller (Forschungszentrum Jülich, Germany) for the kind gift of pure DHHA; and the beamline staffs of the European Synchrotron Radiation Facility and the National Synchrotron Light Source for the generous assignment of beam time. This work was sponsored in part by Deutsche Forschungsgemeinschaft Grant BL 587/1 (to W.B.) and the Protein Structure Initiative of the National Institutes of Health (Grant P50 GM62413 to L.T.).
Author contributions: W.B., L.T., P.B., P.J., and D.V.M. designed research; W.B., A.P.K., T.S., Y.K., P.B., P.J., and D.V.M. performed research; W.B., A.P.K., Y.K., L.T., P.B., P.J., and D.V.M. analyzed data; and W.B., L.T., L.S.T., and D.V.M. wrote the paper.
Abbreviations: DapF, diaminopimelate epimerase; DHHA, trans-2,3-dihydro-3-hydroxyanthranilic acid; 3OHAA, 3-hydroxyanthranilic acid; PCA, phenazine-1-carboxylic acid.
Data deposition: The atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 1U1V (selenium derivative, sulfate complex), 1U1W (3OHAA complex), and 1U1X (DHAA complex)].
Note Added in Proof: While this manuscript was in review, a similar report was published (33).
References
- 1.Laursen, J. B. & Nielsen, J. (2004) Chem. Rev. 104, 1663–1685. [DOI] [PubMed] [Google Scholar]
- 2.Thomashow, L. S. & Weller, D. M. (1988) J. Bacteriol. 170, 3499–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin-A-Woeng, T. F., Bloemberg, G. V. & Lugtenberg, B. J. (2003) New Phytol. 157, 503–523. [DOI] [PubMed] [Google Scholar]
- 4.Mazzola, M., Cook, R. J., Thomashow, L. S., Weller, D. M. & Pierson, L. S. (1992) Appl. Environ. Microbiol. 58, 2616–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ran, H., Hassett, D. J. & Lau, G. W. (2003) Proc. Natl. Acad. Sci. USA 100, 14315–14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau, G. W., Ran, H., Kong, F., Hassett, D. J. & Mavrodi, D. (2004) Infect. Immun. 72, 4275–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierson, L. S., III, Gaffney, T., Lam, S. & Gong, F. (1995) FEMS Microbiol. Lett. 134, 299–307. [DOI] [PubMed] [Google Scholar]
- 8.Mavrodi, D. V., Ksenzenko, V. N., Bonsall, R. F., Cook, R. J., Boronin, A. M. & Thomashow, L. S. (1998) J. Bacteriol. 180, 2541–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mavrodi, D. V., Bonsall, R. F., Delaney, S. M., Soule, M. J., Phillips, G. & Thomashow, L. S. (2001) J. Bacteriol. 183, 6454–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin-A-Woeng, T. F. C., Thomas-Oates, J. E., Lugtenberg, B. J. J. & Bloemberg, G. V. (2001) Mol. Plant–Microbe Interact. 14, 1006–1015. [DOI] [PubMed] [Google Scholar]
- 11.McDonald, M., Mavrodi, D. V., Thomashow, L. S. & Floss, H. G. (2001) J. Am. Chem. Soc. 123, 9459–9460. [DOI] [PubMed] [Google Scholar]
- 12.Mavrodi, D. V., Bleimling, N., Thomashow, L. S. & Blankenfeldt, W. (2004) Acta Crystallogr. D 60, 184–186. [DOI] [PubMed] [Google Scholar]
- 13.Kabsch, W. (1993) J. Appl. Crystallogr. 26, 795–800. [Google Scholar]
- 14.Otwinoski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 15.Schneider, T. R. & Sheldrick, G. M. (2002) Acta Crystallogr. D 58, 1772–1779. [DOI] [PubMed] [Google Scholar]
- 16.de la Fortelle, E. & Bricogne, G. (1997) Methods Enzymol. 276, 472–497. [DOI] [PubMed] [Google Scholar]
- 17.Perrakis, A., Sixma, T. K., Wilson, K. S. & Lamzin, V. S. (1997) Acta Crystallogr. D 53, 448–455. [DOI] [PubMed] [Google Scholar]
- 18.Vagin, A. & Teplyakov, A. (1997) J. Appl. Crystallogr. 30, 1022–1025. [Google Scholar]
- 19.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 20.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 21.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 23.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000) Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piotto, M., Saudek, V. & Sklenar, V. (1992) J. Biomol. NMR 2, 661–665. [DOI] [PubMed] [Google Scholar]
- 25.Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. (2000) J. Mol. Biol. 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 26.Cirilli, M., Zheng, R., Scapin, G. & Blanchard, J. S. (1998) Biochemistry 37, 16452–16458. [DOI] [PubMed] [Google Scholar]
- 27.Grassick, A., Sulzenbacher, G., Roig-Zamboni, V., Campanacci, V., Cambillau, C. & Bourne, Y. (2004) Proteins 55, 764–767. [DOI] [PubMed] [Google Scholar]
- 28.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raponi, M. & Arndt, G. M. (2002) Nucleic Acids Res. 30, 2546–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iriyama, C., Matsuda, S., Katsumata, R. & Hamaguchi, M. (2001) J. Hum. Genet. 46, 289–292. [DOI] [PubMed] [Google Scholar]
- 31.Matsuda, S., Katsumata, R., Okuda, T., Yamamoto, T., Miyazaki, K., Senga, T., Machida, K., Thant, A. A., Nakatsugawa, S. & Hamaguchi, M. (2000) Cancer Res. 60, 13–17. [PubMed] [Google Scholar]
- 32.Esnouf, R. M. (1997) J. Mol. Graph. Model. 15, 132–134. [DOI] [PubMed] [Google Scholar]
- 33.Parsons, J. F., Song, F., Parsons, L., Calabrese, K., Eisenstein, E. & Ladner, J. E. (2004) Biochemistry 43, 12427–12435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.