Abstract
The bacterial origins of DNA replication have been isolated from Pseudomonas aeruginosa and Pseudomonas putida. These origins comprise a second class of bacterial origins distinct from enteric-type origins: both origins function in both Pseudomonas species, and neither functions in Escherichia coli; enteric origins do not function in either pseudomonad. Both cloned sequences hybridize to chromosomal fragments that show properties expected of replication origins. These origin plasmids are highly unstable, are present at low copy number, and show mutual incompatibility properties. DNA sequence analysis shows that both origins contain several 9-base-pair (bp) E. coli DnaA protein binding sites; four of these are conserved in position and orientation, two of which resemble the R1 and R4 sites of the E. coli origin. Conserved 13-bp direct repeats adjacent to the analogous R1 site are also found. No GATC sites are in the P. aeruginosa origin and only four are in the P. putida origin; no other 4-bp sequence is present in high abundance. Both origins are found between sequences similar to the E. coli and Bacillus subtilis dnaA, dnaN, rpmH, and rnpA genes, a gene organization identical to that for B. subtilis and unlike that of E. coli. A second autonomously replicating sequence was obtained from P. aeruginosa that has some properties of bacterial origins.
Full text
PDF
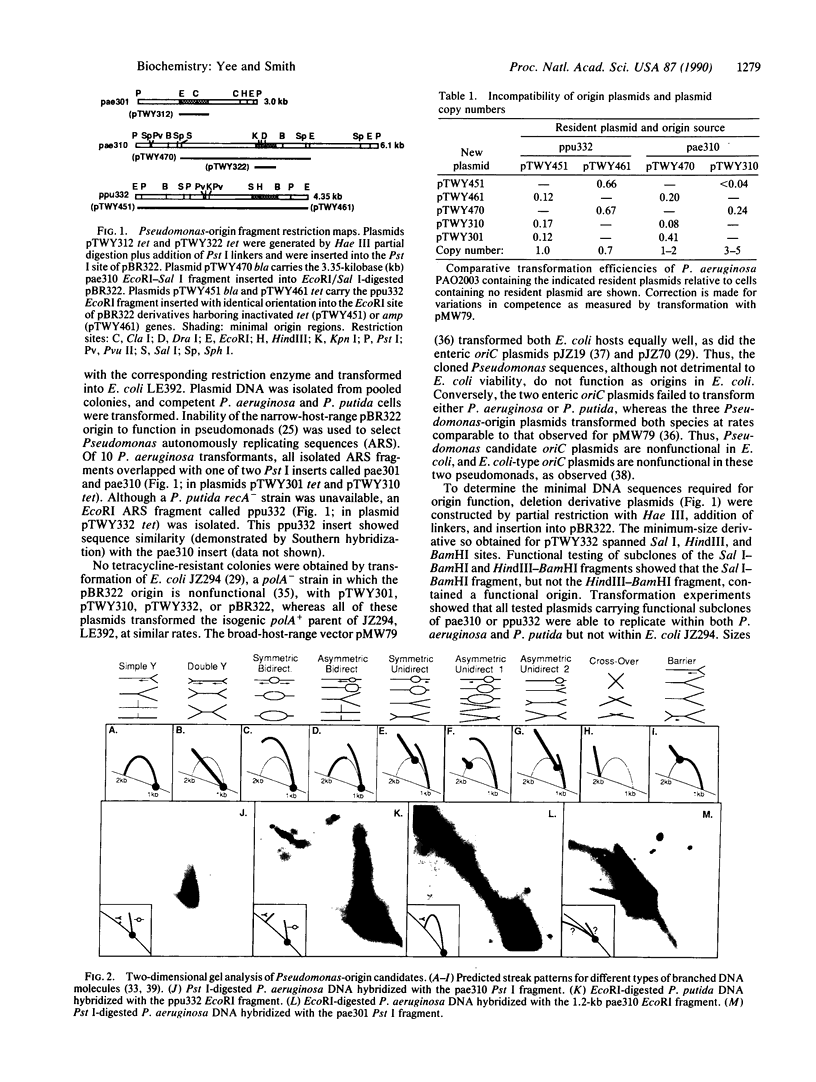



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Timmis K. N. Host: vector systems for gene cloning in Pseudomonas. Curr Top Microbiol Immunol. 1982;96:47–67. doi: 10.1007/978-3-642-68315-2_4. [DOI] [PubMed] [Google Scholar]
- Bakker A., Smith D. W. Methylation of GATC sites is required for precise timing between rounds of DNA replication in Escherichia coli. J Bacteriol. 1989 Oct;171(10):5738–5742. doi: 10.1128/jb.171.10.5738-5742.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeyron T., Kean K., Forterre P. DNA adenine methylation of GATC sequences appeared recently in the Escherichia coli lineage. J Bacteriol. 1984 Nov;160(2):586–590. doi: 10.1128/jb.160.2.586-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilofsky H. S., Burks C. The GenBank genetic sequence data bank. Nucleic Acids Res. 1988 Mar 11;16(5):1861–1863. doi: 10.1093/nar/16.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. A replication fork barrier at the 3' end of yeast ribosomal RNA genes. Cell. 1988 Nov 18;55(4):637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Brooks J. E., Blumenthal R. M., Gingeras T. R. The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 1983 Feb 11;11(3):837–851. doi: 10.1093/nar/11.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler P. M., Krishnapillai V. Isolation and properties of recombination-deficient mutants of Pseudomonas aeruginosa. Mutat Res. 1974 Apr;23(1):15–23. doi: 10.1016/0027-5107(74)90155-9. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Fujita M. Q., Yoshikawa H., Ogasawara N. Structure of the dnaA region of Pseudomonas putida: conservation among three bacteria, Bacillus subtilis, Escherichia coli and P. putida. Mol Gen Genet. 1989 Feb;215(3):381–387. doi: 10.1007/BF00427033. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Funnell B. E., Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984 Oct;38(3):889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- George D. G., Barker W. C., Hunt L. T. The protein identification resource (PIR). Nucleic Acids Res. 1986 Jan 10;14(1):11–15. doi: 10.1093/nar/14.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding N. E., Cleary J. M., Smith D. W., Michon J. J., Brusilow W. S., Zyskind J. W. Chromosomal replication origins (oriC) of Enterobacter aerogenes and Klebsiella pneumoniae are functional in Escherichia coli. J Bacteriol. 1982 Dec;152(3):983–993. doi: 10.1128/jb.152.3.983-993.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. Temperature-sensitive mutants for the replication of plasmids in Escherichia coli: requirement for deoxyribonucleic acid polymerase I in the replication of the plasmid ColE 1 . J Bacteriol. 1973 Jun;114(3):1116–1124. doi: 10.1128/jb.114.3.1116-1124.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A., Henckes G., Vannier F., Séror S. J. Chromosomal initiation in Bacillus subtilis may involve two closely linked origins. Mol Gen Genet. 1987 Jun;208(1-2):37–44. doi: 10.1007/BF00330419. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Carraway M., Frey A. Z., Brown L., Arraj J. A. Insertion mutations in the dam gene of Escherichia coli K-12. Mol Gen Genet. 1983;192(1-2):288–289. doi: 10.1007/BF00327681. [DOI] [PubMed] [Google Scholar]
- Marinus M. G. DNA methylation in Escherichia coli. Annu Rev Genet. 1987;21:113–131. doi: 10.1146/annurev.ge.21.120187.000553. [DOI] [PubMed] [Google Scholar]
- Matsui M., Oka A., Takanami M., Yasuda S., Hirota Y. Sites of dnaA protein-binding in the replication origin of the Escherichia coli K-12 chromosome. J Mol Biol. 1985 Aug 5;184(3):529–533. doi: 10.1016/0022-2836(85)90299-2. [DOI] [PubMed] [Google Scholar]
- Messer W., Bellekes U., Lother H. Effect of dam methylation on the activity of the E. coli replication origin, oriC. EMBO J. 1985 May;4(5):1327–1332. doi: 10.1002/j.1460-2075.1985.tb03780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Moriya S., Fukuoka T., Ogasawara N., Yoshikawa H. Regulation of initiation of the chromosomal replication by DnaA-boxes in the origin region of the Bacillus subtilis chromosome. EMBO J. 1988 Sep;7(9):2911–2917. doi: 10.1002/j.1460-2075.1988.tb03149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya S., Ogasawara N., Yoshikawa H. Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. III. Nucleotide sequence of some 10,000 base pairs in the origin region. Nucleic Acids Res. 1985 Apr 11;13(7):2251–2265. doi: 10.1093/nar/13.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- O'Neill E. A., Bender R. A. Klebsiella pneumoniae origin of replication (oriC) is not active in Caulobacter crescentus, Pseudomonas putida, and Rhodobacter sphaeroides. J Bacteriol. 1988 Aug;170(8):3774–3777. doi: 10.1128/jb.170.8.3774-3777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N., Mizumoto S., Yoshikawa H. Replication origin of the Bacillus subtilis chromosome determined by hybridization of the first-replicating DNA with cloned fragments from the replication origin region of the chromosome. Gene. 1984 Oct;30(1-3):173–182. doi: 10.1016/0378-1119(84)90118-5. [DOI] [PubMed] [Google Scholar]
- Ogden G. B., Pratt M. J., Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988 Jul 1;54(1):127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- Parker B., Marinus M. G. A simple and rapid method to obtain substitution mutations in Escherichia coli: isolation of a dam deletion/insertion mutation. Gene. 1988 Dec 20;73(2):531–535. doi: 10.1016/0378-1119(88)90517-3. [DOI] [PubMed] [Google Scholar]
- Russell D. W., Zinder N. D. Hemimethylation prevents DNA replication in E. coli. Cell. 1987 Sep 25;50(7):1071–1079. doi: 10.1016/0092-8674(87)90173-5. [DOI] [PubMed] [Google Scholar]
- Skarstad K., von Meyenburg K., Hansen F. G., Boye E. Coordination of chromosome replication initiation in Escherichia coli: effects of different dnaA alleles. J Bacteriol. 1988 Feb;170(2):852–858. doi: 10.1128/jb.170.2.852-858.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. W., Garland A. M., Herman G., Enns R. E., Baker T. A., Zyskind J. W. Importance of state of methylation of oriC GATC sites in initiation of DNA replication in Escherichia coli. EMBO J. 1985 May;4(5):1319–1326. doi: 10.1002/j.1460-2075.1985.tb03779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séror-Laurent S. J., Henckes G. An RNA-DNA copolymer whose synthesis is correlated with the transcriptional requirement for chromosomal initiation in Bacillus subtilis contains ribosomal RNA sequences. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3586–3590. doi: 10.1073/pnas.82.11.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wood D. O., Hollinger M. F., Tindol M. B. Versatile cloning vector for Pseudomonas aeruginosa. J Bacteriol. 1981 Mar;145(3):1448–1451. doi: 10.1128/jb.145.3.1448-1451.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S., Hirota Y. Cloning and mapping of the replication origin of Escherichia coli. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5458–5462. doi: 10.1073/pnas.74.12.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee T., Inouye M. Two-dimensional DNA electrophoresis applied to the study of DNA methylation and the analysis of genome size in Myxococcus xanthus. J Mol Biol. 1982 Jan 15;154(2):181–196. doi: 10.1016/0022-2836(82)90059-6. [DOI] [PubMed] [Google Scholar]
- Zyskind J. W., Cleary J. M., Brusilow W. S., Harding N. E., Smith D. W. Chromosomal replication origin from the marine bacterium Vibrio harveyi functions in Escherichia coli: oriC consensus sequence. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1164–1168. doi: 10.1073/pnas.80.5.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind J. W., Deen L. T., Smith D. W. Isolation and mapping of plasmids containing the Salmonella typhimurium origin of DNA replication. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3097–3101. doi: 10.1073/pnas.76.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind J. W., Smith D. W. Nucleotide sequence of the Salmonella typhimurium origin of DNA replication. Proc Natl Acad Sci U S A. 1980 May;77(5):2460–2464. doi: 10.1073/pnas.77.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]





