Abstract
The human eye proteome is the latest addition to the HUPO Human Proteome Project (HPP). Semba et al. (The Human Eye Proteome Project: Perspectives on an emerging proteome. Proteomics 2013, 13, 2500–2511) establish a provisional baseline for the proteomes of the many anatomical compartments of the eye, based on literature review. As part of the Biology and Disease-driven HPP, they and their colleagues will generate fresh data and meet the stringent guidelines for protein identification and characterization as established by the HPP.
Keywords: Biology and Disease-driven Human Proteome Project, Biomedicine, Eye proteome, Human Proteome Project
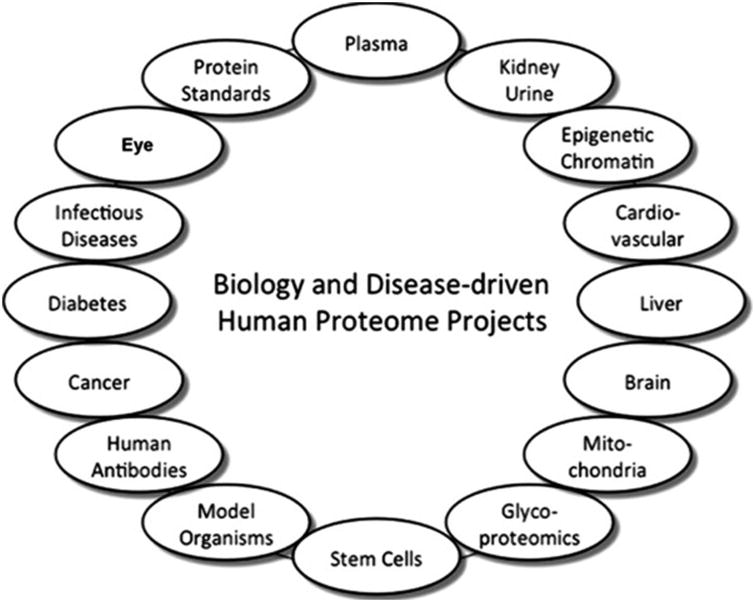
In this issue, Semba et al. [1] lay the foundation for a new component of the HUPO Biology and Disease-driven Human Proteome Project (B/D-HPP), the Human Eye Proteome Project. The goal is to gain insights into the pathophysiology of eye diseases and contribute to new preventive and therapeutic modalities. Figure 1 shows our updateof the B/D-HPP diagram published in January 2013 [2] to include now the eye proteome.
Figure 1.
Current set of 16-investigator teams for the Biology and Disease-driven Human Proteome Project (B/D-HPP), with addition of the Eye Proteome Project.
The B/D-HPP comprises (i) the pre-existing HUPO organ-and biofluid-based proteome projects developed over the past decade—the plasma, liver, brain, kidney/urine, stem cells, cardiovascular, and model organisms proteome projects and the human antibody, glycoproteomics, and protein standards initiatives, and (ii) new teams on diabetes, cancers, infectious diseases, epigenetics/chromatin, mitochondria, and now the eye. Anadditional team on autoimmune disorders is being organized. Details on the HPP and B/D-HPP executive committees and investigator teams are available at www.thehpp.org.
The B/D-HPP and the overall HPP have the ambitious goal of fundamentally transforming biological research by enabling the reliable detection and quantification of every human protein by the broad life science research community. As illustrated by the SRM study of ovarian cancer associated proteins in plasma and urine by Huttenhain et al. [3], targeted MS is now well established with software programs for assay development, robust data analysis tools, spectral library repositories, and SRM Atlas resources, as well as new data-independent acquisition MS methods [2].
Semba et al. [1] have compiled from literature reports using MS a total of 4842 nonredundant protein identifications across the many compartments of the eye, including the cornea (3708), tear film (1698), choroid (897), retina (672), vitreous humor (545), aqueous humor (465), and lens (273). Studies are not available for the ciliary body, sclera, iris, and optic nerve body. Many of these results are from studies of pathological conditions affecting the eye. The authors combined results from UniProt (Universal Protein Resource) and IPI (International Protein Index) with a clustering tool for sequence homology >90% to remove duplicates and isoform redundancy. MS Data Miner was used to generate subpro-teome lists for eye compartments. Unfortunately, most of these studies lack quality measures or confidence parameters for the diverse datasets tallied up in this review.
The authors' Supporting Information Table 2 shows the various criteria used by each of the included studies for protein and peptide identifications. It appears that many studies accepted identifications based on matching only one peptide. Raw spectra are available for only one of the 24 studies. Standardized reprocessing of data through Trans-Proteomic-Pipeline to create a Human-Eye PeptideAtlas is not feasible without the raw data. As the Eye Proteome Project proceeds, it will be highly desirable to create such an Eye Pep-tideAtlas, using the same stringent 1% false discovery rate (FDR) at the protein level (corresponding to about 0.2% FDR at the peptide level) that yielded 1929 canonical proteins for the 2010 build of the Human-Plasma PeptideAtlas [4] and 12 629 proteins for the 2012 build of the overall Human Pep-tideAtlas (G. S. Omenn, paper submitted) [5]. Thus, the 4842 grand total for eye protein identifications should be considered provisional, perhaps similar to the exhaustive list of 9504 proteins for plasma in the original plasma proteome paper [6] or, more likely, the “sequence-unique” list in the 2010 Plasma PeptideAtlas [7].
The average FDR for the datasets included in the current review must be much higher than 1% FDR, as acknowledged by the authors. In fact, the history of protein counts in the Human Plasma Proteome Project is instructive: the first publication presented a core dataset of 3020 protein identifications, based on two or more matching peptides and the author's assertion of “high confidence” [6]. Subsequently, a much more stringent analysis, including a Bonferroni-type adjustment for multiple comparisons, yielded 889 proteins [8]. As part of the B/D-HPP, the Eye Proteome investigators will have assistance from the PeptideAtlas team of the HPP for standardized analyses.
As fresh data are generated for each component of the eye, this team will be able to cross-analyze the Eye PeptideAtlas with the Plasma PeptideAtlas to ascertain whether proteotypic peptides have already been detected in plasma that could serve as accessible biomarkers for the corresponding proteins inside the eye, especially for studies of the prevention, progression, or treatment of eye diseases. As a demonstration of the approach, they have compared the provisional eye proteome with the 2010 build of the Human-Plasma PeptideAtlas and found 1317 proteins common to eye and plasma, 3525 only in the eye, and 611 only in the plasma. As Semba et al. [1] note, future studies will take account of PTMs, alternative splice isoforms, polymorphic mutations (SNPs, single nucleotide polymorphisms), and protein complexes, as will the overall HPP.
Abbreviations
- B/D-HPP
Biology and Disease-driven Human Pro-teome Project
- FDR
false discovery rate
- HPP
Human Proteome Project
Footnotes
The author has declared no conflict of interest.
References
- 1.Semba RD, Enghild JJ, Venkatraman V, Dyrlund TF, Van Eyk JE. The Human Eye Proteome Project: perspective sonane merging proteome. Proteomics. 2013;13:2500–2511. doi: 10.1002/pmic.201300075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebersold R, Bader GD, Edwards AM, van Eyk JE, et al. The biology/disease-driven human proteome project (B/D-HPP): enabling protein research for the life sciences community. J Proteome Res. 2013;12:23–27. doi: 10.1021/pr301151m. [DOI] [PubMed] [Google Scholar]
- 3.Huttenhain R, Soste M, Selevsek N, Rost H, et al. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci Transl Med. 2012;4:142ra194. doi: 10.1126/scitranslmed.3003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrah T, Deutsch EW, Omenn GS, Campbell DS, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006353. M110 006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrah T, Deutsch EW, Hoopmann MR, Hallows JL, et al. The state of the human proteome in 2012 as viewed through PeptideAtlas. J Proteome Res. 2013;12:162–171. doi: 10.1021/pr301012j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omenn GS, States DJ, Adamski M, Blackwell TW, et al. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 7.Farrah T, Deutsch EW, Omenn GS, Campbell DS, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006353. M110 006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.States DJ, Omenn GS, Blackwell TW, Fermin D, et al. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol. 2006;24:333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]