Abstract
Background and Purpose
The aim of the present study was to investigate the effects and possible underlying mechanisms of dioscin against pancreatic cancer in vitro and in vivo.
Experimental Approach
In vitro actions of dioscin on viability of ASPC‐1 and PANC‐1 cells, and in vivo effects to suppress the tumour growth of cell xenografts in nude mice were assessed. In addition, microRNA microarray analysis determined which microRNAs were affected by dioscin. The mechanisms underlying the actions of dioscin against pancreatic cancer were elucidated in terms of Akt1 and other proteins related to aopoptosis.
Key Results
Dioscin markedly induced apoptosis and significantly suppressed the tumour growth of ASPC‐1 and PANC‐1 cell xenografts, in nude mice. Total of 107 microRNAs with differential changes were found, in which miR‐149‐3P targeted with Akt1 was markedly up‐regulated by dioscin. Further studies showed that dioscin significantly down‐regulated Akt1 levels, and thus induced cell apoptosis by increasing the levels of Bax, Apaf‐1, cleaved caspase‐3/9, cleaved PARP, suppressing Bcl‐2 levels, and causing cytochrome c release. The effects of an inhibitor of miR‐149‐3P and of siRNA of testicular Akt1 suggested that dioscin showed excellent activity against pancreatic cancer via miR‐ 149‐3P‐mediated inhibition of Akt1 signalling pathway.
Conclusions and Implications
Collectively, these findings confirmed the potent effects of dioscin against pancreatic cancer and also provided novel insights into the mechanisms of the compound as a potential candidate for the treatment of pancreatic cancer.
Abbreviations
- AKT1
protein kinase B
- AO
acridine orange
- Apaf-1
apoptotic protease activating facter‐1
- Bax
Bcl‐2‐associated X protein
- Bcl‐2
B‐cell leukemia 2 protein
- Caspase‐3
cysteinyl aspartate spcific proteinase‐3
- Caspase‐9
cysteinyl aspartate spcific proteinase‐9
- DAPI
4',6'‐Diamidino‐2‐phenylindole
- Dio
dioscin
- EB
ethidium bromide
- GCB
gemcitabine
- H&E
hematoxylin‐eosin
- miRNAs
microRNAs
- MTT
3‐(4,5‐ dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolinum bromide
- mut
mutant
- PARP
poly (ADP‐ribose) polymerase
- PI
Propidium Iodide
- siRNA
small interfering RNA
- TUNEL
in situ terminal deoxynucleotidyl transferase dUTP nick-end labeling
- Wt
wild‐type
Tables of Links
| LIGANDS |
|---|
| Dioscin |
| Gemcitabine |
These Tables list key protein targets and ligands in this article that are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Southan et al., 2016), and are permanently archived in the Concise Guide to PHARMACOLOGY 2015/16 (a,bAlexander et al., 2015a,b).
Introduction
Pancreatic cancer, one of the most lethal solid malignancies, is a frequent cancer‐related cause of death in the Western world (Werner et al., 2013). Although considerable progresses have been made in improving cancer survival rates over the past few decades, the 5 year survival rate remains at only 5% (Siegel et al., 2013). The American Cancer Society estimates that 45 220 Americans were diagnosed with pancreatic cancer in 2013, and 38 460 people died from the disease (Wolfgang et al., 2013). In China, the incidence of pancreatic cancer is 7.28/100 000, ranking the seventh among all cancers according to the reports of the National Central Cancer Registry of China. Only in 2011, the estimated number of newly diagnosed pancreatic cancer cases and deaths were 80 344 and 72 723 respectively (He et al., 2015). Current chemotherapy protocols can produce modest survival benefits to treat pancreatic cancer (Morran et al., 2014). Treatment with gemcitabine has modest clinical benefits and yields a marginal survival advantage for patients. However, the median survival of patients with metastatic pancreatic cancer remains less than 6 months (Burris et al., 1997). Thus, development of effective therapeutic methods to treat pancreatic cancer is important.
The occurrence of pancreatic cancer is complex. Mammalian cells express three evolutionarily conserved, highly homologous serine/threonine kinases Akt isoforms (Akt1–3). Akt is hyperactivated in the majority of human cancers, which is related to apoptosis resistance and increased cell proliferation, growth and energy metabolism (Hay, 2005). Akt isoforms are frequently activated in pancreatic cancer, and the hyperactivation of Akt1 has been implicated in pancreatic cancer formation (Albury et al., 2015), suggesting that there may be a therapeutic opportunity to treat pancreatic cancer through inhibiting AktT1. However, the data available on the exact mechanisms of action of these isoforms on pancreatic cancer are limited.
MicroRNAs (miRNAs) play important roles in regulating the translation and degradation of mRNAs (Lau et al., 2001; Lee and Ambros, 2001). miRNAs are involved in many biological processes including cell death, cell proliferation, stress resistance and fat metabolism (Ambros, 2003). With the emergence of miRNAs as the key players in the occurrence of diseases, development of sensitive, rapid and quantitative methods of detection of miRNA is of great interest. Chip‐based devices have been used to detect ultra‐trace analytes (Hou et al., 2014), and a nucleic acid‐based microarray has been widely used for high‐throughput detecting and profiling miRNA targets (Chen et al., 2010; Liu et al., 2012; Roy et al., 2016).
Currently, some drugs including oxaliplatin, irinotecan, leucovorin and 5‐ fluorouracil have achieved survival benefits for pancreatic cancer patients compared with gemcitabine. However, the increased toxicities limit their clinical applications (Neesse et al., 2014). Thus, exploration of novel drugs with high efficacy and low toxicity for the treatment of pancreatic cancer is urgent.
As the essence of Chinese traditional culture, traditional Chinese medicines (TCMs) with low toxicity and high efficiency have been widely used to protect health and control diseases in China for thousands of years (Xu et al., 2014). Some natural products including resveratrol, curcumin and bufalin extracted from medicinal herbs have active effects against pancreatic cancer (Liu et al., 2013; Kanai, 2014; Li et al., 2014). Thus, exploration of effective natural products from medicinal plants to treat pancreatic cancer is reasonable.
Dioscin (Supporting Information Figure S1), a typical natural product, is an active ingredient in some medicinal plants (Lu et al., 2012). Pharmacological studies have shown that dioscin can ameliorate cerebral ischaemia/reperfusion injury (Tao et al., 2015), induce autophagy (Hsieh et al., 2013) and regulate hyperlipidaemia (Li et al., 2010). It also has anti‐inflammatory (Wang et al., 2007), anti‐fungal (Cho et al., 2013), anti‐virus (Aquino et al., 1991), anti‐liver fibrosis (Zhang et al., 2015; Gu et al., 2016), anti‐obesity (Liu et al., 2015) and hepatoprotective activities (Lu et al., 2012; Zhao et al., 2012). In addition, dioscin shows active effects against colon cancer, lung cancer, hepatocarcinoma, esophageal cancer, laryngeal cancer and glioblastoma multiforme (Wang et al., 2012a,b; Wei et al., 2013; Chen et al., 2014; Si et al., 2016). Furthermore, dioscin can down‐regulate the level of peroxiredoxin 1 and induce ROS‐mediated apoptosis in MiaPaCa‐2 cells (Zhao et al., 2014), suggesting that dioscin might exert potent effects against pancreatic cancer. However, earlier experiments have not examined the effects of dioscin on the human pancreatic cancer cell lines ASPC‐1 and PANC‐1. Moreover, the in‐depth mechanisms and in vivo anti‐pancreatic cancer activity of the compound have not yet been studied.
Therefore, the aim of the present study was to investigate the effects and the possible mechanisms of dioscin against pancreatic cancer in vitro and in vitro. The findings could provide novel insights or lead to the development of a potent candidate for preventing and treating pancreatic cancer.
Methods
Cell culture
The human HPDE6‐C7 cell line was purchased from Saiqi Biological Engineering Co., Ltd. (Shanghai, China). The human ASPC‐1 and PANC‐1 cell lines were purchased from Shanghai Institutes for Biological Sciences (Shanghai, China). These three cell lines were cultured in high‐glucose DMEM supplemented with 10% FBS, 100 units mL−1 penicillin and 100 μg·mL−1 streptomycin. Cultures were maintained at 37°C in a humidified incubator containing 5% CO2 for experiments.
MTT assay and morphological changes
The cells were seeded at a density of 5 × 104 cells mL−1 in 96‐well plates in growth medium supplemented with 10% serum at 37°C with 5% CO2 overnight and incubated. Next, 100 μL of medium was removed, and 100 μL of sample solution with various concentrations of dioscin (1.4, 2.9 and 5.8 μM) was added. Then, the cells were incubated for 6, 12 or 24 h. Subsequently, 15 μL of MTT stock solution was added, and the plates were incubated for another 4 h. Following this 4 h incubation at 37°C, 100 μL of DMSO was added to dissolve the formazan crystals. The absorbance was measured at 490 nm with a microplate reader (Thermo, USA), and the data were normalized to control unwanted sources of variation. In addition, cell morphology was imaged using a phase contrast microscope (Nikon, Japan).
Staining wth fluorescent dyes
The ASPC‐1 and PANC‐1 cells were cultured in 6‐well plates for 24 h and then treated with different concentrations of dioscin (1.4, 2.9 and 5.8 μM) for 24 h. After incubation, the cells were washed twice with PBS. Then, 1 mL of a solution containing the same volume of acridine orange (AO; 1.0 μg·mL−1) and ethidium bromide (EB; 1.0 μg·mL−1) was added to the cells according to Leite et al., (1999). The stained cells were observed under a fluorescence microscope (OLYMPUS, Japan). DAPI staining was performed after dioscin treatment as described above. The cells were washed twice with PBS, fixed with 10% formaldehyde for 10 min at room temperature, and washed again in PBS for three times. The cells were then stained with DAPI (1.0 μg·mL−1) solution for 20 min at 37°C. Finally, the images were obtained using a fluorescence microscope (OLYMPUS, Japan).
Apoptosis assay
After treatment with different concentrations of dioscin (1.4, 2.9 and 5.8 μM) for 24 h, the cells and supernatant were collected. The collected cells were washed twice with ice‐cold PBS, and stained with Annexin V‐FITC and propidium iodide (PI) in binding buffer at room temperature for 30 min in the dark according to the manufacturer's instructions. The samples were analysed using flow cytometry (Becton‐Dickinson, USA).
Caspase inhibitor experiments
The ASPC‐1 and PANC‐1 cells were randomly divided into control groups, control inhibitor groups and dioscin groups (here the cells were treated with 5.8 μM of dioscin for 24 h). The caspase inhibitor group (cells treated with 50 μM of Z‐VAD‐FMK for 30 min), and the caspase inhibitor plus dioscin group (cells treated with 50 μM of Z‐VAD‐FMK for 30 min and then incubated with 5.8 μM of dioscin for 24 h). The cell viability was determined according to the MTT method described above, and the data were normalized to control unwanted sources of variation.
Transfection of ASPC‐ and PANC‐1 cells
The luciferase genes were transduced into ASPC‐1 and PANC‐1 cells using the pLEX system with lentiviral vectors containing a puromycin resistance gene (Genechem Co., Ltd., China). The cells were seeded in 6‐well plates, cultured with DMEM with 10% FBS for 24 h, and transduced with 50 μg/mL of lentivirus in each well for 12 h. The stably transduced ASPC‐1 or PANC‐1 cells were obtained by puromycin selection in the presence of 2 μg·mL−1 of puromycin for 2 weeks. The surviving colonies were cultured independently. The ASPC‐1‐luc and PANC‐1‐luc cells were harvested, and then the transduced cells were screened through bioluminescence. The ASPC‐1‐luc and PANC‐1‐luc cells with the highest luciferase activities were used for the in vivo study.
Animal models and experimental protocols
All animal care and experimental procedures were approved by the Animal Care and Use Committee of Dalian Medical University and performed in strict accordance with the People's Republic of China Legislation Regarding the Use and Care of Laboratory Animals. Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny et al., 2010; McGrath & Lilley, 2015).
The BALB/c nude mice (15–20 g;4–6 weeks old) were obtained from the Experimental Animal Center at Dalian Medical University (Dalian, China) (SCXK: 2013–0003). All animals were housed in a controlled environment at 23 ± 2°C under a 12 h dark/light cycle with free access to food and water. Optimized ASPC‐1‐luc and PANC‐1‐luc cells (5 × 105) were suspended in 0.2 mL of PBS, which were injected subcutaneously into the underarm regions of the mice.
We began the treatment on day 5 post‐inoculation when the tumorigenic rate in each group was 100%. The models of BALB/c nude mice injected with ASPC‐1‐luc and PANC‐1‐luc cells did not vary with individual differences. Therefore, we adopted the randomized method instead of blinding, which has been usually applied in highly subjective clinical trials. In this paper, the mice were randomly divided into control, gemcitabine ‐treated, low‐dose (40 mg·kg−1) of dioscin‐treated and high‐dose (80 mg·kg−1) of dioscin‐treated groups (n = 5). In the animal experiments, the concentrations of dioscin were prepared as solutions of 4.0 and 8.0 mg·mL−1, and gemcitabine was prepared as a solution of 5.0 mg·mL−1 The animals in control and gemcitabine‐treated groups were treated with 0.5% CMC‐Na and 50 mg·kg−1 of gemcitabine (10 mL·kg−1), and the mice in dioscin‐treated groups were treated with 40 or 80 mg·kg−1 of dioscin (10 mL·kg−1). The drug was administered by gavage once daily for 26 days. As reported by Workman et al., (2010), the tumour burden did not exceed the recommended maximum diameter (1.5 cm in therapeutic studies). The tumour volume was measured every 3 days and calculated using the following formula: V = A × B2/2, where A and B represent the maximum and minimum diameters. The tumour inhibition (%) was calculated based on the formula (tumour volume of the control group‐tumour volume of the treatment group)/tumour volume of the control group ×100. At the end of the test, bioluminescent images of the mice were obtained using an IVIS 200 Imaging System (IVIS 200, Xenogen Corp., Alameda, USA) through the signals emitted from the transfected cells. Then, the animals were killed, and the tumours were photographed and weighed.
Histopathological and immunofluorescence assays
Formalin‐fixed tumour tissues were embedded in paraffin and cut into 5 μm sections. The sections were stained with haematoxylin–eosin (H&E). Images of the stained sections were obtained using a light microscope (Nikon Eclipse TE2000‐U, Japan) with 200× magnification. Immunofluorescence staining for Akt1 in tissue sections or cells was performed using anti‐Akt1 antibody in a humidified box at 4°C overnight, and followed by incubation with an Alexa fluorescein‐labelled secondary antibody for 1 h at 37°C. The cell nuclei were stained with DAPI (5.0 μg·mL−1). Immunostained samples were imaged by fluorescence microscopy (Olympus, Japan) at 200× magnification.
miRNA microarray assay
RNA extraction
The ASPC‐1 cells were treated with dioscin (5.8 μM) for 24 h. The total RNA samples from dioscin‐treated and untreated cells were isolated with TRIzol reagent (Invitrogen, USA) and purified using RNeasy mini kit (QIAGEN) according to the manufacturer's instructions. RNA quality and quantity were measured using a Nanodrop spectrophotometer (ND‐1000, Nanodrop Technologies, Rockland, DE, USA), and RNA integrity was assessed by gel electrophoresis.
miRNA labelling and array hybridization
RNA labelling and array hybridization were carried out according to Exiqon's manual. After quality control, the miRCURY™ Hy3™/Hy5™ Power labelling kit (Exiqon, Vedbaek, Denmark) was used for miRNA labelling as follows. (i) Approximately 1.0 μL of RNA in 2.0 μL of water was combined with 1.0 μL of CIP and CIP buffer (Exiqon, Vedbaek, Denmark), and the mixture was incubated at 37°C for 30 min. (b) The reaction was terminated by incubation at 95°C for 5 min. Then, 3.0 μL of labelling buffer, 2.0 μL of DMSO, 2.0 μL of labelling enzyme, and 1.5 μL of fluorescent label (Hy3™) were added to the mixture. The labelling reaction was incubated at 16°C for 1 h. (iii) The reaction was terminated by incubation at 65°C for 15 min. After stopping the labelling procedure, the Hy3™‐labelled samples were hybridized using the miRCURYTM LNA Array (v.18.0) (Exiqon, Vedbaek, Denmark) according to the array manual as follows. (i) A total of 25.0 μL of the mixture consisting of the Hy3™‐labelled samples with 25.0 μL hybridization buffer was first denatured at 95°C for 2 min and incubated on ice for 2 min. (ii) Then, the samples were hybridized to the microarray for 16–20 h at 56°C in a 12‐Bay Hybridization System (Hybridization System‐Nimblegen Systems, Inc., Madison, WI, USA). (iii) Following hybridization, the slides were obtained and washed several times using a Wash buffer kit (Exiqon, Vedbaek, Denmark). Finally, the slides were scanned using an Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA).
Analysis of array data
The scanned images were then imported into GenePix Pro 6.0 software (Axon) for data extraction and grid alignment. Replicated miRNAs were averaged, and miRNAs with intensities ≥30 in all samples were selected for calculation of the normalization factor. The expressed data were normalized using the median normalization. After normalization, significantly differentially expressed miRNAs between two groups were identified through P‐values and fold changes. Finally, hierarchical clustering was performed to produce the distinguishable miRNA expression profiling of the samples.
Quantitative real‐time PCR assay
The total miRNA samples were extracted from ASPC‐1 cells, PANC‐1 cells and tumour tissues using SanPrep Column microRNA Mini‐Preps Kit (Sangon Biological Engineering Technology & Services Co., Ltd., China) following the manufacturer's protocol. The purity of the extracted miRNA was determined, and the RT‐PCR was carried out using a microRNA First Strand cDNA Synthesis Kit following the manufacturer's instructions with a TC‐512 PCR system (TECHNE, UK). Quantitative real‐time PCR assay was performed with a MicroRNAs Quantitation PCR Kit (Sangon Biological Engineering Technology & Services Co., Ltd., China) and ABI 7500 Real Time PCR System (Applied Biosystems, USA). For the data of each sample, the Ct value of the target gene was normalized to that of U6 (Sangon Biological Engineering Technology & Services Co., Ltd., China). The unknown template was calculated using the standard curve for quantitative analysis. Eventually, the expression level of each gene was performed and the data were normalized to control unwanted sources of variation.
Dual luciferase reporter assay
The plasmids containing the wild‐type miR‐149‐3P‐ Akt1 response element (wt‐ Luc‐Akt1) and the corresponding mutant (mut‐Luc‐Akt1) were purchased from GenePharma Corp. (Shanghai, China). Plasmid DNA (wt‐Luc‐Akt1, mut‐Luc‐Akt1or control vector) and miR‐149‐3P mimics or miR‐149‐3P negative control were co‐ transfected into ASPC‐1 and PANC‐1 cells. The miR‐149‐3P mimic sequences are listed in Supporting Information Table S1. When appropriate, the cells were incubated with dioscin (5.8 μM) or without dioscin for 24 h after transfection. The luciferase activity was assessed with a Double‐Luciferase Reporter Assay Kit (TransGen Biotec, China) using the Dual‐Light Chemiluminescent Reporter Gene Assay System (Berthold, Germany), which was normalized to Renilla luciferase activity.
Western blotting assay
The total, mitochondrial, and cytoplasmic proteins from ASPC‐1 cells, PANC‐1 cells and tumour tissues were extracted following standard protocols according to the manufacturer's protocol, and the protein concentration was determined using a BCA protein assay kit. Proteins were loaded onto SDS‐PAGE (10–15%) gels, separated electrophoretically, and transferred to PVDF membranes (Millipore, USA). After blocking with 5% dried skim milk for 3 h at room temperature, the membranes were incubated overnight at 4°C with primary antibodies (listed in Supporting Information Table S2). After incubation with horseradish peroxidase‐conjugated antibodies for 2 h at room temperature, an enhanced chemiluminescence method was used for detection, and images were acquired using a Bio‐Spectrum Gel Imaging System (UVP, USA). Bands were normalized with GAPDH as an internal control.
TUNEL assay
The TUNEL assay was performed using the In Situ Cell Death Detection Kit (TMR Red, Roche, NJ, USA) according to the manufacturer's instructions. Briefly, the ASPC‐1 and PANC‐1 cells were treated with or without dioscin after transfection with miR‐149‐3P inhibitor or Akt1 siRNA. The fluorescein (green)‐labelled dUTP solution was then added. The cells were incubated at 37°C for 2 h and washed in PBS. Finally, the images were obtained using fluorescence microscopy (Olympus BX63, Japan), and the apoptotic cells were evaluated as the average number of positive cells (green spots) in the fluorescent images.
Transfection with MiR‐149‐3P inhibitor or Akt1 siRNA
Transfection was performed to down‐regulate the expression levels of miR‐149‐3P and Akt1. The miR‐149‐3P inhibitor and Akt1 siRNA sequences are listed in Supporting Information S3. Briefly, the miR‐149‐3P‐targeted inhibitor and control inhibitor were separately dissolved in Opti‐MEM. The solutions were then equilibrated for 5 min at room temperature. Each solution was combined with Lipofectamine 2000 transfection reagent according to the manufacturer's protocol. Then, the solution was mixed gently and allowed to form inhibitor liposomes for 20 min. The ASPC‐1 and PANC‐1 cells were transfected with the transfection mixture in antibiotic‐free cell medium. Cell viability, cell apoptosis, and the expression levels of Akt1, Bcl‐2, Bax, cytochrome c, Apaf‐1, cleaved caspase‐3/9 and cleaved PARP were detected after 24 h of transfection in the absence or presence of dioscin (5.8 μM) for an additional 24 h. In addition, the Akt1 siRNA transfection test was also carried out using the same procedure as described above.
Data and statistical analysis
The data and statistical analysis in this study comply with the recommendations on experimental design and analysis in pharmacology (Curtis et al., 2015). The data are expressed as the mean ± SD. Some data were normalized to control unwanted sources of variation, according to the following standard procedures. First, any outliers in different groups were removed from the data. Then, the data was normalized to bring all of the variation into proportion with one another. Technically though, the coefficients associated with each variable will scale appropriately to adjust for the disparity in the variable sizes. Furthermore, statistical analysis was performed with GraphPad Prism 5.0 software (San Diego, CA, USA) and only when a minimum of n = 5 independent samples was acquired. It was performed with one‐way ANOVA followed by Tukey's post hoc test when comparing multiple groups; with unpaired t‐test when comparing only two different groups. Post hoc tests were performed when ANOVAs indicated that a significant difference existed between groups. Statistical significance was considered to be P < 0.05.
Materials
Dioscin was purchased from Shanghai Tauto Biochemical Technology Co., Ltd. (Shanghai, China), and was dissolved in 0.1% DMSO for in vitro experiments or in 0.5% carboxymethylcellulose sodium (CMC‐Na) solution for in vivo experiments. Tissue Protein Extraction Kit was obtained from KEYGEN Biotech. Co., Ltd. (Nanjing, China). The bicinchoninic acid (BCA) Protein Assay Kit, Apoptosis Analysis Kits, Annexin V‐FITC Apoptosis Detection Kit were all purchased from Beyotime Institute of Biotechnology (Jiangsu, China). 3‐(4,5‐dimethylthiazol‐2‐yl)‐ 2,5‐diphenyltetra‐zolinum bromide (MTT) was obtained from Roche Diagnostics (Basel, Switzerland). The caspase inhibitor (Z‐VAD‐FMK) was obtained from Selleck Chemicals (Houston, TX, USA). Acridine orange (AO) and ethidium bromide (EB) fluorescent dyes, DAPI, Tris, SDS, CMC‐Na were purchased from Sigma (St. Louis, MO, USA). MiR‐149‐3P inhibitor, miR‐149‐3P mimics, Akt1 siRNA, SanPrep Column MicroRNA Mini‐Preps Kit, MicroRNA First Strand cDNA Synthesis Kit and MicroRNAs Quantitation PCR Kit were purchased from Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). Gemcitabine purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China) was used as the positive control in vivo experiments.
Results
Effects of dioscin on the viability and morphology of cancer cells
The human HPDE6‐C7, ASPC‐1, and PANC‐1 cells were treated with different concentrations of dioscin for different treatment times. The MTT results (Figure 1A) show the inhibitory effects of dioscin on ASPC‐1 and PANC‐1 cells in a dose‐ and time‐dependent manner. Dioscin (5.8 μM for 24 h) significantly decreased cell viability of ASPC‐1 and PANC‐1 cells to about 40%. However, in HPDE6‐C7 cells, a normal human pancreatic ductal epithelial cell line, the same conditions of incubation with dioscin reduced cell viability to about 70%. As shown in Figure 1B, the bright‐field images show the visible signs of cell death caused by dioscin. With AO/EB staining, the viable cells were bright green, while the apoptotic cells show orange fluorescence, which increased with the increased concentrations of dioscin (Figure 1C). DAPI staining showed that the chromatin in cell nuclei was condensed, and the nuclear apoptotic bodies were formed in dioscin‐treated groups (Figure 1D, as indicated by the arrow). The damage of the cancer cells caused by dioscin was also visible, using electron microscopy (Supporting Information Figure S2). The bright‐field images, AO/EB and DAPI staining of human HPDE6‐C7 cells treated by dioscin (1.4, 2.9 and 5.8 μM) are shown in Supporting Information Figure S3.
Figure 1.
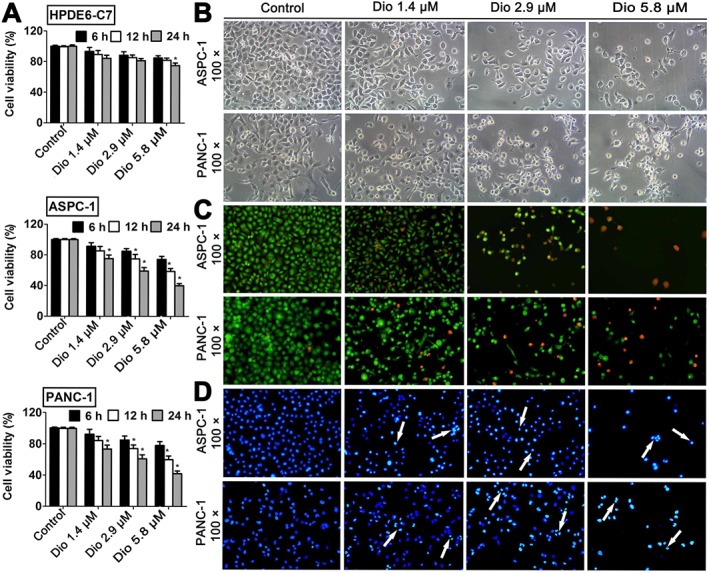
Effects of dioscin on ASPC‐1, PANC‐1 and HPDE6‐C7 cells. (A) Effects of dioscin on viability of ASPC‐1, PANC‐1 and HPDE6‐C7 cells, measured by the MTT assay. (B) Effects of dioscin (1.4, 2.9 and 5.8 μM) for 24 h on ASPC‐1 and PANC‐1 cell morphology and structures (bright‐field image, 100× magnification). (C) Fluorescence images of ASPC‐ 1 and PANC‐1 cells stained by AO/EB (100× magnification). (D) Fluorescence images of ASPC‐1 and PANC‐1 cells stained by DAPI (100× magnification). The data are presented as the mean ± SD (n = 5). *P < 0.05, significantly different from control groups.
Dioscin induces apoptosis of cancer cells
As shown in Figure 2A,B, dioscin induced apoptosis in human ASPC‐1 and PANC‐1 cells, compared with the control groups. This increased apoptosis after 24h incubation with dioscin was clearly concentration‐dependent. In addition, compared with the control groups, dioscin markedly decreased the viabilities of ASPC‐1 and PANC‐1 cells (P < 0.05; Figure 2C). However, the cell viabilities in Z‐VAD‐FMK and dioscin plus Z‐VAD‐FMK groups were not decreased (P > 0.05).
Figure 2.
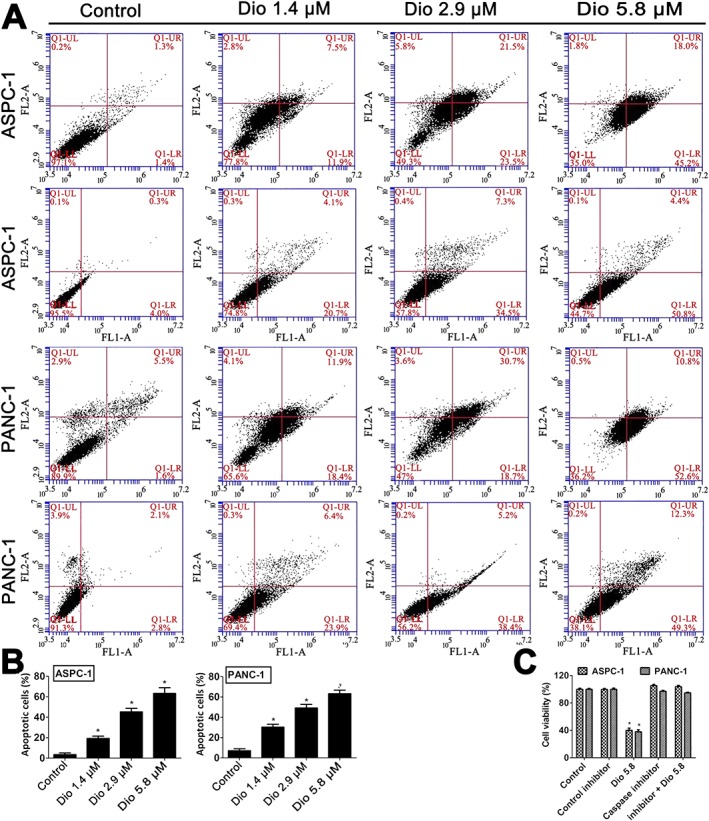
Effects of dioscin on apoptosis of ASPC‐1 and PANC‐1 cells. (A,B) Effects of dioscin on apoptosis of ASPC‐1 and PANC‐1 cells using flow cytometry analysis. (C) Effects of dioscin on the viability of ASPC‐1 and PANC‐1 cells using MTT assay, with or without the caspase inhibitor, Z‐VAD‐FMK, in vitro. The data are presented as the mean ± SD (n = 5). *P < 0.05, significantly different from control groups.
Dioscin inhibits tumour growth of cell xenografts in nude mice
As shown in Figure 3A,B and Supporting Information Figures S4 and S7, significant differences in tumour volume were found based on bioluminescence imaging and the tumour images after 26 days of treatment. The mean tumour volumes (Figure 3C), after treatment with dioscin (40 and 80 mg·kg−1) or gemcitabine, were decreased in nude mice transplanted with ASPC‐1 cells and in those transplanted with PANC‐1 cells. The tumour weights were also significantly decreased after treatment (Figure 3D). Moreover, the numbers of tumour cells, based on H&E staining, were lower in dioscin‐treated groups, compared with the large number of tumour cells were observed in the control groups, (Figure 3E). Overall, these results suggested that dioscin inhibited tumour growth in vivo.
Figure 3.
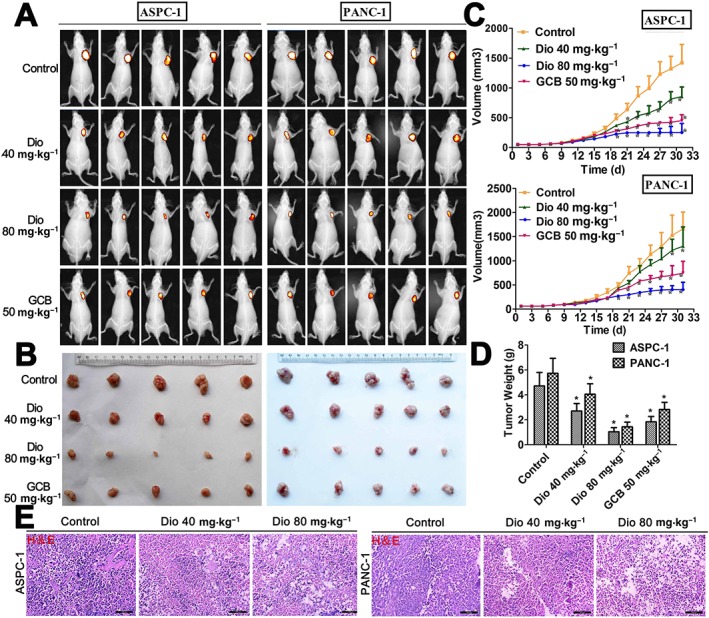
In vivo anti‐cancer effects of dioscin in nude mice. (A) Bioluminescence imaging of the mice. The raw images are shown in Supporting Information Figures S4 and S5. (B) Images of the tumours collected from the mice. The raw images are shown in Supporting Information Figures S6 and S7. (C) Effects of dioscin on tumour volume. The tumour volume was calculated using the formula: V = A × B2/2. (D) Effects of dioscin on tumour weight. (E) Effects of dioscin on tumour histopathology based on H&E staining (200× magnification). The data are presented as the mean ± SD (n = 5). *P < 0.05, significantly different from control groups.
Differentially expressed miRNAs caused by dioscin
A total of 107 differentially expressed miRNAs with at least twofold changes and P< 0.05, compared with the control group were identified in the dioscin‐ treated group using microRNA microarray analysis (the scan chips are shown in Supporting Information Figures S8 and S9, and the differentially expressed miRNAs are listed in Supporting Information Table S4). Of these, 48 miRNAs were down‐regulated and 59 miRNAs were up‐regulated. The heat map (Figure 4A,B) indicated the results of a two‐way hierarchical clustering of the samples and 107 differentially expressed miRNAs, which displayed the relative expression levels of miRNAs identified by microarray analyses. Specifically, six up‐regulated and five down‐regulated miRNAs were further validated by real‐time PCR assay. As shown in Figure 5A, dioscin significantly increased the levels of miR‐4795‐5p, miR‐4255, miR‐4299, miR‐4533, miR‐149‐3p, miR‐638, and markedly decreased the levels of miR‐4638‐5p, miR‐4284, miR‐668‐3p, miR‐4288 and miR‐191‐5p in ASPC‐1 and PANC‐1 cells. In particular, the levels of miR‐149‐3P were significantly up‐regulated by dioscin (5.8 μM) with 9.5‐ and 9.0‐fold in ASPC‐1 and PANC‐1 cells respectively. The interaction networks between the 11 differentially expressed miRNAs and their target genes were predicted by TargetScan (http://www.Targetscan.org/vert60/) and MiRanda (http://www.microrna. org/microrna/home.do). Only those genes and miRNAs with ≥2 binding sites identified by the two algorithms were considered to be the predicted target genes of the differentially expressed miRNAs. The results in Figure 5B revealed that these miRNAs, modulated by dioscin, could regulate many target genes. A target gene can also be regulated by several miRNAs, suggesting that the effects of dioscin against pancreatic cancer could occur through manipulating complex miRNA‐mediated pathways.
Figure 4.
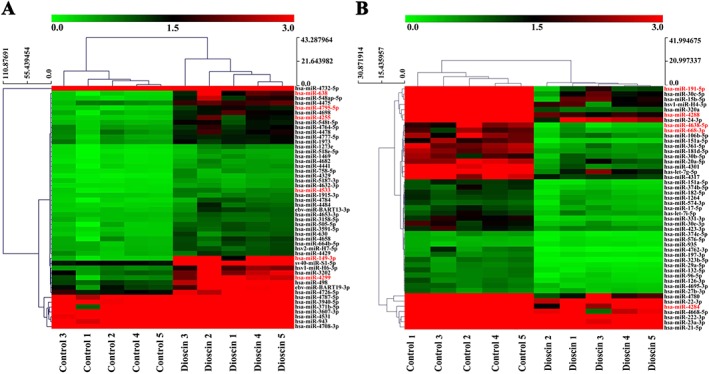
Hierarchical clustering of the differentially expressed miRNAs induced by dioscin in ASPC‐1 cells. Each row represented an individual miRNA, and each column represented a sample. The dendrogram at the top of the image displayed the similarity of expression between these samples at the same stage. The red colour indicated high relative expression level, and green indicated low relative expression level. Fold changes >2.0 and P‐values <0.05 were considered significant (n = 5). The miRNA marked in red to the right of the hierarchical clustering was selected for verification. (A) Up‐regulated miRNAs in ASPC‐1 cells identified by microarrays. (B) Down‐regulated miRNAs in ASPC‐1 cells identified by microarrays.
Figure 5.
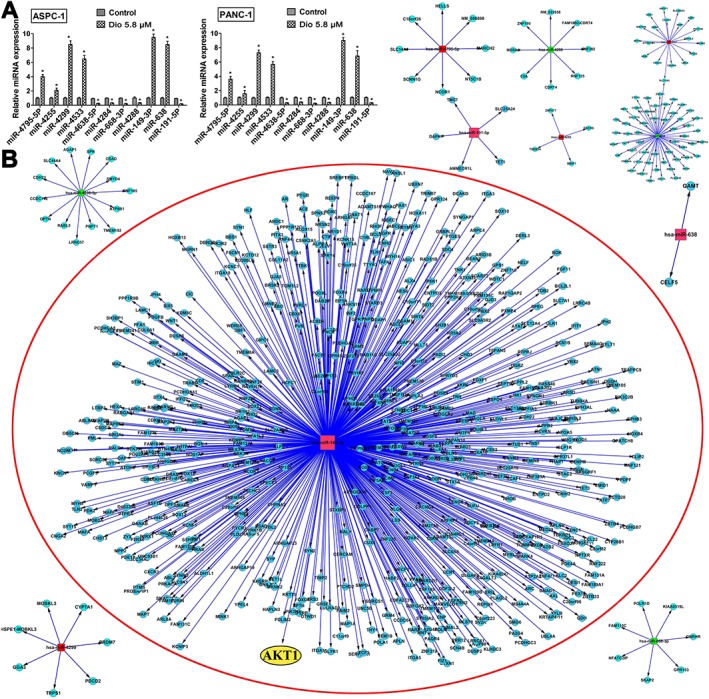
Validation and interaction networks of the differentially expressed miRNAs. (A) Confirmation of miRNA expression levels by qRT‐PCR. The data are presented as the mean ± SD (n = 5). *P < 0.05, significantly different from control groups. (B) The interaction networks of the 11 differentially expressed miRNAs and their target genes. The up‐regulated miRNAs were marked in red, and the down‐regulated miRNAs were marked in green. Akt1 is marked in yellow in a red circle.
MiR‐149‐3P targets Akt1
MiR‐149‐3P, which plays an important role in the progression of several solid malignant tumours (Pan et al., 2012; Lin et al., 2010), was selected for investigating the possible mechanism of dioscin against pancreatic cancer. Based on the interaction network shown in Figure 5B, we found that Akt1 was the target gene of miR‐149‐3P. Dual luciferase reporter assays were performed, and RNA sequence alignment showed that the 3″‐UTR of Akt1 mRNA contained a complementary site for the seed region of miR‐149‐3P. Akt1 mut, a mutant construct with substitutions in the complementary region, was used as a negative control (Figure 6A). Luciferase activity was significantly repressed by miR‐149‐3P overexpression compared with negative control group. However, these effects were not observed with the mutated Akt1‐3′‐UTR (Figure 6B), suggesting that dioscin decreased Akt1 expression at least partly in a miR‐149‐3P‐dependent manner.
Figure 6.
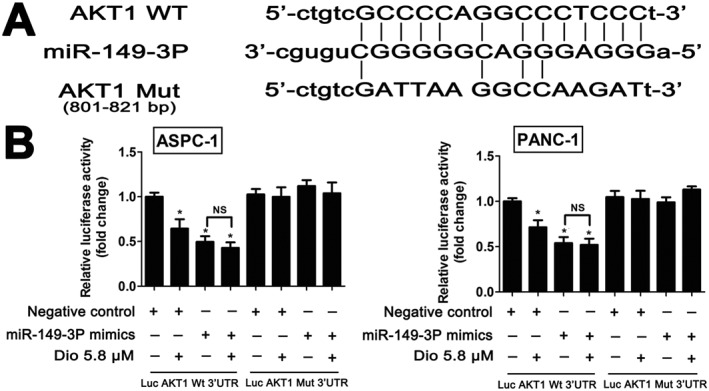
Akt1 is a direct target of miR‐149‐3P in ASPC‐1 and PANC‐1 cells. (A) RNA sequence alignment showed that the 3′‐UTR of Akt1mRNA contained a complementary site for the seed region of miR‐149‐3P. Akt1 mut was a mutant with substitutions in the complementary region used as a negative control. (B) Luciferase reporter assays of Akt1 and miR‐149‐3P. The data are presented as the mean ± SD (n = 5). *P < 0.05, significantly different from control groups.
Dioscin up‐regulates the levels of miR‐149‐3P in vitro and in vivo
As shown in Figure 7A, the quantitative real‐time PCR assays indicated that dioscin (1.4, 2.9 or 5.8 μM) significantly up‐regulated the expression levels of miR‐149‐3P in ASPC‐1 cells and in PANC‐1 cells, compared with control groups. In addition, dioscin (40 and 80 mg·kg−1) also markedly increased the levels of miR‐149‐3P in vivo in ASPC‐1 tumour tissue, and in PANC‐1 tumour tissue.
Figure 7.
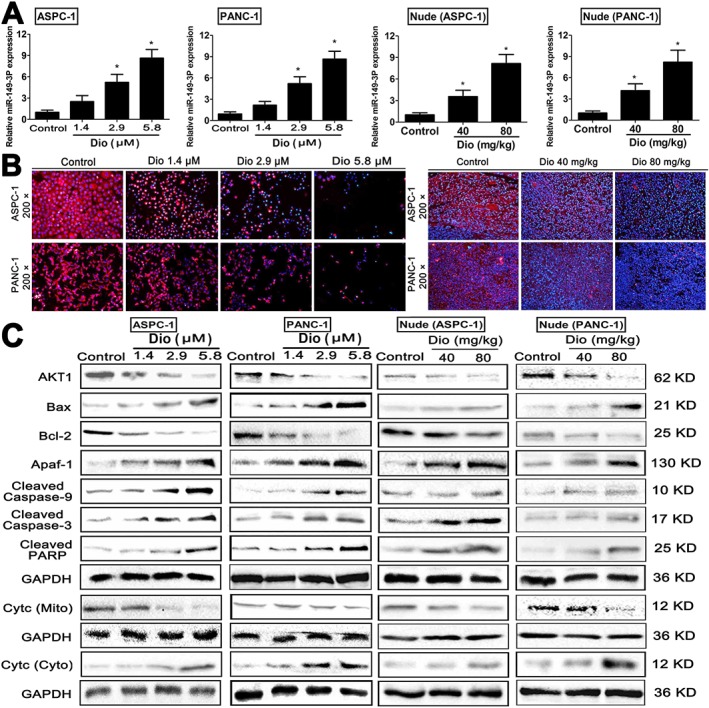
Effects of dioscin on miR‐149‐3P/ Akt1 signalling pathway. (A) Effects of dioscin on the expression levels of miR‐149‐3P in vitro and in vivo based on quantitative real‐time PCR assay. (B) Effects of dioscin on Akt1 levels based on immunofluorescence staining in vitro and in vivo (200× magnification). (C) Effects of dioscin on the levels of Akt1, Bax, Bcl‐2, Apaf‐1, cleaved caspase‐3/9, cleaved PARP, and cytochrome c release in vitro and in vivo. Data are presented as the mean ± SD (n = 5). *P < 0.05, significantly different from control groups.
Dioscin regulates the Akt1 signalling pathway in vitro and in vivo
As shown in Figure 7B, dioscin clearly suppressed the expression levels of Akt1 based on immunofluorescence assays in vitro and in vivo. Then, the expression levels of AKT1 and some downstream signalling molecules were assessed by Western blots. As shown in Figure 7C, the levels of Bax, Apaf‐1, cleaved caspase‐3/9 and cleaved PARP were markedly increased, and the levels of Akt1 and Bcl‐2 were significantly decreased by dioscin, compared with the control groups. In addition, dioscin significantly enhanced the release of cytochrome c (Supporting Information Figure 10A–D).
An inhibitor of MiR‐149‐3P blocks the effect of dioscin on cell viability
To investigate the mechanisms of dioscin against pancreatic cancer, we hypothesized that the anti‐cancer effect of dioscin may primarily result from down‐ regulation of the Akt1 signalling pathway, via up‐regulation of miR‐149‐3P. To test this hypothesis, ASPC‐1 and PANC‐1 cells were transfected with a miR‐149‐3P inhibitor. As shown in Figure 8A, transfection with the miR‐149‐3P inhibitor alone did not induce cell apoptosis, compared with dioscin‐treated (5.8 μM) groups (the bright‐field images are provided in Supporting Information Figure S1). Moreover, no significant differences were found between miR‐149‐3P inhibitor groups and dioscin‐treated (5.8 μM) groups after transfection (P > 0.05). In addition, the Akt1 levels in ASPC‐1 and PANC‐1 cells were notably increased after treating with the miR‐149‐3P inhibitor, compared with control groups, suggesting that miR‐149‐3P inhibited Akt1 expression. Furthermore, the levels of Bcl‐2 and cytochrome c (in mitochondria) were notably increased, and the levels of Bax, Apaf‐1, cleaved caspase‐9/3, cleaved PARP and cytochrome c (in cytosol) were markedly decreased after transfection compared with control groups. There were no significant differences between miR‐149‐3P inhibitor groups and dioscin‐treated groups after transfection (Figure 8B and Supporting Information Figure S12). These results suggested that miR‐149‐3P inhibitor transfection completely reversed the inhibitory effects of dioscin on the Akt1 signalling pathway.
Figure 8.
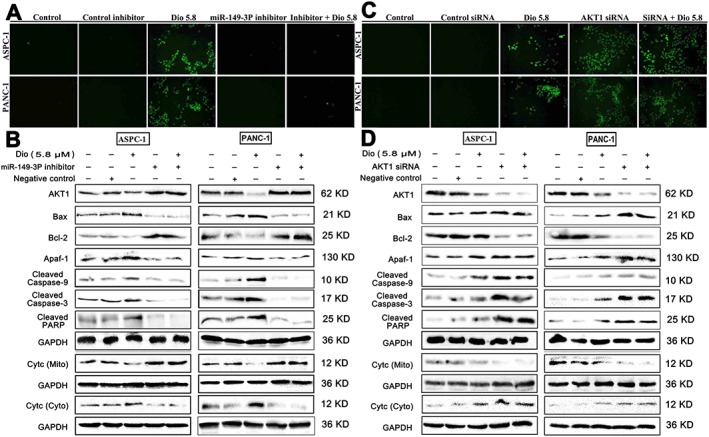
Effects of dioscin on miR‐149‐3P/ Akt1 signalling pathway using miR‐149‐3P inhibitor and Akt1 siRNA assays. (A) Effects of dioscin on the apoptosis of ASPC‐1 and PANC‐1 cells using TUNEL assay with or without transfecting miR‐149‐ 3P inhibitor in vitro. (B) Effects of dioscin on the levels of Akt1, Bax, Bcl‐2, Apaf‐1, cleaved caspase‐3/9, cleaved PARP and cytochrome c release with or without transfecting miR‐149‐3P inhibitor in vitro. (C) Effects of dioscin on the apoptosis of ASPC‐1 and PANC‐1 cells using TUNEL assay with or without transfecting Akt1 siRNA in vitro. (D) Effects of dioscin on the levels of Akt1, Bax, Bcl‐2, Apaf‐1, cleaved caspase‐ 3/9, cleaved PARP and cytochrome c release with or without transfecting Akt1 siRNA in vitro.
The siRNA for Akt1 potentiates the inhibitory effect of dioscin on cell viability
To explore whether Akt1 played a crucial role in the inhibitory effect of dioscin, we used transfection with AKT1 siRNA. As shown in Figure 8C, transfection with Akt1 siRNA markedly induced cell apoptosis compared with control siRNA group (bright‐field images are provided in Supporting Information Figure S3). Moreover, no significant differences between Akt1 siRNA groups and dioscin‐treated (5.8 μM) groups were found after transfection (P > 0.05). As shown by the Western blots in Figure 8D, the levels of Akt1, Bcl‐2 and cytochrome c (in mitochondria) were notably decreased, and the levels of Bax, Apaf‐1, cleaved caspase‐9/3, cleaved PARP and cytochrome c (in cytosol) were markedly increased after transfection compared with control groups. There were no significant differences between Akt1 siRNA group and dioscin‐treated groups after transfection (Supplementary Figure S14).
Discussion
Pancreatic cancer is a highly lethal, solid malignancy with few effective therapies (Biankin et al., 2012). Dioscin, a natural product, has beneficial effects on colon cancer, glioblastoma multiforme and lung cancer in our previous studies (Wei et al., 2013; Chen et al., 2014). Therefore, we explored whether dioscin could exert active effects against pancreatic cancer. In this study, dioscin significantly inhibited cell viability and induced cell apoptosis in two human pancreatic cancer cell lines ASPC‐1 and PANC‐1 cells. In vivo, dioscin significantly suppressed the tumour growth of ASPC‐1 and PANC‐1 cell xenografts in nude mice. These results suggested that dioscin has potent effects against pancreatic cancer in vitro and in vivo.
The occurrence of pancreatic cancer is complex, and many biological molecules and signalling pathways are critically important in this disease. Accumulating evidence shows that dysregulated miRNA expression is a common feature of human tumours (Croce, 2009). MiRNAs can act either as oncogenes or as tumour suppressors by suppressing some key genes related to cancer development and progression (Xu et al., 2013). Thus, miR‐196b and miR‐196a expression levels were markedly increased in pancreatic ductal adenocarcinoma (PDAC) (Szafranska et al., 2007). Schmittgen's group found that miR‐301 and miR‐376a levels were also up‐regulated in PDAC (Lee et al., 2007). In contrast, the levels of miR‐142‐P, miR‐345 and miR‐139 were markedly down‐regulated (Paranjape et al., 2009). Currently, several techniques and methods have been used to screen and detect differentially expressed miRNAs caused by chemicals or drugs to investigate molecular mechanisms. Microarray technology, a sensitive, rapid and quantitative high‐throughput method, has been widely used for detecting and profiling miRNAs. In this study, total of 107 miRNAs with differential changes in ASPC‐1 cells caused by dioscin were identified, in which miR‐149‐3P was significantly up‐regulated, with the highest fold change, by dioscin. Recent studies have suggested that miR‐149 has an important role in various diseases and functions as both a tumour suppressor (Lin et al., 2010) and an oncogene (Jin et al., 2011) in the development of several types of solid tumours. The data in the present work suggested that the anti‐pancreatic cancer effect of dioscin may be related to these differentially expressed miRNAs.
MiRNAs exert their biological activities by regulating their downstream target genes. The interaction networks of the identified miRNAs and their target genes in our work showed that Akt1 could be a target gene of miR‐149‐3P, which was consistent with the reports of Akt1 and miR‐149 in glioma and HeLa cancer cells (Lin et al., 2010; Pan et al., 2012). Also, Akt1 can inhibit apoptosis and promote cell survival. contributing to its oncogenic potential. through many signalling pathways including PI3K (Wang et al., 2016), ERK1/2 (Li et al., 2016) and JNK (Qi et al., 2016). However, the interaction between miR‐149‐3P and Akt1 had not been confirmed in pancreatic cancer. Thus, we needed to show that dioscin regulated the miR‐149‐3P/AKT1 pathway to inhibit pancreatic cancer, and to provide evidence for the mechanistic link between the decreased Akt1‐mediated cell proliferation and the increased levels of miR‐149‐3P after dioscin. We found that the expression levels of miR‐149‐3P were up‐regulated by dioscin, which directly targeted with Akt1, based on the dual‐luciferase reporter assay. In addition, after suppression of miR‐149‐3P, the levels of AKT1 were increased and cell apoptosis was consequently decreased.
In the present work, dioscin, in vitro and in vivo, significantly increased the expression levels of Bax, Apaf‐1, cleaved caspase‐3/9, cleaved PARP, decreased the expression levels of Akt1, Bcl‐2, and enhanced the release of cytochrome c. Therefore, we would suggest that the anti‐cancer effect of dioscin may result primarily from inhibition of the Akt1 pathway, mediated by miR‐149‐3P (Figure 9).
Figure 9.
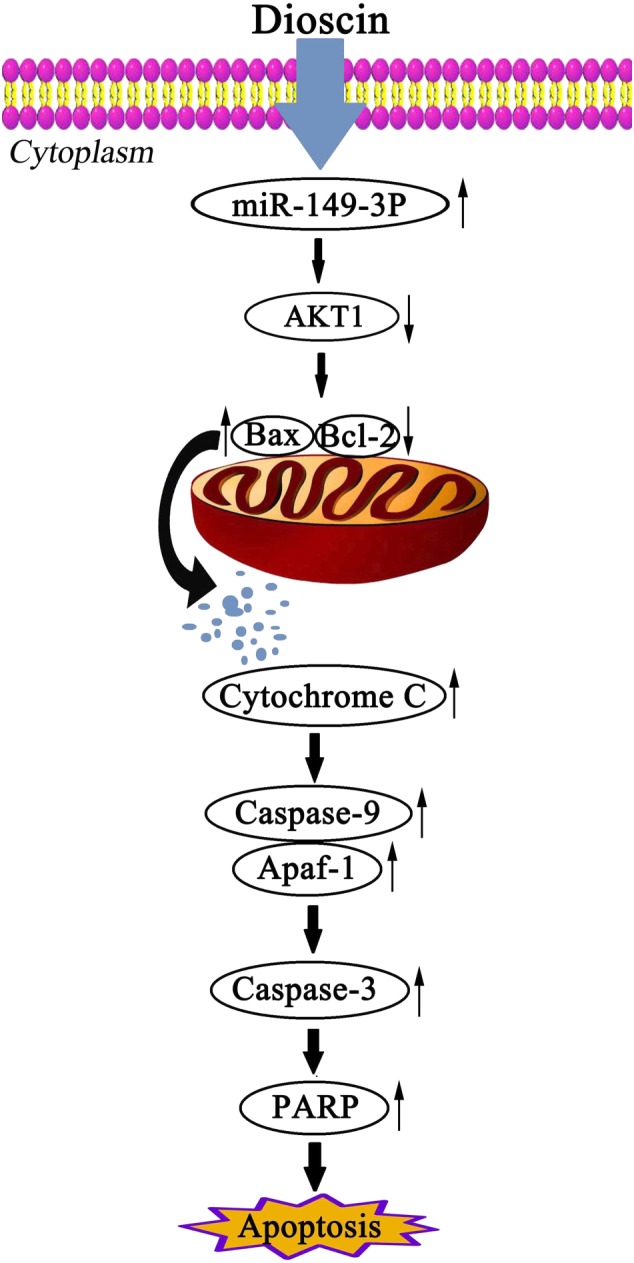
Diagram of the mechanism of action of dioscin against pancreatic cancer. Dioscin decreased the expression level of Akt1 by up‐regulating miR‐149‐3P to cause cell apoptosis.
To further validate the effect of dioscin against pancreatic cancer through miR‐149‐3P‐mediated inhibition of the Akt1 pathway, the miR‐149‐3P inhibitor and Akt1 siRNA were used. The results showed that dioscin significantly increased the expression levels of Bax, Apaf‐1, cleaved caspase‐3/9, cleaved PARP; decreased the expression levels of AKT1, Bcl‐2, and enhanced the release of cytochrome c. However, these alterations induced by dioscin were abolished by the miR‐149‐3P inhibitor or the Akt1 siRNA. In addition, Akt1 siRNA aggravated the inhibitory effect of dioscin on cell viability. These results suggested that dioscin inhibited the Akt1 signalling pathway by up‐regulating levels of miR‐149‐3P.
Dioscin is one major active ingredient of some medicinal herbs including Discorea nipponica Makino and Rhizoma dioscoreae, and some TCMs including Liuwei Dihuang decoction (LW), Di'ao Xinxuekang (Di'ao XXK) and Shuyu Zaogan tablets, which have been widely used to treat various diseases including dementia, osteoporosis and diabetics (Zhang et al., 2009; Yu et al., 2014; Zhou et al., 2016). Furthermore, Di'ao XXK has been allowed to register in the European Union market in 2012. Based on our investigation, dioscin showed potent effects against pancreatic cancer in vivo and in vitro by inhibiting the Akt1 signal pathway via up‐regulation of miR‐149‐3P. However, these data were obtained from cells and animals, not from human patients because dioscin is a natural product and is not a clinically used compound. Thus, at the present time, no clinical data for dioscin can be provided. However, the results of the present work may serve to expand the clinical applications of the related medicinal plants and TCMs to treat pancreatic cancer. In addition, these findings provide novel insights into the mechanisms of dioscin against pancreatic cancer, which should be developed as a potential candidate to treat this disease. Further work is needed to fully elucidate the mechanisms and clinical applications of the compound.
Author contributions
S.‐L.L. and P.‐J.Y. designed and conceived all experiments. S.‐L.L. performed the in vitro experiments. S.‐L.L. executed the animal experiments. X.‐L.N. and Y.‐L.H. analysed the data. Q.Y., H.X., X.‐Y.W. and Z.‐Y.Y. prepared the compound. S.‐L.L., X.‐L.N. and P.‐J.Y. wrote the paper. P.‐J.Y. and L.‐K.X. supervised the study.
Conflict of interest
The authors declare no conflicts of interest.
Declaration of transparency and scientific rigour
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Table S1 The primer sequences used for real‐time PCR assay in the present work.
Table S2 The antibodies used in the present work.
Table S3 The sequences used in the present work.
Table S4 Differentially expressed microRNAs caused by dioscin in ASPC‐ 1 cells using microRNA microarray analysis.
Figure S1 The chemical structure of dioscin.
Figure S2 TEM micrographs of ASPC‐1 and PANC‐1 cells without or with dioscin (5.8 μM) for 24 h in different magnifications (8000 × in the left and 30000 × in the right).
Figure S3 Effects of different concentrations of dioscin (1.4, 2.9 and 5.8 μM) for 24 h on HPDE6‐C7 cell morphology and structure (100× magnification).
Figure S4 Raw bioluminescence images of mice inoculated with ASPC‐1 cells (n = 5).
Figure S5 Raw tumor images of mice inoculated with ASPC‐1 cells (n = 5).
Figure S6 Raw bioluminescence images of mice inoculated with PANC‐1 cells (n = 5).
Figure S7 Raw tumors images of mice inoculated with PANC‐1 cells (n = 5).
Figure S8 Detection of miRNAs by microarrays. Total RNAs extracted from control groups were covalently labeled with Hy3 (green channel) and hybridized to the array.
Figure S9 Detection of miRNAs by microarrays. Total RNAs extracted from dioscin‐treated group (5.8 μM) were covalently labeled with Hy3 (green channel) and hybridized to the array.
Figure S10 (A‐D) Analytical results the protein levels of Akt1, Bax, Bcl‐2, Apaf‐1, Cleaved caspase‐3/9, cleaved PARP and Cytochrome c treated by dioscin in vitro and in vivo. Data are presented as the mean ± S.D. (n = 5). *P < 0.05 compared with control groups.
Figure S11 Effects of dioscin on cellular morphology and structure of ASPC‐1 and PANC‐1 cells by bright image (×100, magnification) investigation with or without transfecting miR‐149‐3P inhibitor in vitro.
Figure S12 Analytical results the protein levels of Akt1, Bax, Bcl‐2, Apaf‐1, Cleaved caspase‐3/9, cleaved PARP and Cytochrome c after treated with miR‐149‐3P inhibitor in ASPC‐1 and PANC‐1 cells. Data are presented as the mean ± S.D. (n = 5). *P < 0.05 compared with control inhibitor group; NS, not significant.
Figure S13 Effects of dioscin on cellular morphologies and structures of ASPC‐1 and PANC‐1 cells by bright image (×100, magnification) investigation with or without transfecting Akt1 siRNA in vitro.
Figure S14 Analytical results the protein levels of Akt1, Bax, Bcl‐2, Apaf‐1, Cleaved caspase‐3/9, cleaved PARP, Cytochrome c after treated with Akt1 siRNA in ASPC‐1 and PANC‐1 cells. Data are presented as the mean ± S.D. (n = 5). *P < 0.05 compared with control siRNA group; NS, not significant.
Acknowledgements
This work was supported by the Program for Liaoning Innovative Research Team in University (LT2013019) and the Foundation of Innovation Team of Education Ministry (IRT13049).
Si, L. , Xu, L. , Yin, L. , Qi, Y. , Han, X. , Xu, Y. , Zhao, Y. , Liu, K. , and Peng, J. (2017) Potent effects of dioscin against pancreatic cancer via miR‐149‐3P‐mediated inhibition of the Akt1 signalling pathway. British Journal of Pharmacology, 174: 553–568. doi: 10.1111/bph.13718.
References
- Albury TM, Pandey V, Gitto SB, Dominguez L, Spinel LP, Talarchek J et al. (2015). Constitutively active Akt1 cooperates with KRas (G12D) to accelerate in vivo pancreatic tumor onset and progression. Neoplasia 17: 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SP, Kelly E, Marrion N, Peters JA, Benson HE, Faccenda E et al. (2015a). The concise guide to PHARMACOLOGY 2015/16: Overview. Br J Pharmacol 172: 5729–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Fabbro D, Kelly E, Marrion N, Peters JA, Benson HE et al. (2015b). The Concise Guide to PHARMACOLOGY 2015/16: Enzymes. Br J Pharmacol 172: 6024–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V (2003). MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113: 673–676. [DOI] [PubMed] [Google Scholar]
- Aquino R, Conti C, Simone FD, Orsi N, Pizza C, Stein ML (1991). Antiviral activity of constituents of Tamus communis. J Chemother 3: 305–309. [DOI] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL et al. (2012). Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491: 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris HA, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR et al. (1997). Improvements in survival and clinical benefit with gemcitabine as first‐line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol 15: 2403–2413. [DOI] [PubMed] [Google Scholar]
- Chen H, Xu LN, Yin LH, Xu YW, Han X, Qi Y et al. (2014). iTRAQ‐based proteomic analysis of dioscin on human HCT‐116 colon cancer cells. Proteomics 14: 51–73. [DOI] [PubMed] [Google Scholar]
- Chen XQ, Gong J, Zeng H, Chen N, Huang R, Huang Y et al. (2010). MicroRNA145 targets BNIP3 and suppresses prostate cancer progression. Cancer Res 70: 2728–2738. [DOI] [PubMed] [Google Scholar]
- Cho J, Choi H, Lee J, Kim MS, Sohn HY, Lee DG (2013). The antifungal activity and membrane‐disruptive action of dioscin extracted from Dioscorea nipponica . Biochim Biophys Acta 1828: 1153–1158. [DOI] [PubMed] [Google Scholar]
- Croce CM (2009). Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10: 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MJ, Bond RA, Spina D, Ahluwalia A, Alexander SPA, Giembycz MA et al. (2015). Experimental design and analysis and their reporting: new guidance for publication in BJP. Br J Pharmacol 172: 3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu LN, Tao XF, Xu YW, Han X, Qi Y, Xu LN et al. (2016). Dioscin alleviates BDL‐ and DMN‐induced hepatic fibrosis via Sirt1/Nrf2‐mediated inhibition of p38 MAPK pathway. Toxicol Appl Pharmacol 292: 19–29. [DOI] [PubMed] [Google Scholar]
- Hay N (2005). The Akt–mTOR tango and its relevance to cancer. Cancer Cell 8: 179–183. [DOI] [PubMed] [Google Scholar]
- He Y, Zheng R, Li D, Zeng H, Zhang S, Chen W (2015). Pancreatic cancer incidence and mortality patterns in China, 2011. Chin J Cancer Res 27: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Zhang HC, Yang Q, Li MZ, Song YL, Jiang L (2014). Bio‐inspired photonic‐crystal microchip for fluorescent ultratrace detection. Angew Chem Int Ed Engl 53: 5791–5795. [DOI] [PubMed] [Google Scholar]
- Hsieh MJ, Tsai TL, Hsieh YS, Wang CJ, Chiou HL (2013). Dioscin‐induced autophagy mitigates cell apoptosis through modulation of PI3K/Akt and ERK and JNK signaling pathways in human lung cancer cell lines. Arch Toxicol 87: 1927–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Hu WL, Jiang CC, Wang JX, Han CC, Chu P et al. (2011). MicroRNA‐149, a p53‐ responsive microRNA, functions as an oncogenic regulator in human melanoma. Proc Natl Acad Sci U S A 108: 15840–15845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M (2014). Therapeutic applications of curcumin for patients with pancreatic cancer. World J Gastroenterol 20: 9384–9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG (2010). Animal research: Reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol 160: 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL et al. (2007). Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer 120: 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Ambros V (2001). An extensive class of small RNAs in Caenorhabditis elegans . Science 294: 862–864. [DOI] [PubMed] [Google Scholar]
- Leite M, Quinta‐Costa M, Leite PS, Guimarães JE (1999). Critical evaluation of techniques to detect and measure cell death–study in a model of UV radiation of the leukaemic cell line HL60. Anal Cell Pathol 19: 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Huang W, Wen YQ, Gong GH, Zhao QB, Yu G (2010). Anti‐thrombotic activity and chemical characterization of steroidal saponins from Dioscorea zingiberensis C.H. Wright. Fitoterapia 81: 1147–1156. [DOI] [PubMed] [Google Scholar]
- Li MY, Yu XJ, Guo H, Sun LM, Wang AJ, Liu QJ et al. (2014). Bufalin exerts antitumor effects by inducing cell cycle arrest and triggering apoptosis in pancreatic cancer cells. Tumour Biol 35: 2461–2471. [DOI] [PubMed] [Google Scholar]
- Li W, Ma X, Li N, Liu H, Dong Q, Zhang J et al. (2016). Resveratrol inhibits Hexokinases II mediated glycolysis in non‐small cell lung cancer via targeting Akt signaling pathway. Exp Cell Res 349: 320–327 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Lin YC, Yu AL (2010). miR‐149* induces apoptosis by inhibiting Akt1 and E2F1 in human cancer cells. Mol Carcinog 49: 719–727. [DOI] [PubMed] [Google Scholar]
- Liu M, Xu YW, Han X, Yin LH, Xu LN, Qi Y et al. (2015). Dioscin alleviates alcoholic liver fibrosis by attenuating hepatic stellate cell activation via the TLR4/MyD88/NF‐κB signaling pathway. Sci Rep 5: 18038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Liang H, Xia Q, Li P, Kong H, Lei P et al. (2013). Resveratrol induces apoptosis of pancreatic cancers cells by inhibiting miR‐21 regulation of BCL‐2 expression. Clin Transl Oncol 15: 741–746. [DOI] [PubMed] [Google Scholar]
- Liu SP, Guo WX, Shi J, Li N, Yu XY, Xue J et al. (2012). MicroRNA‐135a contributes to the development of portal vein tumor thrombus by promoting metastasis in hepatocellular carcinoma. J Hepatol 56: 389–396. [DOI] [PubMed] [Google Scholar]
- Lu BN, Xu YS, Xu LN, Cong XN, Yin LH, Li H et al. (2012). Mechanism investigation of dioscin against CCl4‐induced acute liver damage in mice. Environ Toxicol Pharmacol 34: 127–135. [DOI] [PubMed] [Google Scholar]
- McGrath JC, Lilley E (2015). Implementing guidelines on reporting research using animals (ARRIVE etc.): new requirements for publication in BJP. Br J Pharmacol 172: 3189–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran DC, Wu J, Jamieson NB, Mrowinska A, Kalna G, Karim SA et al. (2014). Targeting mTOR dependency in pancreatic Cancer. Gut 63: 1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neesse A, Frese KK, Chan DS, Bapiro TE, Howat WJ, Richards FM et al. (2014). SPARC independent drug delivery and antitumour effects of nab‐paclitaxel in genetically engineered mice. Gut 63: 974–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan SJ, Zhan SK, Pei BG, Sun QF, Bian LG, Sun BM (2012). MicroRNA‐149 inhibits proliferation and invasion of glioma cells via blockade of AKT1 signaling. Int J Immunopathol Pharmacol 25: 871–881. [DOI] [PubMed] [Google Scholar]
- Paranjape T, Slack FJ, Weidhaas JB (2009). MicroRNAs: tools for cancer diagnostics. Gut 58: 1546–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi DS, Tao JH, Zhang LQ, Wang M, Qu R, Zhang LQ et al. (2016). Neuroprotection of Cilostazol against ischemia/reperfusion‐induced cognitive deficits through inhibiting JNK3/caspase‐3 by enhancing Akt1. Brain Res 1653: 67–74 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Roy S, Soh JH, Ying JY (2016). A microarray platform for detecting disease‐specific circulating miRNA in human serum. Biosens Bioelectron 75: 238–246. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A (2013). Cancer statistics, 2013. CA Cancer J Clin 63: 11–30. [DOI] [PubMed] [Google Scholar]
- Si LL, Zheng LL, Xu LN, Yin LH, Han X, Qi Y et al. (2016). Dioscin suppresses human laryngeal cancer cells growth via induction of cell‐cycle arrest and MAPK‐ mediated mitochondrial‐derived apoptosis and inhibition of tumor invasion. Eur J Pharmacol 774: 105–117. [DOI] [PubMed] [Google Scholar]
- Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SP et al. (2016). The IUPHAR/BPS guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res 44: D1054–D1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A et al. (2007). MicroRNA expression alterations are linked to tumorigenesis and non‐neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 26: 4442–4452. [DOI] [PubMed] [Google Scholar]
- Tao XF, Sun XC, Yin LH, Han X, Xu LN, Qi Y et al. (2015). Dioscin ameliorates cerebral ischemia/reperfusion injury through the downregulation of TLR4 signaling via HMGB‐1 inhibition. Free Radic Biol Med 84: 103–115. [DOI] [PubMed] [Google Scholar]
- Wei YL, Xu YS, Han X, Qi Y, Xu LN, Xu YW et al. (2013). Anti‐cancer effects of dioscin on three kinds of human lung cancer cell lines through inducing DNA damage and activating mitochondrial signal pathway. Food Chem Toxicol 59: 118–128. [DOI] [PubMed] [Google Scholar]
- Wang Z, Cheng Y, Wang N, Wang DM, Li YW, Han F et al. (2012a). Dioscin induces cancer cell apoptosis through elevated oxidative stress mediated by down‐ regulation of peroxiredoxins. Cancer Biol Ther 13: 138–147. [DOI] [PubMed] [Google Scholar]
- Wang SP, Wu X, Tan M, Gong J, Tan W, Bian BL et al. (2012b). Fighting fire with fire: poisonous Chinese herbal medicine for cancer therapy. J Ethnopharmacol 140: 33–45. [DOI] [PubMed] [Google Scholar]
- Wang Y, Che CM, Chiu JF, He QY (2007). Dioscin (saponin)‐induced generation of reactive oxygen species through mitochondria dysfunction: a proteomic‐based study. J Proteome Res 6: 4703–4710. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li D, Luo J, Tian G, Zhao LY, Liao D (2016). Intrinsic cellular signaling mechanisms determine the sensitivity of cancer cells to virus‐induced apoptosis. Sci Rep 6: 37213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner J, Combs SE, Springfeld C, Hartwig W, Hackert T, Büchler MW (2013). Advanced‐stage pancreatic cancer: therapy options. Nat Rev Clin Oncol 10: 323–333. [DOI] [PubMed] [Google Scholar]
- Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ et al. (2010). Guidelines for the welfare and use of animals in cancer research. Br J Cancer 102: 1555–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK et al. (2013). Recent progress in pancreatic. cancer. CA Cancer J Clin 63: 318–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GL, Xie M, Yang XY, Song Y, Yan C, Yang Y et al. (2014). Spectrum‐effect relationships as a systematic approach to traditional chinese medicine research: current status and future perspectives. Molecules 19: 17897–17925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XJ, Fan ZY, Kang L, Han JQ, Jiang CY, Zheng XF et al. (2013). Hepatitis B virus X protein represses miRNA‐148a to enhance tumorigenesis. J Clin Invest 123: 630–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Hu S, Li G, Xue J, Li Z, Liu X et al. (2014). Comparative effectiveness of Di'ao Xin Xue Kang capsule and Compound Danshen tablet in patients with symptomatic chronic stable angina. Sci Rep 4: 7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XL, Han X, Yin LH, Xu LN, Qi Y, Xu YW et al. (2015). Potent effects of dioscin against liver fibrosis. Sci Rep 5: 9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZH, Chen ZJ, Sheng YC, Guo YF, Xie W et al. (2009). Systematic evaluation of Shuyu Zaogan tablets for coronary heart disease. World Clinical Drugs 32: 159–164. [Google Scholar]
- Zhao X, Ren H, Gao S, Hao JH (2014). Effects of dioscin on apoptosis in pancreatic cancer MiaPaCa‐2 cells and its mechanism. Zhonghua Zhong Liu Za Zhi 36: 5–10. [PubMed] [Google Scholar]
- Zhao XM, Cong XN, Zheng LL, Xu LN, Yin LH, Peng JY (2012). Dioscin, a natural steroid saponin, shows remarkable protective effect against acetaminophen‐induced liver damage in vitro and in vivo . Toxicol Lett 214: 69–80. [DOI] [PubMed] [Google Scholar]
- Zhou W, Cheng X, Zhang Y (2016). Effect of Liuwei Dihuang decoction, a traditional Chinese medicinal prescription, on the neuroendocrine immunomodulation network. Pharmacol Ther 162: 170–178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 The primer sequences used for real‐time PCR assay in the present work.
Table S2 The antibodies used in the present work.
Table S3 The sequences used in the present work.
Table S4 Differentially expressed microRNAs caused by dioscin in ASPC‐ 1 cells using microRNA microarray analysis.
Figure S1 The chemical structure of dioscin.
Figure S2 TEM micrographs of ASPC‐1 and PANC‐1 cells without or with dioscin (5.8 μM) for 24 h in different magnifications (8000 × in the left and 30000 × in the right).
Figure S3 Effects of different concentrations of dioscin (1.4, 2.9 and 5.8 μM) for 24 h on HPDE6‐C7 cell morphology and structure (100× magnification).
Figure S4 Raw bioluminescence images of mice inoculated with ASPC‐1 cells (n = 5).
Figure S5 Raw tumor images of mice inoculated with ASPC‐1 cells (n = 5).
Figure S6 Raw bioluminescence images of mice inoculated with PANC‐1 cells (n = 5).
Figure S7 Raw tumors images of mice inoculated with PANC‐1 cells (n = 5).
Figure S8 Detection of miRNAs by microarrays. Total RNAs extracted from control groups were covalently labeled with Hy3 (green channel) and hybridized to the array.
Figure S9 Detection of miRNAs by microarrays. Total RNAs extracted from dioscin‐treated group (5.8 μM) were covalently labeled with Hy3 (green channel) and hybridized to the array.
Figure S10 (A‐D) Analytical results the protein levels of Akt1, Bax, Bcl‐2, Apaf‐1, Cleaved caspase‐3/9, cleaved PARP and Cytochrome c treated by dioscin in vitro and in vivo. Data are presented as the mean ± S.D. (n = 5). *P < 0.05 compared with control groups.
Figure S11 Effects of dioscin on cellular morphology and structure of ASPC‐1 and PANC‐1 cells by bright image (×100, magnification) investigation with or without transfecting miR‐149‐3P inhibitor in vitro.
Figure S12 Analytical results the protein levels of Akt1, Bax, Bcl‐2, Apaf‐1, Cleaved caspase‐3/9, cleaved PARP and Cytochrome c after treated with miR‐149‐3P inhibitor in ASPC‐1 and PANC‐1 cells. Data are presented as the mean ± S.D. (n = 5). *P < 0.05 compared with control inhibitor group; NS, not significant.
Figure S13 Effects of dioscin on cellular morphologies and structures of ASPC‐1 and PANC‐1 cells by bright image (×100, magnification) investigation with or without transfecting Akt1 siRNA in vitro.
Figure S14 Analytical results the protein levels of Akt1, Bax, Bcl‐2, Apaf‐1, Cleaved caspase‐3/9, cleaved PARP, Cytochrome c after treated with Akt1 siRNA in ASPC‐1 and PANC‐1 cells. Data are presented as the mean ± S.D. (n = 5). *P < 0.05 compared with control siRNA group; NS, not significant.


