Abstract
Chromosomal rearrangements have a relevant role in organismic evolution. However, little is known about the mechanisms that lead different phylogenetic clades to have different chromosomal rearrangement rates. Here, we investigate the causes behind the wide karyotypic diversity exhibited by mammals. In particular, we analyzed the role of metabolic, reproductive, biogeographic and genomic characteristics on the rates of macro- and microstructural karyotypic diversification (rKD) using comparative phylogenetic methods. We found evidence that reproductive characteristics such as larger litter size per year and longevity, by allowing a higher number of meioses in absolute time, favor a higher probability of chromosomal change. Furthermore, families with large geographic distributions but containing species with restricted geographic ranges showed a greater probability of fixation of macrostructural chromosomal changes in different geographic areas. Finally, rKD does not evolve by Brownian motion because the mutation rate depends on the concerted evolution of repetitive sequences. The decisive factors of rKD evolution will be natural selection, genetic drift and meiotic drive that will eventually allow or not the fixation of the rearrangements. Our results indicate that mammalian karyotypic diversity is influenced by historical and adaptive mechanisms where reproductive and genomic factors modulate the rate of chromosomal change.
Introduction
Chromosomal rearrangements play a relevant role in evolution. Chromosomal mutations can be macrostructural, when they produce an obvious alteration of chromosome morphology and/or number caused, for instance, by pericentric inversions or Robertsonian fusions/fissions (White, 1978a; King, 1995). However, they can be also microstructural rearrangements because of heterochromatin amplification, small deletions, duplications, inversions or translocations that can only be detected through chromosome banding or painting techniques (see, for example, Stanyon and Graphodatsky, 2012).
The evolutionary relevance of chromosomal rearrangements is suggested by the close association between speciation and chromosomal evolution (Bush et al., 1977; White, 1978a). Newly arisen spontaneous chromosomal rearrangements may eventually produce transient or stable polymorphisms within a population. As genomic changes are likely to be under selective pressure, such polymorphisms may become fixed (that is, homozygous) if they provide some evolutionary advantage to carriers (Lande, 1984). Nevertheless, neutral or even underdominant rearrangements may also become fixed through stochastic processes in small isolated populations, and eventually disperse by migration (Lande, 1984; Dobigny et al., 2015). It was traditionally thought that chromosomal rearrangements could act as strong post-mating isolation mechanisms and even trigger speciation because of decreased fertility of hybrids between populations differing in one or several rearrangements, and thus fixation of chromosomal polymorphisms could contribute to the long-term process of speciation (White, 1978b; King, 1995). However, the hypothesis of chromosomal speciation because of meiotic-derived sterility was controversial and difficult to verify and was frequently based on indirect evidence, although in many cases, chromosome divergence could be a consequence rather than a cause of speciation. Furthermore, previous studies correlating speciation with chromosomal evolution between mammalian clades lacked a comparative phylogenetic approach, using taxonomy as a stand for phylogeny (see, for example, Bush et al., 1977). Nevertheless, interest in the speciation–chromosomal diversification association has remained present and some authors (see, for example, Rieseberg, 2001) have proposed novel models of speciation based on recombination effects and reduction in gene flow instead of classic underdominance in hybrids because of meiotic segregation impairment. Although preliminary works have mainly explored the role of chromosomal rearrangements in speciation, the causes of differential chromosomal variation among clades and across evolutionary time has received much less attention (Martinez et al., 2015; Ross et al., 2015). Today, many tools are at our disposal to apply robust phylogenetic analyses to our data sets, making comparative studies more powerful than in the past. In this sense, our paper represents a unique and fresh contribution to the problem of chromosomal evolution.
Mutational rates and chromosomal rearrangements are frequently associated to repetitive DNA sequences (Garagna et al., 2001). Translocations and inversions require the breakage of both DNA strands; however, there are regions of the genome where these events are more frequent. In fact, chromosomal rearrangements are not randomly distributed along the genome but concentrated in recombination hot spots (Bailey et al., 2004). These hot spots are usually associated with tandemly repeated sequences, frequently grouped in the form of heterochromatin (Garagna et al., 2001; Bailey et al., 2004), and generally characterized by rapid and convergent evolution (Dover, 2002). Similarly, repetitive sequences are under concerted evolution and can be distributed among different chromosomes by means of unequal crossing over, transposition, retrotransposition and/or slippage (Dover, 2002). Hence, it can be predicted that repetitive sequences can affect the rate of chromosomal change, whereas mechanisms such as meiotic drive, genetic drift and natural selection will affect the fixation rate of chromosomal rearrangements (White, 1978b; Villena and Sapienza, 2001; Molina et al., 2014); thus, we hypothesize that the rate of karyotypic diversification (rKD) would show a non-Brownian evolutionary mechanism (Non-neutral Hypothesis).
Several hypotheses have been proposed to explain the differential rates of genomic evolution that are associated with the rKD of the different taxonomic groups. These hypotheses involve metabolic, reproductive, biogeographic and genomic characteristics of species. The Metabolic Rate Hypothesis states that organisms with high metabolic rates in relation to body mass show high genomic mutation rates, probably because of higher oxidative damage of DNA (Martin and Palumbi, 1993; Allen et al., 2006). The Longevity Hypothesis proposes that larger and long-lived organisms have a lower mutational rate by possessing sophisticated repair mechanisms (Seluanov et al., 2009; Munshi-South and Wilkinson, 2010). There is evidence that larger and more longevous organisms invest more in DNA repair because they have to maintain a larger number of cells for a longer period of time. Thus, the efficiency of DNA repair mechanisms seems to increase with body size and longevity (Bromham, 2011). The Reproduction Rate Hypothesis suggests that species with a higher number of offspring per unit of time will also test more meioses per unit of time, allowing for a higher chance of mutations, and would accumulate more errors during DNA replication (Laird et al., 1969; Li et al., 1996). The Geographic Range Hypothesis states that taxa with wide geographic distributions, but with small isolated populations, will have a higher probability that genetic changes, adaptive or not, become fixed either through natural selection or genetic drift (White, 1978b; Lande, 1984; Hendry, 2004; Charlesworth et al., 2003; Hooper and Price 2015; Ross et al., 2015). Clearly, these hypotheses are not mutually exclusive.
Although a large amount of karyotypic information of vertebrates is available today (see, for example, O'Brien et al., 2006; Arai, 2011), it is noteworthy that few phylogenetic comparative studies have analyzed the causes of chromosomal diversification and its role in speciation (see, for example, Organ et al., 2008; Hooper and Price, 2015; Martinez et al., 2015). In the present study, we used comparative analyses of mammalian chromosomes to test several metabolic, reproductive and biogeographic hypotheses concerning rKD evolution (Table 1).
Table 1. Five hypotheses associated with the rate of genomic evolution and their predictions for the rate of karyotypic diversification (rKD) of mammals.
| Hypotheses | Prediction | References |
|---|---|---|
| Non-neutral Hypothesis: Multiple causes and mechanisms such as meiotic drive, genetic drift and natural selection affect rKD. | rKD does not evolve neutrally, and will exhibit a significant phylogenetic signal. | 1, 2, 3, 4 |
| Metabolic Rate Hypothesis: Metabolic rates, when controlled for body mass, affect genomic mutation rates. | Lineages with higher metabolic rate in relation to body mass will show higher rKD. | 5, 6, 7 |
| Longevity Hypothesis: The efficiency of DNA repair mechanisms increases with body mass and longevity. | Lineages with larger and long-lived organisms will have lower rKD. | 8, 9, 10 |
| Reproduction Rate Hypothesis: More offspring per unit of time allows a higher chance of accumulating errors during DNA replication. | Lineages with higher litter size per year and earlier sexual maturity will have higher rKD. | 11, 12 |
| Geographic Range Hypothesis: Taxa with wider geographic distributions will have higher probability of fixation of genetic changes in different parts of their range. | Lineages with larger geographic distribution will have higher rKD. | 13, 14 |
References: (1) Charlesworth et al. (2003); (2) Dover (2002); (3) Villena and Sapienza (2001); (4) Molina et al. (2014); (5) Martin and Palumbi (1993); (6) Allen et al. (2006); (7) Barja and Herrero (2000); (8) Seluanov et al. (2009); (9) Munshi-South and Wilkinson (2010); (10) Bromham (2011); (11) Laird et al. (1969); (12) Li et al. (1996); (13) White (1978a, 1978b); (14) Hendry (2004).
Mammals are a model group to test these hypotheses of chromosomal diversification. First, they are the best-known organisms regarding cytogenetics at macro- and microstructural levels. Second, there is a vast amount of knowledge on their biological characteristics and geographic distributions. Finally, robust phylogenetic hypotheses exist that allow performing comparative analyses. In particular, we predict that (1) rKD does not evolve neutrally, (2) lineages with higher metabolic rate in relation to body mass will show higher rKD, (3) lineages with larger and long-lived individuals will have a lower rKD, (4) lineages with higher litter size per year and earlier sexual maturity will have higher rKD and (5) lineages with larger geographic distributions will have higher rKD (Table 1).
Materials and methods
Our analyses were conducted at two levels regarding karyotypic information. First, we collected information on karyotype descriptions (diploid and fundamental numbers) for 1137 mammalian species belonging to 107 families based on data from Villena and Sapienza (2001) and O'Brien et al. (2006) (Supplementary Appendix A and Supplementary Table S1). We also obtained data of chromosome painting and chromosome banding of 208 mammalian species from different bibliographic sources (Supplementary Appendix A and Supplementary Table S2). From this information, two indices of karyotypic diversity were estimated. (1) Because Roberstonian fusions/fissions and pericentric inversions alter the chromosome number and the fundamental number, respectively, the number of karyotypes was quantified for each family and those with differences in chromosome number and/or fundamental number were considered distinct. A rate of macrostructural karyotypic diversification (rKDmacro) was calculated for each mammalian family dividing the number of different karyotypes by the divergence time of the most recent common ancestor of the family. Thus, the rate quantifies chromosome changes per million years (c/my). A linear regression indicated that rKDmacro is not determined by the percentage of karyotyped species per family (P=0.207), indicating that in our data set this mutation index is robust to sampling bias. Divergence times were obtained from The Tree of Life Web Project (Maddison et al., 2007, available at: http://tolweb.org/tree/) (Supplementary Appendix A and Supplementary Table S3). One possible limitation of our index could be that families with the same time of origin and same species richness but with different speciation rates (for example, one family quickly diversifying close to its time of origin and another having diversified more recently) will have the same rate of karyotypic diversification. To test the possible impact of this limitation on our analyses we estimated the speciation rates through the BAMM 2.5 software (Rabosky et al., 2014) and compared whether sister families (with the same time of origin) have very different diversification rates. Our results showed that sister families have, on average, differences in speciation rate that are similar or lower than those of non-sister families (Supplementary Appendix B and Supplementary Table S1). Thus, we considered that the impact of this possible limitation of our index was low given the phylogenetic amplitude we used. (2) On the other hand, most micro-rearrangements are underestimated by the rKDmacro that only considers macrostructural gross changes. Thus, a second index was calculated using data of chromosomal changes detected through chromosome painting and chromosome banding. For constructing this index we used only those works in the available cytogenetic literature in which the karyotypic changes were polarized with respect to an ancestral karyotype. To elaborate the karyotype hypotheses we first created a presence/absence matrix of chromosomal fragments detected from G-banding and chromosome painting based on published results. Then, we used parsimony analyses combined with external groups to determine ancestral syntenies and possible associations between karyotypes (more details in Wienberg and Stanyon, 1997, 1998; Graphodatsky et al., 2012). Thereby, the construction of ancestral kayotypes allowed us to detect and polarize chromosomal changes over evolutionary time. To quantify chromosomal changes, we considered the deletions, insertions, fusions and fissions in relation to the ancestral karyotype. In this way, we quantified the number of chromosomal changes per species with respect to the ancestral karyotype over the time of origin of this ancestor. We called this index the microstructural rate of karyotypic diversification (rKDmicro) (Supplementary Appendix A and Supplementary Table S2).
Our rKDmicro index has a negative correlation with ancestral age of lineages (r2=0.04, b=−0.011, P<0.002) where more recent lineages show a higher rate of chromosomal diversification. This results possibly reflects a bias resulting from a higher number of chromosome painting studies in phylogenetically closely related species (for example, belonging to the same genus) performed when authors already expect marked or interesting chromosomal differences.
To analyze whether the rates of macro- and microstructural karyotypic diversification were correlated with metabolic and/or reproductive characteristics, we obtained data of body mass, metabolic rate, longevity, litter per year and sexual maturity age. For the rKDmacro analyses, mean values of all variables were obtained for each family. To analyze whether geographic distribution factors affected chromosomal diversification rates we calculated the distributional range of each family and each species from PanTHERIA database (Jones et al., 2009). However, for rKDmacro, the distributional range is a rather crude predictor: comparable latitudinal ranges could include few species with large distributions, or many species with small ones. Thus, we calculated the mean distributional range for species from each family to be used as predictor in correlation analyses (Supplementary Appendix A and Supplementary Tables S2 and S3). Litters per year and sexual maturity age are directly correlated with the number of gametes produced and involved in fertilization, and both variables depend on the longevity of organisms. Therefore, we also included the interaction of litter per year and sexual maturity age, with longevity in the construction of models. Trait data were obtained from PanTHERIA database (Jones et al., 2009) and completed with four additional data sets (Pacifici et al., 2013; Tacutu et al., 2013; Verde Arregoitia et al., 2013; Wilman et al., 2014). Traits considered for the analyses with rKDmacro and rKDmicro had 6.4% and 34.6% of missing data, respectively. Having complete data is desirable in order to perform multiple regression models without losing too many degrees of freedom, and thus imputations methods in trait data sets are increasingly being used (Verde Arregoitia et al., 2013). In order to improve the precision of the imputation, we used the whole mammalian trait data set (25 traits, 5416 species) to impute our five traits of interest (body mass, metabolic rate, longevity, litters per year and sexual maturity age) with the missForest function (Stekhoven and Bühlmann, 2012). This function has been shown to perform better than other imputation methods and produces less biased results than those generated with incomplete data sets (Penone et al., 2014).
To analyze the mode of evolution of both rKD indices we used models for evolution of continuous characters (Pagel, 1999) with the GEIGER package (Harmon et al., 2008) in R 3.0.2 (https://www.r-project.org/). Maximum likelihood values for five evolutionary models were obtained: Brownian model (Blomberg et al., 2003), Speciational (kappa) model, deceleration and acceleration (delta) model (Pagel, 1999), Ornstein–Uhlenbeck process with a single optimum (Butler and King, 2004) and a model that assumes the absence of phylogenetic effect.
As across species evolutionary questions require control of phylogenetic dependence (Felsenstein, 1985), the relationship between both rKDs and the different variables was analyzed through phylogenetic generalized least squares regression (PGLS, Grafen, 1989); Pagel's λ (Pagel, 1999) (phylogenetic signal in the model residuals) was estimated using a maximum likelihood method, performed with the Caper package implemented in R 3.0.2. The models were performed using a phylogenetic covariance matrix assuming either the Brownian Motion (BM) or the Ornstein–Uhlenbeck model of evolution. The Brownian Motion model showed a better fit in the PGLS (Supplementary Appendix A and Supplementary Table S4), and thus we used the covariance matrix assuming Brownian Motion for PGLS. The best PGLS and evolutionary model was selected by Akaike's information criterion (AIC): when ΔAIC>2 we considered as indicative of strong support for the best model over other candidate models (Burnham and Anderson, 2002).
For all analyses of rKDmacro we used a recent robust phylogenetic hypothesis based on a molecular supermatrix (35 603 base pairs) from Meredith et al. (2011). This phylogenetic hypothesis includes 164 species, representing 96 to 99% of mammalian families (Meredith et al., 2011). Meanwhile, the phylogenetic hypothesis of Meredith et al. (2011) did not include the great majority of species that we utilized to calculate rKDmicro. Thereby, for the analyses with rKDmicro, we used the phylogenetic hypothesis proposed by Rolland et al. (2014). This phylogeny has shown consistent results in the estimation of diversification rates (Rolland and Salamin, 2016). For instance, it did not exhibit differences in diversification rates when compared with other phylogenetic hypotheses such as those of Faurby and Svenning (2015) and Hedges et al. (2015).
To confirm that our imputation of traits does not affect the results, we repeated the analyses using only the complete data, that is, removing species with missing values. These results did not change our main conclusions and are presented in the Supplementary material (Supplementary Appendix A, and Supplementary Table S5). In order to graphically visualize the evolution of rKDmacro and rKDmicro in the course of the diversification of mammalian families and species, we performed the reconstruction of the ancestral state with the Maximum Likelihood method for continuous traits assuming a Brownian Model of evolution. To interpolate the internal state along the length of the branches, we used Felsenstein's equation (Felsenstein, 1985) with Phytools packages (Revell, 2013) in R 3.0.2. The functions and arguments from R environment that were used in our analyses were provided in Supplementary Appendix C.
Results
The analysis of the mode of evolution of rKDmacro revealed that the best explanatory model was Ornstein–Uhlenbeck with a single optimum (σ=0.83, α=1.43) (Table 2). On the other hand, rKDmicro adjusted better to a Kappa speciational model (σ=0.053, κ=0) (Table 3). For both rates of karyotypic diversification, the Brownian Motion model showed one of the worse fits, supporting the Non-neutral Hypothesis.
Table 2. Summary of parameters and model selection for rate of macrostructural karyotypic diversification (rKDmacro).
| Trait | Model | Description | Parameters | Log-Lik | k | AIC | ΔAIC |
|---|---|---|---|---|---|---|---|
| rKDmacro | Brownian | Neutral | σ2=0.36 | −89.06 | 2 | 182.13 | 17.06 |
| Delta | Time dependent | σ2=0.15 δ=2.99 | −83.70 | 3 | 173.39 | 8.32 | |
| Kappa | Speciational | σ2=0.25 κ=0.65 | −87.07 | 3 | 180.14 | 15.07 | |
| OU | Constrained | σ2=0.83 α=1.43 | −79.53 | 3 | 165.07 | 0 | |
| White | Non-phylogenetic | σ2=0.28 | −84.46 | 2 | 172.92 | 7.85 |
Abbreviations: AIC, Akaike's information criterion; Log-Lik, log-likelihood; OU, Ornstein–Uhlenbeck.
The best models are shown in bold.
Table 3. Summary of parameters and model selection for rate of microstructural karyotypic diversification (rKDmicro).
| Trait | Model | Description | Parameters | Log-Lik | k | AIC | ΔAIC |
|---|---|---|---|---|---|---|---|
| rKDmicro | Brownian | Neutral | σ2=0.018 | −181 | 2 | 367 | 186 |
| Delta | Time dependent | σ2=0.006 δ=3 | −155 | 3 | 316 | 135 | |
| Kappa | Speciational | σ2=0.053 κ=0 | −87.4 | 3 | 181 | 0 | |
| OU | Constrained | σ2=0.04 α=0.08 | −115 | 3 | 236 | 55 | |
| White | Non-phylogenetic | σ2=0.23 | −140 | 2 | 285 | 104 |
Abbreviations: AIC, Akaike's information criterion; Log-Lik, log-likelihood; OU, Ornstein–Uhlenbeck.
The best models are shown in bold.
Rates of macrostructural chromosomal evolution varied two orders of magnitude among families. The lower value was observed in the Cynocephalidae (Dermoptera) with 0.012 c/my, whereas the highest corresponded to the rodent family Cricetidae (Rodentia) with 2.562 c/my (Figure 1a, Supplementary Appendix A and Supplementary Table S3). The use of PGLS did not reveal any correlation of rKDmacro with body mass, nor with metabolic rate. On the contrary, sexual maturity age showed a negative correlation marginally significant with rKDmacro (t=−1.85, P=0.066, d.f.=105), providing support for the Reproductive Rate Hypothesis (Table 4). In addition, we obtained a highly significant negative correlation between rKDmacro and mean species geographic range (t=−2.43, P=0.016, d.f.=105) and a positive correlation with family geographic range (t=8.14, P<0.001, d.f.=105) (Table 4). In the multiple regression models, the combined effect of family geographic range (positive correlation) and mean species geographic range (negative correlation) was the best model, strongly supporting the Geographic Range Hypothesis (P<0.001, d.f.=103) (Table 4) that showed ΔAIC >4 over other candidate models.
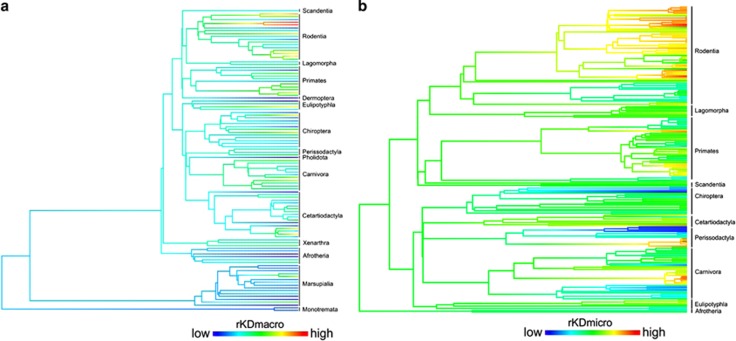
Figure 1.
Ancestral character reconstructions of two indices of karyotypic diversification rates (rKD) of mammals, assuming a Brownian Motion. Color of branches represents rKD values from low (blue) to high (red). (a) Rates of karyotypic diversification (rKDmacro) for 107 families (1137 species) based on the phylogenetic hypothesis of Meredith et al. (2011). (b) Rates of karyotypic diversification (rKDmicro) for 208 species based on the phylogeny of Rolland et al. (2014).
Table 4. Results of the correlation analyses of PGLS of rate of macrostructural karyotypic diversification (rKDmacro) with metabolic, reproductive and geographic variables.
| Response | Model | Characteristic | Traits | t | P | d.f. | λ | AIC | ΔAIC |
|---|---|---|---|---|---|---|---|---|---|
| rKDmacro | Simple | Metabolic | Body mass | −1.34 | 0.182 | 105 | 0.66 | 164.04 | 56.06 |
| Metabolic rate | −1.37 | 0.172 | 105 | 0.66 | 163.96 | 55.98 | |||
| Reproductive | Longevity | −1.09 | 0.277 | 105 | 0.64 | 164.65 | 56.67 | ||
| Litters per year | 0.79 | 0.428 | 105 | 0.61 | 165.22 | 57.24 | |||
| Sexual maturity age | −1.85 | 0.066 | 105 | 0.65 | 162.41 | 54.43 | |||
| Geographic | Family geographic range | 8.14 | <0.001 | 105 | 0.69 | 113.48 | 5.5 | ||
| Mean species geographic range | −2.43 | 0.016 | 105 | 0.66 | 160.00 | 52.02 | |||
| Multiple | Metabolic | Longevity × body mass | 0.609 | 103 | 0.65 | 167.97 | 59.99 | ||
| Reproductive | Litters per year × longevity | 0.730 | 103 | 0.65 | 168.51 | 60.53 | |||
| Sexual maturity age × longevity | 0.226 | 103 | 0.61 | 165.37 | 57.39 | ||||
| Geographic | Family geographic range × mean species geographic range | <0.001 | 103 | 0.76 | 107.98 | 0 |
Abbreviations: AIC, Akaike's information criterion; PGLS, phylogenetic generalized least squares regression.
The best models are shown in bold.
At the species level, Mus platyhrix (Muridae, Rodentia) and Microtus oeconomus (Cricetidae, Rodentia) showed the highest rKDmicro with 2.54 and 2.28 c/my respectively (Figure 1b, Supplementary Appendix A and Supplementary Table S2). In contrast, the bat Tadarida teniotis (Molossidae, Chiroptera) showed the lowest rate of chromosomal evolution with only 0.014 c/my (Figure 1b, Supplementary Appendix A and Supplementary Table S2). In the PGLS analysis of rKDmicro we observed a significant correlation only with the reproductive variables. In the simple models, rKDmicro was only negatively correlated with longevity (t=−2.28, P=0.023, d.f.=207), whereas the model with lower AIC value was obtained for the interaction between longevity (negative correlation) and litter size per year (positive correlation) (P=0.023, d.f.=205) (Table 5), strongly supporting the Longevity and Reproductive Rate Hypothesis.
Table 5. Results of the correlation analyses of PGLS of rate of microstructural karyotypic diversification (rKDmicro) with metabolic, reproductive and geographic variables.
| Response | Model | Characteristic | Traits | t | P | d.f. | λ | AIC | ΔAIC |
|---|---|---|---|---|---|---|---|---|---|
| rKDmicro | Simple | Metabolic | Body mass | −0.20 | 0.839 | 207 | 0.92 | 73.77 | 5.17 |
| Metabolic rate | 0.06 | 0.948 | 207 | 0.92 | 73.80 | 5.2 | |||
| Reproductive | Longevity | −2.28 | 0.023 | 207 | 0.92 | 68.60 | 0 | ||
| Litters per year | 0.32 | 0.743 | 207 | 0.92 | 73.70 | 4.9 | |||
| Sexual maturity age | −1.21 | 0.224 | 207 | 0.92 | 72.34 | 3.74 | |||
| Geographic | Geographic range | −0.51 | 0.607 | 207 | 0.92 | 73.54 | 4.94 | ||
| Multiple | Metabolic | Longevity × body mass | 0.132 | 205 | 0.92 | 72.11 | 3.31 | ||
| Reproductive | Litters per year × longevity | 0.031 | 205 | 0.93 | 68.92 | 0.32 | |||
| Sexual maturity age × longevity | 0.084 | 205 | 0.92 | 71.08 | 2.48 |
Abbreviations: AIC, Akaike's information criterion; PGLS, phylogenetic generalized least squares regression.
The best models are shown in bold.
Discussion
Modes of evolution
Biological and biogeographical characters influence the karyotypic diversification in mammals. Mammals exhibit the largest diversity of diploid numbers within the vertebrates (Villena and Sapienza, 2001). Several mechanisms have been proposed to explain mammalian chromosomal evolution: (1) genetic drift, (2) divergent selection and (3) meiotic drive (White, 1978b; Villena and Sapienza, 2001; Molina et al., 2014). These mechanisms facilitate or inhibit the fixation of new chromosomal rearrangements. However, until present, the ultimate causes that lead to higher or lower rates of chromosomal mutation, the source of the variation on which these mechanisms act, had not been revealed.
Our results indicate that rKDmacro and rKDmicro are highly variable traits that do not evolve through neutral evolution. For rKDmacro, an Ornstein–Uhlenbeck model under stabilizing selection with a single optimum (Butler and King, 2004), and moderate selection strength, was chosen as the best alternative model. This is consistent with a possible adaptive role of macrostructural rearrangements such as pericentric inversions and Robertsonian fusions/fissions that can restrict recombination of locally adaptive alleles (Guerrero and Kirkpatrick, 2014). An increase in an initially low rKDmacro may be advantageous because deleterious mutations will be selectively eliminated from the population, whereas rare beneficial mutations may become rapidly fixed. On the other hand, if the mutation rate is initially high, its increase could be disadvantageous by producing an extremely high number of deleterious mutations that can overcome the capacity of normalizing selection to eliminate them. As a result of this process, there is a tendency of rKDmacro toward an adaptive optimum where fitness is maximized at a given mutation rate (Gerrish et al., 2013).
In contrast, rKDmicro showed a very good agreement with a punctuational evolutionary model, indicating the existence of periods of stasis of chromosomal change and abrupt burst of chromosomal mutations over short periods of time (Pagel, 1999). Repetitive sequences usually grouped in the form of heterochromatin, change swiftly in amount and position and are an important component of genome size (Gregory, 2005). Several mechanisms such as unequal crossing-over, slippage, transposition and retrotransposition can lead to increases and decreases of these sequences that have non-Mendelian dynamics (Dover, 2002). Phylogenetically close groups, populations of the same species and even the same individual along its life may have great differences with respect to location and quantity of heterochromatin (Ventura et al., 2004; Pieńkowska-Schelling et al., 2008). Chromosomal rearrangements essentially depend on these repeated sequences that are recombination hot spots, consisting of GC-rich sequences (Bailey et al., 2004). This is supported by the observation that groups with high recombination rates such as Muridae (Rodentia) have large quantities of GC repeats in pericentromeric regions as a result of the generation of neocentromeres after centric fission events (Webber and Ponting, 2005). This is clearly observed when rKDmicros are mapped onto the mammalian phylogeny (Figure 1b): contrasting patterns are observed within the same phylogenetic group. These divergent patterns in phylogenetically close groups that share similar life-history characteristics and distribution are a good indicator that karyotypic intrinsic characteristics (for example, heterochromatin localization) modulate chromosomal rearrangement. Thus, rKDmicro shows evolution dependent on repetitive sequences (White, 1978b; Garagna et al., 2001; Dover, 2002), although the determinant factors will be natural selection, genetic drift and/or meiotic drive that will allow or impede the fixation of chromosomal changes.
Geographic and life-history correlates
The influence of metabolic rate on the rate of mutation has been originally proposed for mitochondrial DNA (Martin and Palumbi, 1993; Allen et al., 2006). Although it is widely accepted that animals with high metabolic rates and small body size show higher mutation rates for mitochondrial DNA, we did not find any evidence to support the Metabolic Rate Hypothesis in relation to the mammalian rates of karyotypic diversification.
In principle, one can expect that the rate of chromosomal rearrangements should be correlated with reproductive characteristics. Reproduction is affected by many life-history traits such as sexual maturity age, number of litters per year and longevity. However, literature on this topic is scarce. In a recent series of reviews of karyotypic evolution of mammals, few references to reproductive characteristics or mating systems were made (Stanyon and Graphodatsky, 2012). The rationale of the Reproductive Rate Hypothesis is that chromosomal rearrangements are mutations that must be transmitted during reproduction through gametes to the next generation in order to have a chance to persist in a population. Gametes are produced by meiosis where DNA synthesis and repair occur at interphase and during crossing-over where chromosomal mutations may occur. Thus, the larger the number of meiosis tested through reproduction, the higher the probability of a chromosomal rearrangement being transmitted to the next generation.
However, our results indicate that both indices of chromosomal diversification show somewhat different responses to reproductive predictors. Although rKDmacro and rKDmicro show correlation with reproductive characteristics, significance is higher for the second index. In addition, rKDmicro shows a significant correlation with the litters per year × longevity interaction, and a marginally significant correlation with litters per year × sexual maturity age. Thus, reproductive factors seem to be more relevant to the evolutionary accumulation of chromosomal micro-rearrangements that are accounted for by rKDmicro. The reasons for this difference may be attributed to the differential behavior of macro- and microstructural chromosomal changes during meiosis: chromosomal mutations such as Robertsonian fusions/fissions or large inversions that visibly alter karyotypic structure usually present meiotic problems at the synaptic and orientation/segregation stages in heterozygosis (King, 1995). This abnormal behavior can produce gametic imbalance and loss of fertility, thus compromising their transmission to future generations and their establishment in a population (King, 1995). Micro-rearrangements on the contrary, although representing cryptic structural hybridity, are not expected to necessarily produce severe meiotic impairment (for example, small inversions may remain unsynapsed during pachytene and the problem of crossing-over within the inverted region is avoided) and their expectations of transmission are increased.
Our results might be explained as follows: more litters per year imply that more gametes (potential carriers of a chromosomal rearrangement) are involved, thus increasing the probability of a chromosomal mutation being transmitted and eventually established in a population. As explained before, those mutations that cause less meiotic problems (mainly micro-rearrangements) stand a better chance of persisting in time. As the number of litters per year and sexual maturity age are variables that also depend on longevity, two species with the same sexual maturity age and rates of reproduction but different longevities should show different probabilities of establishment of new chromosomal variants. The former explains the significant and marginally significant correlations of rKDmicro with litters per year × longevity and sexual maturity age × longevity respectively (Table 5). These correlations also make sense if it is considered that the genome is a life-history variable whose characteristics, such as mutation rate, scale with other life-history traits (Bromham, 2011). Thus, our results support the Reproductive Rate Hypothesis.
Although chromosomal changes are not a condition for speciation, it is widely acknowledged that chromosomal change may contribute to the establishment of reproductive isolation favoring the differentiation of incipient species (Faria and Navarro, 2010). Our analyses showed that a high rKDmacro is associated with families with wide geographical range but contain species with restricted distributions (for example, Muridae and Cricetidae), supporting the Geographic Range Hypothesis. This is made explicit by the highly significant correlation of rKDmacro with family geographic range × mean species geographic range (Table 4). The local chromosomal characteristics may diverge from their initial state through the action of adaptation and/or genetic drift. Different environments produce differential selective pressures, and thus if a chromosome rearrangement confers an adaptive advantage to carriers in a given environment, it could spread and eventually become fixed in a population (Bonvicino et al., 2001; Hendry, 2004; Kirkpatrick and Barton, 2006; Hooper and Price, 2015). In addition, species with fragmented distributions and small populations favor the chance fixation of chromosomal rearrangements by drift, and gene flow between populations that is a determinant factor in the fixation of local adaptive characteristics (Hooper and Price, 2015; Martinez et al., 2015). Highly vagile species tend to have more migrants between populations and, as a consequence, a lower probability of fixation of adaptive characteristics (Olson-Manning et al., 2012; Martinez et al., 2015). However, if the selective pressure on the trait is very strong, it can become fixed even in the presence of high gene flow (Coyne and Orr, 2004). This seems to be the case of some macrostructural chromosome changes. Rearrangements such as Robertsonian fusions and inversions can have very pronounced effects on meiotic recombination indices, sometimes protecting adaptive supergenes that can be beneficial in certain environments (Bidau et al., 2001; Lowry and Willis, 2010). Under this perspective, a larger geographical distribution of a family can expose its species to different selective pressures allowing the fixation of different rearrangements in different environments.
However, it is accepted that most chromosomal changes are not adaptive, but even in that case they may become fixed in different populations that could sometimes lead to speciation by postzygotic reproductive isolation (Rieseberg, 2001; Faria and Navarro, 2010). However, for this to occur it seems essential that populations with different fixed rearrangements are geographically isolated (Lande, 1985; Britton-Davidian et al., 2000). Populations are more susceptible of diverging genetically in allopatry, and under certain circumstances, accumulate chromosomal rearrangements (Faria and Navarro, 2010). In peripheral areas of species distributions, populations are more prone to differentiate. In these regions populations have the opportunity of colonizing and exploiting new environments or niches (Bush et al., 1977). If underdominant or neutral chromosomal rearrangements occur in these incipient species, they will only differentiate and expand if their distributions are strongly geographically isolated (Wright, 1951; Lande, 1985; Charlesworth et al., 2003). If not, they will coalesce with their more numerous parental population through hybridization, and the new mutation will be rapidly lost because it does not offer any adaptive advantage that could permit its permanence (Rieseberg, 2001). Characteristics of species such as low vagility may favor this diversification mechanism because this condition may have allowed exclusive karyotypes to become isolated and fixed in different localities that can eventually lead to speciation. Our results also support this situation where families that comprise species with restricted geographic range and possibly low vagility, like Ctenomyidae or Soricidae, show higher karyotypic diversity (Table 4).
Geographic range does not predict the degree of chromosomal diversification of rKDmicro as it occurs at the family level. This is expected as the present geographic distribution of a given species is the result of historical factors that probably lack any relation with its chromosomal evolution despite the fact that the same species could have started as a chromosomally differentiated population. At the family level, a large past geographic distribution in a heterogeneous environment could have favored chromosomal diversification as explained above. Conversely, a high rate of chromosomal diversification in the family's ancestors might have favored the expansion of its geographic range by allowing the colonization of new habitats.
In summary, our results showed that chromosomal evolution is non-neutral and is correlated with reproductive rate and geographic range, as expected by our predictions (Table 1). Nevertheless, mammalian karyotype diversification is extremely complex and it is influenced by historical and adaptive processes where reproductive and genetic factors modulate the rate of chromosomal changes. Mechanisms of chromosomal evolution have always been controversial and phylogenetically based large-scale studies such as the present one may help understand the factors that modulate chromosomal diversity across evolutionary time.
Data archiving
No data to deposit.
Acknowledgments
We are grateful to Alex Pyron, Juan Pablo Zurano and Gustavo Paterno for their criticism on a previous draft of this manuscript. CNPq supported PAM, TFA, RVF and CRF (PQ 305304/2013-5), and CAPES-PNPD supported UPJ.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
The authors declare no conflict of interest.
Supplementary Material
References
- Allen AP, Gillooly JF, Savage VM, Brown JH. (2006). Kinetic effects of temperature on rates of genetic divergence and speciation. Proc Natl Acad Sci USA 103: 9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai R. (2011) Fish Karyotypes. Springer Japan: Tokyo. [Google Scholar]
- Bailey JA, Baertsch R, Kent WJ, Haussler D, Eichler EE. (2004). Hotspots of mammalian chromosomal evolution. Genome Biol 5: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barja G, Herrero A. (2000). Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J 14: 312–318. [DOI] [PubMed] [Google Scholar]
- Bidau CJ, Giménez MD, Palmer CL, Searle JB. (2001). The effects of Robertsonian fusions on chiasma frequency and distribution in the house mouse (Mus musculus domesticus) from a hybrid zone in northern Scotland. Heredity 87: 305–313. [DOI] [PubMed] [Google Scholar]
- Blomberg SP, Garland T, Ives AR. (2003). Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57: 717–745. [DOI] [PubMed] [Google Scholar]
- Bonvicino CR, Dandrea PS, Borodin PM. (2001). Pericentric inversion in natural populations of Oligoryzomys nigripes (Rodentia: Sigmodontinae). Genome 44: 791–796. [DOI] [PubMed] [Google Scholar]
- Britton-Davidian J, Catalan J, da Graça Ramalhinho M, Ganem G, Auffray JC, Capela R et al. (2000). Rapid chromosomal evolution in island mice. Nature 403: 158. [DOI] [PubMed] [Google Scholar]
- Bromham L. (2011). The genome as a life-history character: why rate of molecular evolution varies between mammal species. Philos Trans R Soc Lond B Biol Sci 366: 2503–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. (2002) Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. 2nd edn. Springer Verlag: New York. [Google Scholar]
- Bush GL, Case SM, Wilson AC, Patton JL. (1977). Rapid speciation and chromosome evolution in mammals. Proc Natl Acad Sci USA 74: 3942–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MA, King AA. (2004). Phylogenetic comparative analysis: a modeling approach for adaptive evolution. Am Nat 164: 683–695. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. (2003). Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. (2004) Speciation. Sinauer Associates, Inc.: Sunderland, MA, USA. [Google Scholar]
- Dobigny G, Britton-Davidian J, Robinson TJ. (2015). Chromosomal polymorphism in mammals: an evolutionary perspective. Biol Rev doi:10.1111/brv.12213. [DOI] [PubMed]
- Dover G. (2002). Molecular drive. Trends Genet 18: 587–589. [DOI] [PubMed] [Google Scholar]
- Faria R, Navarro A. (2010). Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol 25: 660–669. [DOI] [PubMed] [Google Scholar]
- Faurby S, Svenning JC. (2015). A species-level phylogeny of all extant and late Quaternary extinct mammals using a novel heuristic-hierarchical Bayesian approach. Mol Phylogenet Evol 84: 14–26. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Phylogenies and the comparative. Am Nat 125: 1–15. [Google Scholar]
- Garagna S, Marziliano N, Zuccotti M, Searle JB, Capanna E, Redi CA. (2001). Pericentromeric organization at the fusion point of mouse Robertsonian translocation chromosomes. Proc Natl Acad Sci USA 98: 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish PJ, Colato A, Sniegowski PD. (2013). Genomic mutation rates that neutralize adaptive evolution and natural selection. J R Soc Interface 10: 20130329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graphodatsky A, Ferguson-Smith MA, Stanyon R. (2012). A short introduction to cytogenetics studies in Mammals with reference to present volume. Cytogenet Genome Res 137: 83–96. [DOI] [PubMed] [Google Scholar]
- Grafen A. (1989). The Phylogenetic regression. Philos Trans R Soc Lond B Biol Sci 326: 119–157. [DOI] [PubMed] [Google Scholar]
- Gregory TR. (2005). Synergy between sequence and size in large-scale genomics. Nat Rev Genet 6: 699–708. [DOI] [PubMed] [Google Scholar]
- Guerrero RF, Kirkpatrick M. (2014). Local adaptation and the evolution of chromosome fusions. Evolution 68: 2747–2756. [DOI] [PubMed] [Google Scholar]
- Harmon LJ, Weir TJ, Brock CD, Glor RE, Challenger W. (2008). GEIGER: investigating evolutionary radiations. Bioinformatics 24: 129–131. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. (2015). Tree of life reveals clock-like speciation and diversification. Mol Biol Evol 32: 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry AP. (2004). Selection against migrants contributes to the rapid evolution of ecologically dependent reproductive isolation. Evol Ecol Res 6: 1219–1236. [Google Scholar]
- Hooper DM, Price TD. (2015). Rates of karyotypic evolution in Estrildid finches differ between island and continental clades. Evolution 69: 890–903. [DOI] [PubMed] [Google Scholar]
- Jones KE, Bielby J, Cardillo M, Fritz SA, O'Dell J, Orme CDL et al. (2009). PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90: 2648. [Google Scholar]
- King M. (1995) Species Evolution: The Role of Chromosome Change. Cambridge University Press: Cambridge. [Google Scholar]
- Kirkpatrick M, Barton N. (2006). Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird CD, McConaughy BL, McCarthy BJ. (1969). Rate of fixation of nucleotide substitutions in evolution. Nature 224: 149–154. [DOI] [PubMed] [Google Scholar]
- Lande R. (1984). The expected fixation rate of chromosomal inversions. Evolution 743–752. [DOI] [PubMed]
- Lande R. (1985). The fixation of chromosomal rearrangements in a subdivided population with local extinction and colonization. Heredity 54: 323–332. [DOI] [PubMed] [Google Scholar]
- Li WH, Ellsworth DL, Krushkal J, Chang BH, Hewett-Emmett D. (1996). Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis. Mol Phylogenet Evol 5: 182–187. [DOI] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. (2010). A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol 8: E1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison DR, Schulz K, Maddison WP. (2007). The Tree of Life Web Project. Zootaxa 1668: 19–40. [Google Scholar]
- Martin AP, Palumbi SR. (1993). Body size, metabolic rate, generation time, and the molecular clock. Proce Natl Acad Sci USA 90: 4087–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez PA., Zurano JP, Amado TF, Penone C, Betancur-R R, Bidau CJ et al. (2015). Chromosomal diversity in tropical reef fishes is related to body size and depth range. Mol Phylogenet Evol 93: 1–4. [DOI] [PubMed] [Google Scholar]
- Meredith RW, Janečka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC et al. (2011). Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334: 521–524. [DOI] [PubMed] [Google Scholar]
- Molina WF, Martinez PA, Bertollo LAC, Bidau CJ. (2014). Evidence for meiotic drive as an explanation for karyotype changes in fishes. Mari Genomics 15: 29–34. [DOI] [PubMed] [Google Scholar]
- Munshi-South J, Wilkinson GS. (2010). Bats and birds: exceptional longevity despite high metabolic rates. Ageing Res Rev 9: 12–19. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Menninger JC, Nash WG. (2006) Mammalian Chromosomes. John Wiley & Sons, Inc.: Hoboken, NJ, USA. [Google Scholar]
- Olson-Manning CF, Wagner MR, Mitchell-Olds T. (2012). Adaptive evolution: evaluating empirical support for theoretical predictions. Nat Rev Genet 13: 867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ CL, Moreno RG, Edwards SV. (2008). Three tiers of genome evolution in reptiles. Integr Comp Biol 48: 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici M, Santini L, Di Marco M, Baisero D, Francucci L, Grottolo Marasini G et al. (2013). Generation length for mammals. Nat Conserv 5: 89–94. [Google Scholar]
- Pagel M. (1999). Inferring the historical patterns of biological evolution. Nature 401: 877–884. [DOI] [PubMed] [Google Scholar]
- Penone C, Davidson AD, Shoemaker KT, Di Marco M, Rondinini C, Brooks TM et al. (2014). Imputation of missing data in life-history trait datasets: which approach performs the best? Methods Ecol Evol 280: 20131092. [Google Scholar]
- Pieńkowska-Schelling A, Schelling C, Zawada M, Yang F, Bugno M, Ferguson-Smith M. (2008). Cytogenetic studies and karyotype nomenclature of three wild canid species: maned wolf (Chrysocyon brachyurus), bat-eared fox (Otocyon megalotis) and fennec fox (Fennecus zerda). Cytogenet Genome Res 121: 25–34. [DOI] [PubMed] [Google Scholar]
- Revell LJ. (2013). Two new graphical methods for mapping trait evolution on phylogenies. Methods Ecol Evol 4: 754–759. [Google Scholar]
- Rieseberg LH. (2001). Chromosomal rearrangements and speciation. Trends Ecol Evol 16: 351–358. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, Grundler M, Anderson C, Title P, Shi JJ, Brown JW et al. (2014). BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol Evol 5: 701–707. [Google Scholar]
- Rolland J, Condamine FL, Jiguet F, Morlon H. (2014). Faster speciation and reduced extinction in the tropics contribute to the mammalian latitudinal diversity gradient. PLoS Biol 12: e1001775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland J, Salamin N. (2016). Niche width impacts vertebrate Diversification. Global Ecol Biogeogr 25: 1252–1263. [Google Scholar]
- Ross L, Blackmon H, Lorite P, Gokhman VE, Hardy NB. (2015). Recombination, chromosome number and eusociality in the Hymenoptera. J Evol Biol 28: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Chen Z, Hine C, Sasahara THC, Antonio AC, Ribeiro M et al. (2009). Telomerase activity coevolves with body mass not lifespan. Aging Cell 6: 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanyon R, Graphodatsky A. (2012). Evolutionary dynamics of mammalian karyotypes. Cytogenet Genome Res 137: 83–285. [DOI] [PubMed] [Google Scholar]
- Stekhoven DJ, Bühlmann P. (2012). MissForest—nonparametric missing value imputation for mixed-type data. Bioinformatics 28: 112–128. [DOI] [PubMed] [Google Scholar]
- Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, Taranukha D et al. (2013). Human Ageing Genomic Resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res 41: D1027–D1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, Teti M et al. (2004). Recurrent sites for new centromere seeding. Genome Res 14: 1696–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde Arregoitia L, Blomberg SP, Fisher DO. (2013). Phylogenetic correlates of extinction risk in mammals: species in older lineages are not at greater risk. Proc R Soc B 280: 20131092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villena FP, Sapienza C. (2001). Female meiosis drives karyotypic evolution in mammals. Genetics 159: 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber C, Ponting CP. (2005). Hotspots of mutation and breakage in dog and human chromosomes. Genome Res 15: 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJD. (1978. a). Chain processes in chromosomal speciation. Syst Zool 27: 285. [Google Scholar]
- White MJD. (1978. b) Modes of Speciation. WH Freeman: San Francisco. [Google Scholar]
- Wienberg J, Stanyon R. (1997). Comparative painting of mammalian chromosomes. Curr Opin Genet Dev 7: 784–791. [DOI] [PubMed] [Google Scholar]
- Wienberg J, Stanyon R. (1998). Comparative chromosome painting of primate genomes. ILAR J 39: 77–91. [DOI] [PubMed] [Google Scholar]
- Wilman H, Belmaker J, Simpson J, de la Rosa C, Rivadeneira MM, Jetz W. (2014). EltonTraits 1.0: species-level foraging attributes of the world's birds and mammals. Ecology 95: 2027–2027. [Google Scholar]
- Wright S. (1951). The genetical structure of populations. Ann Eugen 15: 323–354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.