Abstract
Triple‐negative breast cancer (TNBC) accounts for 15–20% of all newly diagnosed breast cancers, and is enriched for germline mutation of BRCA. In Asian patients diagnosed with breast cancer, 268 deleterious mutations of BRCA1 and 242 of BRCA2 have been identified so far, including a reported BRCA1 frameshift mutation (rs80350973), apparently found only in Asian people, with a low prevalence of 0.3–1.7% in different breast cancer cohorts. Here, we reported the high prevalence (7.2%) of rs80350973 among 125 Chinese patients with TNBC, which implies its mutational predilection for certain breast cancer subtypes. Although its low prevalence had not indicated any particular clinical significance in previous studies, our results associated rs80350973 mutation with cell checkpoint malfunction, and was found to be more common in TNBC patients with high Ki‐67 indices (P = 0.004). As Ki‐67 overexpression is a predictor of poor prognosis in TNBC, inclusion of this mutation into genetic assessments may improve the clinical management of Chinese patients with TNBC.
Keywords: BRCA, breast cancer, Chinese, epidemiology
Introduction
Triple‐negative breast cancer (TNBC) is defined by little or no expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). This clinical subtype accounts for 15–20% of all newly diagnosed breast cancers (BCs) 1. TNBCs possess highly unstable genomes; they are frequently diagnosed in young women and have a worse prognosis than other BC subtypes 2, 3. Furthermore, no promising regimen has so far been found for TNBC. Conventional chemotherapy remains the one effective treatment modality, but its long‐term clinical outcomes are unsatisfactory 4, 5. Earlier studies of germline BRCA1 mutations in TNBC have shown wide variation in their prevalence, with a range of 10–40%. These mutations are associated with family history 6, 7, risk of recurrence 8, 9, and sensitivity to DNA‐damaging agents 10, 17.
Among Asians, 268 deleterious mutations of BRCA1 and 242 of BRCA2 so far have been documented in patients diagnosed with BC 12. Several studies of Chinese patients with TNBC found that the prevalence of BRCA1 germline mutations varied from 18.6% to 36.8% 12. The high frequency of certain mutations in BRCA1/2 has been widely studied, to optimize genetic testing strategies for those at high risk for BC. On the other hand, clinical trials suggest that BRCA1/2 mutations in TNBCs could respond better to platinum‐based chemotherapy or other DNA‐damaging therapies 13, 14. Notably, current studies mostly focus on identifying novel mutations of BRCA1/2 and their prevalence in TNBC. However, little is known about the high prevalence of certain BRCA1/2 mutations and their clinical relevance in TNBC 12. In this study, we screened recurrent germline mutations, mainly against the BRCA1 gene, and sought their clinical relevance in Chinese patients with TNBCs. The results of our work may help to interpret the clinical significance of certain recurrent mutations of BRCA1.
Material and Methods
Study cohort
A total 125 TNBC of 1300 newly diagnosed BC patients were recruited at our hospital, between January 2013 and June 2015. All subjects gave written informed consent. The inclusion criteria were as follows: (1) patients whose disease was pathologically shown to be negative for ER, PR, and HER2/ErbB2, and (2) for whom complete clinical, pathological and follow‐up data were available. The exclusion criteria were as follows: (1) patients without complete follow‐up data, and (2) those who suffered nontumor‐mediated death. Clinical data were obtained from review of medical records and patient interviews by clinical physicians. This study was approved by the medical ethics committee of our hospital. We initially performed whole‐exon mutations screening of 26 patients with TNBC, using panel‐based next‐generation sequencing (NGS) analysis of 55 susceptibility genes; frequencies of some recurrent mutations were later detected in the larger TNBC cohort using mutation site targeting PCR amplification. Amplified products were submitted to Sanger sequencing.
Panel‐based NGS analysis
The first step for NGS technology was use of the TruSeq Custom Amplicon method to design oligo probes that are specific for all coding sequences and intron/exon boundaries of coding exons from the 55 genes that affect BC susceptibility (Table S1), using Illumina Design‐Studio (Illumina, Inc., San Diego, CA). For each 150‐bp sequence of the target region, a pair of oligo probes were synthesized to hybridize with the 5′ and 3′ ends of the sequence at one end (the other end was complementary to the PCR primers). These oligo probes were used to construct a library containing the necessary nucleotide sequences. The target regions were determined by selecting all exons of the 55 susceptibility genes; however, to include sections of the intron‐exon regions, the regions also included 50 nucleotides upstream and downstream of each exon.
Sequencing was performed using the NGS MiSeq Illumina sequencer (Illumina, Inc.). Obtained sequences were aligned to the reference genome (GRCh37/hg19) using MiSeq Reporter software (Illumina, Inc.), which detected discrepancies determining their type, such as deletions, insertions and SNPs. The sequences were analyzed using MiSeq software. As an acceptance threshold value, we selected a Q‐score of 30, which corresponds to a 1: 1000 error rate.
Genotyping
Since rs80350973 is a heterozygous mutation, we design a pair of primers to amplify the germline DNA fragment containing this mutational site; unaligned sequences from the deletion site cause “double peaks” in Sanger sequencing, whereas the wild‐type will not.
In brief, DNA was extracted from peripheral blood using the QIAamp DNA Blood Mini Kit® (Qiagen, Germantown, MD). Samples were submitted to PCR amplification targeting for BRCA1 mutation (rs80350973). The PCR reaction used SYBR® Green Realtime PCR Master Mix (Toyobo Co., Ltd, Kita‐ku, Osaka, JAPAN). Primers against rs80350973 mutation were as follows: forward primer 5′‐AGGACCCTGGAGTCGATTGA‐3′, reverse primer 5′‐GTAAGCTCATTCTTGGGGTCCTGT‐3′. Amplified products were submitted to sequencing in an ABI 3730 automated sequencer using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) as described by the manufacturer. All analyses were performed in duplicate. Discrepancies in Sanger sequencing outcome between the wild type and rs80350973 mutation are shown in Figure S1.
Immunohistochemistry of Ki‐67
In this study, Ki‐67 expression was quantified by a visual grading system and was determined by counting 1,000 tumor cells using the Olympus Image Analyzer (magnification 400×). Ki‐67 immunoreactivity (Ki‐67 index) was recorded as a continuous variable based on the proportion of positive tumor cells (0–100%), regardless of staining intensity. All cases were histopathologically confirmed independently by two experienced pathologists according to ASCO/CAP 2010 criteria.
Statistical analysis
The statistical significance of associations between the BRCA1 frameshift mutation and clinicopathological features of TNBC patients were evaluated using Pearson's χ2 tests or Fisher's exact tests as appropriate. The Mann–Whitney test was used to analyze associations between the Ki‐67 index and BRCA1 frameshift mutations. Disease‐free survival (DFS) was defined as the time from the date of diagnosis to first recurrence (not including second primary malignancies) or death from BC without a recorded relapse. Lengths of DFS for patients with different axillary lymph node status (N0–2 vs. N3), and Ki‐67 index levels (≤0.7 vs.>0.7, and ≤0.14 vs. >0.14) were plotted with Kaplan–Meier curves and compared with log‐rank tests. DFS was compared between rs80350973 mutation carriers and noncarriers using a time‐dependent covariate Cox regression model, which took lymph node status (N0–2 vs. N3) and Ki‐67 indices into consideration. All statistical tests were two‐sided; P < 0.05 was considered significant. Data analyses were calculated using SPSS® 19.0 software (SPSS Inc., Chicago, IL).
Results
Patient characteristics
Clinical data of the 125 included TNBC patients are summarized in Table 1. Their median age at diagnosis was 47 years (range: 26–75 years). Of these 125 patients, 16 (12.8%) were diagnosed when they were younger than 35 years; 11 (8.8%) had family histories of breast or ovarian cancer. Invasive ductal carcinoma was common (90.4%). Their main metastasis sites were brain (n = 11; 8.8%), lung (n = 49; 39.2%), and liver (n = 18; 14.4%).
Table 1.
Clinicopathological characteristics of 125 Triple‐negative breast cancer patients
| Characteristic | n (%) |
|---|---|
| Age at diagnosis (years) | |
| >35 | 109 (87.2) |
| ≤35 | 16 (12.8) |
| Histology of primary tumor | |
| Ductal | 113 (90.4) |
| Lobular | 3 (2.4) |
| Others | 7 (5.6) |
| Unknown | 2 (1.6) |
| Primary tumor size | |
| T1 | 45 (36.0) |
| T2 | 51 (40.8) |
| T3 | 10 (8.0) |
| T4 | 8 (6.4) |
| Unknown | 11 (8.8) |
| Axillary node involvement | |
| N0 | 58 (46.4) |
| N1 | 23 (18.4) |
| N2 | 21 (16.8) |
| N3 | 20 (16.0) |
| Unknown | 3 (2.4) |
| TNM Stage | |
| I | 25 (20.0) |
| II | 49 (39.2) |
| III | 37 (29.6) |
| IV | 7 (5.6) |
| Unknown | 7 (5.6) |
| Family historya | |
| Yes | 11 (8.8) |
| No | 114 (91.2) |
| Sites of metastases at recurrence | |
| Lung | 49 (39.2) |
| Bone | 34 (27.2) |
| Liver | 18 (14.4) |
| Brain | 11 (8.8) |
Family history was defined as ≥1 first‐ or second‐degree relative with breast cancer at age ≤50 years or ≥1 close blood relative with epithelial ovarian cancer at any age.
Germline mutations of 55 genes that affect BC susceptibility
All coding regions of 55 genes that affect BC susceptibility were sequenced in 26 patients with TNBC. Gene‐based case–control association analysis using Asian population information obtained from the 1000 Genomes Project database. Only deleterious mutations were included in this study. Of all 26 patients with TNBC 6 (23.1%) carried four different BRCA1 mutations, three (11.5%) carried three different PALB2 mutations, three (11.5%) carried three different BRCA2 mutations, and the other three (11.5%) carried three different CDH1 mutations. Notably, four patients carried the same CHEK2 mutation (c.1246A>G). Deleterious mutation sites for each BC‐related gene detected in these patients are listed in Table S2.
For each discovery screen, genes with mutations in at least two patients are shown in Table 2. One heterozygous BRCA1 frameshift mutation (rs80350973) was identified in three (11.5%) patients. Two heterozygous missense mutations—CHEK2 (c.1246A>G) and BRIP1 (c.587A>G)—were identified in 4 (15.4%) patients and 2 (7.7%) patients, respectively. One heterozygous in‐frame BARD1 deletion mutation (c.1075_1095del) was identified in two (7.7%) patients. Details of these recurrent mutations are shown in Table 2.
Table 2.
Detail information of recurrent mutations in 26 TNBC patients
| Gene (Version) | Exon | Nucleotide change | Amino acid change | Type of mutation | dbSNP | PolyPhen predictiona | Clinical significance | Number of TNBC cases, n = 26 | Allele frequency in Asianb |
|---|---|---|---|---|---|---|---|---|---|
| BRCA1 (NM_007300.3) | 24 | c.5533_5540del ATTGGGCA | p.Ile1845Aspfs Ter3 | frameshift variant (heterozygous) | rs80357973 | N/A | unknown | 3 (11.5%) | 0.00% |
| BARD1 (NM_000465.2) | 4 | c.1075_1095del TTGCCTGAATGTTCTTCACCA | p.Leu359_Pro365delinsdel | inframe deletion (heterozygous) | N/A | N/A | neutral polymorphisms 23 | 2 (7.7%) | 0.00% |
| BRIP1 (NM_032043.2) | 4 | c.587A>G | p.Asn196Ser | missense variant (heterozygous) | rs550707862 | benign (0.004) | unknown | 2 (7.7%) | 0.00% |
| CHEK2 (NM_001005735.1) | 11 | c.1246A>G | p.Lys416Glu | missense variant (heterozygous) | rs142470496 | probably damaging (0.907) | unknown | 4 (15.4%) | 0.00% |
PolyPhen was used to predicts possible impact of single amino acid substitution on the structure and function of a human protein using straightforward physical and comparative considerations.
The allele frequency of certain recurrent mutations in Asian dependent on the publication data from 1000 Genomes Project data.
N/A, Not applicable; TNBC, Triple‐negative breast cancer.
Validation of germline mutation rs80350973
The rs80350973 mutation was further validated in an additional 100 TNBC patients using PCR and Sanger sequencing. In total, aside from a failed Sanger sequencing as a result of an unqualified DNA sample, this BRCA1 frameshift mutation was detected in 9 (7.2%) of 125 TNBC patients.
Clinical relevance of the rs80350973 mutation
According to the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/variation/55591/) and Breast Cancer Information Core (BIC) (https://research.nhgri.nih.gov/projects/bic/Member/cgi-bin/bic_query_result.cgi?table=brca1_exons&nt=5589&base_change=del%20ATTGGGCA), the rs80350973 mutation was predicted to cause both a frameshift variant and a noncoding variant. As this frameshift mutation occurs in exon 24, it likely disrupts the second BRCT domain of BRCA1 protein (Fig. S2) which affects DNA damage repair and cell‐cycle checkpoint.
Clinicopathological features of the 125 TNBC patients with or without the rs80350973 mutation are shown in Table 3. The rs80350973 mutation carriers and noncarriers did not significantly differ with regard to age at diagnosis, histological type, tumor size, lymph node status, TNM stage, family history, and visceral metastasis status at relapse. However, a higher Ki‐67 index was seen in rs80350973 mutation carriers than in noncarriers (P = 0.004, Table 3). Kaplan–Meier survival analysis showed that patients with N3 stage lymph node status had significantly shorter DFS than did those with N0–2 status (P = 0.024, Fig. 1A). Although the subgroups with high versus low Ki‐67 indices (≤0.14 vs. >0.14, as recommended by the St. Gallen International Expert Consensus of 2011) showed no statistical discrepancy (P = 0.975, Fig. 1B), shorter DFS was seen in the Ki‐67 index >0.7 group than in the ≤0.7 group (P = 0.028, Fig. 1C). Statistical discrepancies in DFS between rs80350973 mutation carriers and noncarriers were not observed with regard to other clinicopathological features. A covariate Cox regression model showed no statistical difference between rs80350973 mutation carriers and noncarriers with regard to DFS (P = 0.335, Fig. 1D).
Table 3.
Comparison of clinicopathological features between BRCA1 rs80350973 mutation carriers and noncarriers
| Characteristic | Noncarriers (n = 116) | rs80350973 carriers (n = 9) | P‐value |
|---|---|---|---|
| Age at diagnosis (years) | |||
| >35 | 102 | 7 | 0.382 |
| ≤35 | 14 | 2 | |
| Histology of primary tumor | |||
| Ductal | 100 | 7 | 0.598 |
| Lobular | 8 | 0 | |
| Others | 7 | 1 | |
| Primary tumor size | |||
| T1 | 44 | 1 | 0.343 |
| T2 | 45 | 6 | |
| T3 | 9 | 1 | |
| T4 | 7 | 1 | |
| Axillary node involvement | |||
| N0 | 51 | 7 | 0.253 |
| N1 | 23 | 0 | |
| N2 | 20 | 1 | |
| N3 | 19 | 1 | |
| TNM Stage | |||
| I | 24 | 1 | 0.426 |
| II | 43 | 6 | |
| III | 35 | 2 | |
| IV | 7 | 0 | |
| Family history | |||
| Yes | 9 | 2 | 0.140 |
| No | 107 | 7 | |
| Visceral metastases at relapsea | |||
| Negative | 61 | 6 | 0.415 |
| Positive | 55 | 3 | |
| Ki‐67 index | |||
| Median | 0.48 | 0.75 | 0.004 |
| Range | 0.00~0.98 | 0.50~0.90 | |
Patient with one or more metastatic sites including lung, liver, or brain at time of relapse was defined as visceral metastasis positive, otherwise, the patients were defined as negative.
Figure 1.
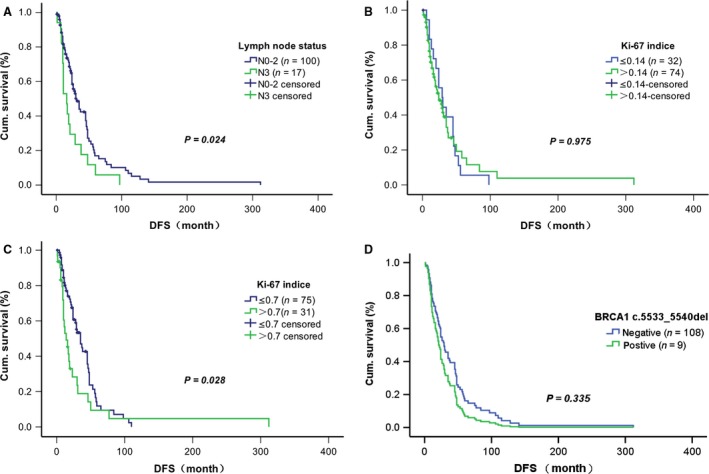
Effects of various factors on disease‐free survival among Chinese patients with Triple‐negative breast cancer. (A) Kaplan–Meier curves of lymph node status (N0–2 vs. N3). (B) Kaplan–Meier curves of Ki‐67 index at ≤0.14 versus >0.14. (C) Kaplan–Meier curves of Ki‐67 index at ≤0.7 versus >0.7. (D) Cox regression estimates by BRCA1 rs80350973 mutational status.
Discussion
Medical consultations for patients with newly diagnosed BCs increasingly use genetic counseling and testing. The National Comprehensive Cancer Network (NCCN) recently recommended BRCA1/2 testing among women with BC diagnoses who met well‐established criteria 15. In 2010, the NCCN guideline suggested that BRCA1/2 testing was indicated for women with TNBC who were younger than 40 years, based on emerging evidence that the triple‐negative phenotype was associated with hereditary cancer syndromes regardless of family history 16, 17. In 2013, the NCCN guidelines officially included women younger than 60 years with TNBC among those for whom genetic testing was appropriate 18.
Earlier studies with enormous cohorts have focused on the prevalence of BRCA1/2 mutations in different patient subgroups classified by race, ethnicity or geographical factors 18, 19, 20, 21. Other TNBC susceptibility genes such as BARD1 and CHEK2 were also detected at relatively high frequency 22, 23 which concords with our findings (Table 2). However, little is known about recurrent germline mutations of BRCA1/2 and its clinical significance in Chinese patients with TNBC. Aside from mutations at specific identifiable sites, their clinical interpretation may be challenging. In this study, we focused on a recurrent germline mutation of BRCA1 and its clinical relevance in Chinese patients with TNBC. We observed that a frameshift BRCA1 mutation (rs80350973) occurs at high frequency (9/125, 7.2%). This mutation was first reported by Suter et al. 24. in 2004, who identified it in two of 645 (0.3%) sporadic BC patients in Shanghai, China, but not in 319 unaffected health controls or in 342 patients with benign breast disease 24. The rs80350973 mutation was also identified in one of 60 (1.7%) Korean women with early‐onset BC (age ≤40 years) 25 and in one of 70 (1.4%) Chinese women with early‐onset BC (age ≤35 years) 26. In 2008, a larger cohort study of Chinese Han nationality was carried out by Li et al. 27., aimed at identifying of recurrent BRCA1/2 mutations. They found the rs80350973 mutation in 4 of 489 (0.8%) women with family histories of BC and/or early‐onset BC. Among these four carriers, two were diagnosed when younger than 35 years, one of whom had a family history of gastric cancer; the other two carriers had family histories of BC. Two rs80350973 mutation carriers (0.5%) were also found among 426 sporadic BC cases; both were diagnosed when older than 35 years, and neither reported family histories of breast or ovary cancer 27.
According to the studies mentioned above, rs80350973 mutation prevalence remained at a low level among Chinese women with sporadic BC (0.3–0.5%), and showed no significant correlation with family cancer history or early age of onset.
Our results show, for the first time, the high prevalence (7.2%) of rs80350973 mutation among 125 Chinese women with TNBC, which indicates its prevalence in this molecular subtype of BC.
So far, studies have shown that this mutation, which is considered deleterious, is only found in Asian people 12, 27. However, its clinical relevance in TNBC remained unclear. According to ClinVar and BIC database, the rs80350973 mutation induces a frameshift mutation in exon 24 of the BRCA1 gene, which could disrupt the BRCT domain (AA1784‐AA1863). This BRCT domain modulates interactions between BRCA1 and proteins that are phosphorylated in response to DNA damage 28; it has also been shown to bind directly to DNA double‐strand breaks (DSB) 29 and affects execution of the cell‐cycle checkpoint 30, 31. Because the BRCT domain could affect proliferation through the cellular checkpoint, disruption of this domain might lead to uncontrolled cell growth. Our data showed that the BRCA1 rs80350973 mutation is only associated with high Ki‐67 index in Chinese patients with TNBC (P = 0.004). A previous study showed that overexpression of Ki‐67 was associated with BRCA1 mutation 32 and was an indicator of poor prognosis in TNBC 33, 34. Thus, we sought to determine if the rs80350973 mutation could contribute to shorter DFS. However, we failed to find a statistical difference in DFS between rs80350973 mutation carriers and noncarriers (P = 0.335).
In conclusion, this study found a high mutational prevalence of rs80350973 in Chinese patients with TNBC, indicating a mutational prevalence of this variant. Bio‐informatics predictions further revealed that this mutation might disrupt the BRCT domain. As the BRCT domain critically affects DSB recognition and execution of checkpoint function, we supposed that rs80350973 mutation contributes to uncontrolled cell growth. This hypothesis is supported by our clinical result that rs80350973 mutation was more frequent in patients with higher Ki‐67 indices. Further functional studies to ascertain the molecular mechanisms behind rs80350973 mutation‐derived biological behaviors (such as cell proliferation and drug sensitivity) are warranted.
Conflict of Interest
No potential conflicts of interest were disclosed.
Supporting information
Figure S1. Identification of BRCA1 rs80350973 mutation in 100 TNBCs using Sanger sequencing.
Figure S2. Location of rs80350973‐induced secondary structure variation in BRCA1 protein.
Table S1. Panel list of 55 genes that affect susceptibility to breast cancer.
Table S2. Exome sequencing of 55 genes that affect susceptibility to breast cancer in 26 patients with TNBC.
Cancer Medicine 2017; 6(3):547–554
References
- 1. Wong‐Brown, M. W. , Meldrum C. J., Carpenter J. E., Clarke C. L., Narod S. A., Jakubowska A., et al. 2015. Prevalence of BRCA1 and BRCA2 germline mutations in patients with triple‐negative breast cancer. Breast Cancer Res. Treat. 150:71–80. [DOI] [PubMed] [Google Scholar]
- 2. Andreopoulou, E. , Schweber S. J., Sparano J. A., and McDaid H. M.. 2015. Therapies for triple negative breast cancer. Expert Opin. Pharmacother. 16:983–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dent, R. , Trudeau M., Pritchard K. I., Hanna W. M., Kahn H. K., Sawka C. A., et al. 2007. Triple‐negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 13:4429–4434. [DOI] [PubMed] [Google Scholar]
- 4. Marme, F. , and Schneeweiss A.. 2015. Targeted therapies in triple‐negative breast cancer. Breast Care (Basel). 10:159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kalimutho, M. , Parsons K., Mittal D., Lopez J. A., S. Srihari , and Khanna K. K.. 2015. Targeted therapies for triple‐negative breast cancer: combating a stubborn disease. Trends Pharmacol. Sci. 36:822–846. [DOI] [PubMed] [Google Scholar]
- 6. Atchley, D. P. , Albarracin C. T., Lopez A., Valero V., Amos C. I., Gonzalez‐Angulo A. M., et al. 2008. Clinical and pathologic characteristics of patients with BRCA‐positive and BRCA‐negative breast cancer. J. Clin. Oncol. 26:4282–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Comen, E. , Davids M., Kirchhoff T., Hudis C., Offit K., and Robson M.. 2011. Relative contributions of BRCA1 and BRCA2 mutations to “triple‐negative” breast cancer in Ashkenazi Women. Breast Cancer Res. Treat. 129:185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen, S. , and Parmigiani G.. 2007. Meta‐analysis of BRCA1 and BRCA2 penetrance. J. Clin. Oncol. 25:1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mavaddat, N. , Peock S., Frost D., Ellis S., Platte R., Fineberg E., et al. 2013. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J. Natl Cancer Inst. 105:812–822. [DOI] [PubMed] [Google Scholar]
- 10. Byrski, T. , Gronwald J., Huzarski T., Grzybowska E., Budryk M., Stawicka M., et al. 2010. Pathologic complete response rates in young women with BRCA1‐positive breast cancers after neoadjuvant chemotherapy. J. Clin. Oncol. 28:375–379. [DOI] [PubMed] [Google Scholar]
- 11. Matulonis, U. A. , Harter P., Gourley C., Friedlander M., Vergote I., Rustin G., et al. 2016. Olaparib maintenance therapy in patients with platinum‐sensitive, relapsed serous ovarian cancer and a BRCA mutation: Overall survival adjusted for postprogression poly(adenosine diphosphate ribose) polymerase inhibitor therapy. Cancer 122:1844–1852. [DOI] [PubMed] [Google Scholar]
- 12. Kwong, A. , Shin V. Y., Ho J. C., Kang E., Nakamura S., Teo S. H., et al. 2016. Comprehensive spectrum of BRCA1 and BRCA2 deleterious mutations in breast cancer in Asian countries. J. Med. Genet. 53:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tutt, A. , Ellis P., Kilburn L., Gilett C., Pinder S., Abraham J., et al. 2015. The TNT trial: A randomized phase III trial of carboplatin (C) compared with docetaxel (D) for patients with metastatic or recurrent locally advanced triple negative or BRCA1/2 breast cancer (CRUK/07/012). Cancer Res. 75:S3–01. doi: 10.1158/1538‐7445.SABCS14‐S3‐01. [Google Scholar]
- 14. Isakoff, S. J. , Mayer E. L., He L., Traina T. A., Carey L. A., Krag K. J., et al. 2015. TBCRC009: a multicenter phase II clinical trial of platinum monotherapy with biomarker assessment in metastatic triple‐negative breast cancer. J. Clin. Oncol. 33:1902–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coit, D. G. , Thompson J. A., Algazi A., Andtbacka R., Bichakjian C. K., Carson W. E. 3rd, et al. 2016. Melanoma, Version 2.2016, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw. 14:450–473. [DOI] [PubMed] [Google Scholar]
- 16. Ready, K. , and Arun B.. 2010. Clinical assessment of breast cancer risk based on family history. J. Natl. Compr. Canc. Netw. 8:1148–1155. [DOI] [PubMed] [Google Scholar]
- 17. Gonzalez‐Angulo, A. M. , Timms K. M., Liu S., Chen H., Litton J. K., Potter J., et al. 2011. Incidence and outcome of BRCA mutations in unselected patients with triple receptor‐negative breast cancer. Clin. Cancer Res. 17:1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greenup, R. , Buchanan A., Lorizio W., Rhoads K., Chan S., Leedom T., et al. 2013. Prevalence of BRCA mutations among women with triple‐negative breast cancer (TNBC) in a genetic counseling cohort. Ann. Surg. Oncol. 20:3254–3258. [DOI] [PubMed] [Google Scholar]
- 19. Gorski, B. 2006. Selected aspects of molecular diagnostics of constitutional alterations in BRCA1 and BRCA2 genes associated with increased risk of breast cancer in the polish population. Hered Cancer Clin. Pract. 4:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Couch, F. J. , Nathanson K. L., and Offit K.. 2014. Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science 343:1466–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Polsler, L. , Fiegl H., Wimmer K., Oberaigner W., Amberger A., Traunfellner P., et al. 2016. High prevalence of BRCA1 stop mutation c.4183C>T in the Tyrolean population: implications for genetic testing. Eur. J. Hum. Genet. 24:258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stevens, K. N. , Vachon C. M., and Couch F. J.. 2013. Genetic susceptibility to triple‐negative breast cancer. Cancer Res. 73:2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De Brakeleer, S. , De Greve J., Desmedt C., Joris S., Sotiriou C., Piccart M., et al. 2016. Frequent incidence of BARD1‐truncating mutations in germline DNA from triple‐negative breast cancer patients. Clin. Genet. 89:336–340. [DOI] [PubMed] [Google Scholar]
- 24. Suter, N. M. , Ray R. M., Hu Y. W., Lin M. G., Porter P., Gao D. L., et al. 2004. BRCA1 and BRCA2 mutations in women from Shanghai China. Cancer Epidemiol. Biomarkers Prev. 13:181–189. [DOI] [PubMed] [Google Scholar]
- 25. Choi, D. H. , Lee M. H., Bale A. E., Carter D., and Haffty B. G.. 2004. Incidence of BRCA1 and BRCA2 mutations in young Korean breast cancer patients. J. Clin. Oncol. 22:1638–1645. [DOI] [PubMed] [Google Scholar]
- 26. Song, C. G. , Hu Z., Wu J., Luo J. M., Shen Z. Z., Huang W., et al. 2006. The prevalence of BRCA1 and BRCA2 mutations in eastern Chinese women with breast cancer. J. Cancer Res. Clin. Oncol. 132:617–626. [DOI] [PubMed] [Google Scholar]
- 27. Li, W. F. , Hu Z., Rao N. Y., Song C. G., Zhang B., Cao M. Z., et al. 2008. The prevalence of BRCA1 and BRCA2 germline mutations in high‐risk breast cancer patients of Chinese Han nationality: two recurrent mutations were identified. Breast Cancer Res. Treat. 110:99–109. [DOI] [PubMed] [Google Scholar]
- 28. Wu, Q. , Jubb H., and Blundell T. L.. 2015. Phosphopeptide interactions with BRCA1 BRCT domains: More than just a motif. Prog. Biophys. Mol. Biol. 117:143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Silver, D. P. , and Livingston D. M.. 2012. Mechanisms of BRCA1 tumor suppression. Cancer Discov. 2:679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clark, S. L. , Rodriguez A. M., Snyder R. R., Hankins G. D., and Boehning D.. 2012. Structure‐function of the tumor suppressor BRCA1. Comput. Struct. Biotechnol. J. 1:pii: e201204005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shakya, R. , Reid L. J., Reczek C. R., Cole F., Egli D., Lin C. S., et al. 2011. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science 334:525–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hassanein, M. , Huiart L., Bourdon V., Rabayrol L., Geneix J., Nogues C., et al. 2013. Prediction of BRCA1 germ‐line mutation status in patients with breast cancer using histoprognosis grade, MS110, Lys27H3, vimentin, and KI67. Pathobiology 80:219–227. [DOI] [PubMed] [Google Scholar]
- 33. Li, H. , Han X., Liu Y., Liu G., and Dong G.. 2015. Ki67 as a predictor of poor prognosis in patients with triple‐negative breast cancer. Oncol. Lett. 9:149–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun, J. , Chen C., Wei W., Zheng H., Yuan J., Tu Y. I., et al. 2015. Associations and indications of Ki67 expression with clinicopathological parameters and molecular subtypes in invasive breast cancer: a population‐based study. Oncol. Lett. 10:1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Identification of BRCA1 rs80350973 mutation in 100 TNBCs using Sanger sequencing.
Figure S2. Location of rs80350973‐induced secondary structure variation in BRCA1 protein.
Table S1. Panel list of 55 genes that affect susceptibility to breast cancer.
Table S2. Exome sequencing of 55 genes that affect susceptibility to breast cancer in 26 patients with TNBC.


