Abstract
Long-read sequencing technology promises to greatly enhance de novo assembly of genomes for nonmodel species. Although the error rates of long reads have been a stumbling block, sequencing at high coverage permits the self-correction of many errors. Here, we sequence and de novo assemble the genome of Drosophila serrata, a species from the montium subgroup that has been well-studied for latitudinal clines, sexual selection, and gene expression, but which lacks a reference genome. Using 11 PacBio single-molecule real-time (SMRT cells), we generated 12 Gbp of raw sequence data comprising ∼65 × whole-genome coverage. Read lengths averaged 8940 bp (NRead50 12,200) with the longest read at 53 kbp. We self-corrected reads using the PBDagCon algorithm and assembled the genome using the MHAP algorithm within the PBcR assembler. Total genome length was 198 Mbp with an N50 just under 1 Mbp. Contigs displayed a high degree of chromosome arm-level conservation with the D. melanogaster genome and many could be sensibly placed on the D. serrata physical map. We also provide an initial annotation for this genome using in silico gene predictions that were supported by RNA-seq data.
Keywords: Drosophila, montium, PacBio, Celera, long reads, genome assembly
Second-generation sequencing (2GS) platforms, such as Illumina sequencing-by-synthesis, have dramatically reduced genome sequencing costs while increasing throughput exponentially (Shendure and Ji 2008). The relatively low cost and massive throughput of 2GS platforms have paved the way for sequencing and de novo assembly of thousands of species’ genomes (Alkan et al. 2011). 2GS methods generate short reads (less than a few hundred base pairs in length) that have limitations for de novo genome assembly, where assembly is performed without the aid of a reference genome (Green 1997; Miller et al. 2008; Nagarajan and Pop 2013; Alkan et al. 2011). With short reads, de novo assembly is an inherently difficult computational problem because repetitive DNA sequences are often much longer than the length of each read (Ukkonen 1992). For instance, it has been estimated that short-read de novo assemblies could be missing up to 20% of sequence information because repeat DNA sequences can increase the number of misassembled and fragmented regions (Schatz et al. 2010; Alkan et al. 2011; Ukkonen 1992). One way to alleviate the problem of repetitive DNA in the de novo assembly process has been to incorporate a second set of mate-pair libraries with very long inserts (> 2 kbp) (Li et al. 2010; Chaisson et al. 2009; Simpson et al. 2009; Alkan et al. 2011; Butler et al. 2008). Mate-pair libraries can resolve repeats (Treangen and Salzberg 2012; Wetzel et al. 2011) and improve scaffolding (van Heesch et al. 2013), but paired-end contamination and insert size misestimation can also lead to misassemblies (Phillippy et al. 2008; Sahlin et al. 2016).
More recently, third-generation (3GS) single-molecule sequencing technologies, such as Pacific Biosciences’ (PacBio) SMRT sequencing and Oxford Nanopore’s MinION sequencing, which currently produce much longer reads of up to 54 kbp (Lee et al. 2014) and > 10 kbp (Quick et al. 2014), respectively, can overcome some of the shortcomings of 2GS de novo assembly (Berlin et al. 2015). Although long-read sequencing technology produces reads with a high error rate, ranging from 82.1% (Chin et al. 2011) to 84.6% accuracy (Rasko et al. 2011), sequencing errors occur at more or less random positions across long reads (Chin et al. 2013) and can be corrected with 2GS short-read data (Koren et al. 2012) or by using excess 3GS reads for self-correction (Chin et al. 2013).
In this paper, we use PacBio long-read sequencing to de novo assemble the genome of the fly, Drosophila serrata, which has been particularly well-studied from an evolutionary standpoint. D. serrata is a member of the D. montium subgroup, which split from the D. melanogaster subgroup ∼40 MYA (Tamura et al. 2004), and consists of an estimated 98 species (Brake and Bächli 2008). At present, only one draft genome assembly (D. kikkawai) is available (Chen et al. 2014) from this species-rich subgroup. D. serrata has a broad geographical distribution, ranging from Papua New Guinea to south eastern Australia and has emerged as a powerful model for addressing evolutionary questions such as the evolution of species borders (Blows and Hoffman 1993; Hallas et al. 2002; Magiafoglou et al. 2002) and climate adaptation (Frentiu and Chenoweth 2010; Latimer et al. 2011; Kellermann et al. 2009). The species has also been used to investigate sexual selection (Hine et al. 2002; Gosden and Chenoweth 2011; Frentiu and Chenoweth 2008; Chenoweth et al. 2015), male mate choice (Chenoweth and Blows 2003; Chenoweth et al. 2007), mate recognition (Higgie et al. 2000), sexual dimorphism (Chenoweth et al. 2008; Yassin et al. 2016), sexual conflict (Delcourt et al. 2009), and indirect genetic effects (Chenoweth et al. 2010b). In addtion, its cuticular hydrocarbons, which serve as contact pheromones (Chung et al. 2014), have been extensively used to develop novel multivariate quantitative genetic approaches for exploring genetic constraints on adaptation (Blows et al. 2004; Chenoweth et al. 2010a; McGuigan et al. 2011b; Rundle et al. 2009).
Despite the importance of D. serrata as a model for evolutionary research, our poor understanding of its genome remains a significant limitation. Linkage and physical genome maps are available (Stocker et al. 2012) and an expressed sequence tag (EST) library has been developed (Frentiu et al. 2009), but the species lacks a draft genome. Here, we report the sequencing and assembly of the D. serrata genome using exclusively PacBio SMRT technology. We also provide an initial annotation of the genome based on in silco gene predictors supported by empirical RNA-seq data. Our de novo genome and its annotation will provide a resource for ongoing population genomic and trait mapping studies in this species as well as facilitate broader studies of genome evolution in the family Drosophilidae.
Materials and Methods
Fly strains and DNA extraction
We sequenced a mix of ∼100 mg of males and females from a single inbred line that originated from Forster, Australia, and had been inbred via full-sib mating for 10 generations before being maintained at a large population size (N ∼ 250 individuals) (McGuigan et al. 2011b). A single further generation of full-sib inbreeding was applied before extraction of DNA. This same inbred line was used for the D. serrata linkage map, was the founding line for previous mutation accumulation studies (Latimer et al. 2015; McGuigan et al. 2011a, 2014a,b), and is fixed for the light female abdominal pigmentation phenotype mapped by Yassin et al. (2016). High molecular weight DNA was extracted from fly bodies (heads were excluded to reduce eye pigment contamination) using a QIAGEN Gentra Puregene Tissue Kit (Cat #158667), which produced fragments > 100 kbp (measured using pulsed-field gel electrophoresis). Two phenol–chloroform extractions were performed at the University of California, Davis at the DNA Technologies Core prior to preparation of a standard sequencing library.
Genome sequencing and assembly
DNA was sequenced using 11 SMRT cells and P6-C4 chemistry on the PacBio RS II platform. In total, this produced ∼13 Gbp spanning 136,119 filtered subreads with a mean read length of 8840 bp and an N50 of 12,220 bp (Supplemental Material, Figure S1). The PacBio genome was assembled using the PBcR pipeline, which implements the MHAP algorithm within the Celera Assembler (version 8.3rc2) (Berlin et al. 2015), and polished with Quiver (GenomicConsensus version 0.9.2 and ConsensusCore version: 0.8.8) (Chin et al. 2013) in three steps: (1) errors were corrected in reads using PBDagCon, which requires at least 50 × genome coverage and utilizes the consensus of oversampled sequences (Chin et al. 2013); (2) overlapping sequences were assembled using MHAP and the Celera Assembler (Berlin et al. 2015); and (3) contigs were polished with Quiver to correct for spurious SNP calls and small indels (Chin et al. 2013). The “sensitive” setting was used for both read correction and genome assembly (Berlin et al. 2015) whereas the default settings were used for polishing with Quiver (Chin et al. 2013). We elected to correct all reads as opposed to the default longest 40 ×. The longest 25 × corrected reads were subsequently used for genome assembly. The PBDagCon correction was performed on a computer with 60 CPU cores and 1 TB of RAM; 58 CPU cores were used for the assembly and the amount of RAM used, although not tracked, was far less than machine capacity. Error correction with PBDagCon took ∼26 days. Assembly of corrected reads using MHAP and the Celera Assembler took ∼19 hr using 28 CPU cores. Our initial runs using the much faster error correction algorithm (HGAP) produced a slightly shorter assembly (194 Mbp compared to 198 Mbp) with a slightly lower N50 (0.88 Mbp vs. 0.95 Mbp). Therefore, we chose to use the more sensitive PBDagCon correction method.
Transcriptome sequencing and assembly
The same inbred fly strain that was used for DNA sequencing was also used for adult mRNA sequencing to annotate the D. serrata genome. Adult males and females were transferred to fresh vials shortly after eclosion and held in groups of ∼25 where they were allowed to mate and lay eggs for 2 d. They were then sexed under light CO2 anesthesia and snap frozen using liquid nitrogen in groups of 10; at the time of freezing, all flies were assumed to be nonvirgins. Total RNA was extracted from each pool of flies using the standard TRIzol protocol. Initial quality assessment of the total RNA using a NanoDrop and gel electrophoresis indicated that the RNA was of high quality, this was later confirmed with a RNA integrity number > 7 (measured uisng a BioAnalyzer). RNA was stored at −80° for several days before being shipped for sequencing.
One male and one female 75 bp paired-end sequencing library was prepared using the TruSeq Stranded mRNA Library prep kit and sequenced on an Illumina NextSeq500 at the Ramaciotti Centre for Genomics, University of New South Wales, Australia. In total, 79 and 88 million reads were produced for males and females, respectively. Quality assessment of the RNA-seq data using FastQC (Andrews 2010) indicated that the reads were of a high quality and therefore no trimming of reads was performed. The transcriptome was de novo assembled for each sex separately using Trinity version 2.1.1 (Grabherr et al. 2011), where all reads were used and the jaccard_clip option was enabled to minimize gene fusion events caused by UTR overlap in high gene density regions.
Annotation
Maker version 2.31.8 (Campbell et al. 2014; Holt and Yandell 2011) was used to annotate the PacBio genome via incorporation of in silico gene models detected by Augustus (Stanke and Morgenstern 2005) and/or SNAP (Johnson et al. 2008), the de novo D. serrata male and female transcriptomes, and protein sequences from 12 Drosophila species genomes (D. ananassae r1.04, D. erecta r1.04, D. grimshawi r1.3, D. melanogaster r6.07, D. mojavensis r1.04, D. persmillis r1.3, D. pseudoobscura pseudoobscura r3.03, D. sechellia 1.3, D. simulans, r2.01, D. virilis r1.03, D. willistoni r1.04, and D. yakuba r1.04) obtained from FlyBase (McQuilton et al. 2012; Attrill et al. 2016). Repeat masking was performed based on D. melanogaster training (Smit et al. 1996). Maker was run with default settings apart from allowing Maker to take extra steps to identify alternate splice variants and correct for erroneous gene fusion events.
Data availability
All sequence data including PacBio and RNA-seq reads have been submitted to public repositories and are available via the D. serrata genome NCBI project accession PRJNA355616. The genome assembly and annotation tracks are available from http://www.chenowethlab.org. We also supply a list of D. melanogaster orthologs in Table S1.
Results and Discussion
To assemble a draft D. serrata genome, we sequenced DNA from a pool of adult males and females (that originated from a single inbred line) to a coverage of ∼65 × using PacBio long-read, SMRT sequencing technology. This produced 136,119 filtered subreads with a mean read length of 8940 bp and a read N50 of 12,200 bp that spanned > ∼13 Gbp (Figure S1). The PacBio reads were assembled using the MHAP algorithm within the Celera Assembler (Miller et al. 2008; Berlin et al. 2015) after self-correction using PBDagCon (Chin et al. 2013). The final genome was polished with a single iteration of Quiver (Chin et al. 2013) and consisted of 1360 contigs spanning > 198 Mbp with a GC content of 39.13% (Table 1). The longest contig was ∼7.3 Mbp and the N50 of all contigs was ∼0.95 Mbp. Flow cytometry studies suggest that species of the montium subgroup commonly have genome lengths over 200 Mbp (Gregory and Johnston 2008) with the estimate for the female D. serrata genome being ∼215 Mbp (0.22 pg). This estimate is in broad agreement with our assembly length of 198 Mbp for the female genome.
Table 1 D. serrata genome assembly statistics.
| Description | Statistic |
|---|---|
| Number of contigs | 1360 |
| Genome size (bp) | 198,298,763 |
| Longest contig (bp) | 7,300,740 |
| < 1 kbp | 0.0% |
| 1–10 kbp | 3.3% |
| 10–100 kbp | 78.8% |
| 100–1000 kbp | 15.3% |
| > 1 Mbp | 2.6% |
| N50 (bp) | 942,627 |
| GC content | 39.13% |
Contig length percentages refer to percent total length in each size bin.
Completeness
Genome completeness was assessed using BUSCO gene set analysis version 2.0, which includes a set of 2799 genes specific to Diptera (Simao et al. 2015). The D. serrata assembly contained 96.2% of the BUSCO genes with 94.1% being complete single-copy (defined as complete when the gene’s length is within 2 SDs of the BUSCO group’s mean length) and 2.5% detected as fragmented. Only 1.3% of the BUSCO genes were not found in the D. serrata assembly (Table 2). Completeness of the D. serrata genome was similar to the reference D. melanogaster genome (version r6.05), which contained 98.7% complete BUSCO genes. As a further point of comparison, we computed BUSCO metrics for a recent PacBio-only assembly of the D. melanogaster ISO1 strain genome using all 790 contigs rather than the 132 that were constructed from > 50 reads only [http://www.cbcb.umd.edu/software/PBcR/MHAP/ (quivered full assembly)], and we also analyzed the only other member of the montium subgroup with a publicly available genome assembly, D. kikkawai, (https://www.hgsc.bcm.edu/arthropods/drosophila-modencode-project; NCBI PRJNA62319). Although these assemblies tended to contain marginally lower numbers of missing BUSCOs, metrics were generally very similar (Table 2), indicating a high level of completeness for the D. serrata assembly.
Table 2. BUSCO gene content assessment for D. serrata and two different D. melanogaster assemblies, version r6.05 from www.flybase.org, and the full ISO 1 PacBio assembly of Berlin et al. (2015) consisting of 790 contigs, also constructed with the PBcR pipeline.
| Category | D. serrata | D. kikkawai | D. melanogaster | |
|---|---|---|---|---|
| r6.05 | PacBio | |||
| Complete Single-copy BUSCOs (%) | 94.1 | 97.1 | 98.2 | 97.7 |
| Duplicated (%) | 2.1 | 1.0 | 0.5 | 0.6 |
| Fragmented BUSCOs (%) | 2.5 | 1.2 | 0.8 | 0.8 |
| Missing BUSCOs (%) | 1.3 | 0.8 | 0.5 | 0.9 |
A total of 2799 BUSCOs were searched that form a set of highly conserved Dipteran genes. PacBio, Pacific Biosciences; BUSCO, Benchmarking Universal Single-Copy Ortholog.
Fragmentation and misassemblies
Although our assembly consisted of 1360 contigs with a N50 of 0.94 Mbp, which was an N50 at the upper end of what might be expected for a short-read assembly, it is much lower than a recent PacBio-only assembly of the D. melanogaster genome (Berlin et al. 2015). There are several reasons why this might be the case. First, we report metrics on all contigs in the assembly rather than excluding those that incorporated < 50 reads, as was the case for the D. melanogaster assembly (Berlin et al. 2015) (132 contigs with an N50 of 13.6 M). Excluding such contigs resulted in a D. serrata assembly of only 273 contigs with a total genome length of 175 Mbp (vs. 198 Mbp) and an N50 of 1.4 Mbp. In this reduced assembly, half of the genome was represented in only 25 contigs, which is closer to the performance seen for D. melanogaster. While contigs with < 50 read support were generally short (median 23.5 kbp and range 6.3–110 kbp) and could be excluded in some cases on the basis of quality, when we examined the D. serrata annotation data, we saw that many of these contigs contained predicted genes that had RNA-seq support, including 14 complete single-copy BUSCOs. Therefore, we have retained all contigs in our assembly.
Second, although our N50 filtered subread length of 12,200 kbp is on a par with the D. melanogaster P5-C3 filtered subread lengths (12.2–14.2 kbp) (Kim et al. 2014), we had approximately half the coverage of the D. melanogaster assembly (65 × vs. 130 ×), which may have reduced our ability to span repetitive regions of the D. serrata genome. To examine this further, we reran the PBcR pipeline with D. melanogaster data from Kim et al. (2014) but downsampled it to 65 ×. We did not see genome contiguity drop to the levels seen for D. serrata (data not shown) and note that similar findings were observed by Chakraborty et al. (2016) (see their Figure 5). Therefore, it seems likely that the D. serrata genome, which is longer than that of D. melanogaster, may also be more complex due to longer repetitive regions. Therefore, adequate repeat-spanning coverage would presumably require additional very long reads to achieve the same assembly contiguity seen for D. melanogaster. A third factor possibly contributing to a higher degree of fragmentation in our assembly is residual heterozygosity, which may have been higher in our D. serrata line than the ISO1 D. melanogaster line.
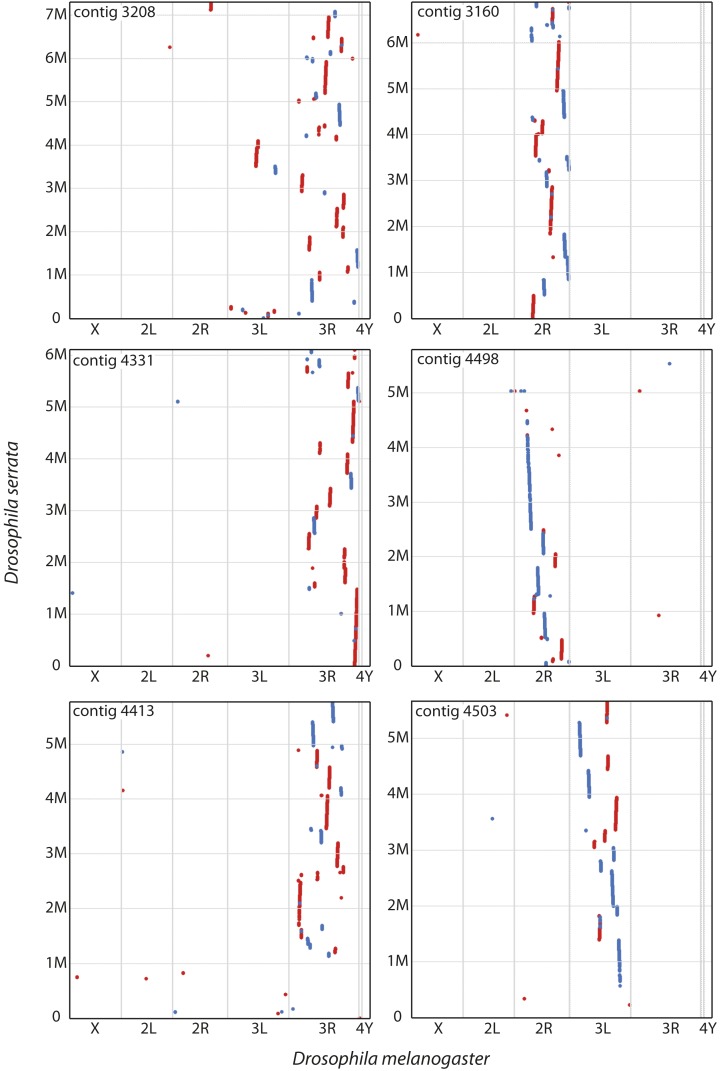
We used several methods to assess the quality of the genome with regards to misassemblies. First, because the D. serrata physical map indicates very strong chromosome arm-level conservation of gene content between D. serrata and D. melanogaster (Stocker et al. 2012), we examined possible misassemblies between chromosomal arms by aligning the six largest contigs (total length ∼37 Mbp) to the D. melanogaster genome using MUMmer (Kurtz et al. 2004). If there were no chromosome arm misplacements, then it was expected that each contig would align to a single D. melanogaster chromosome arm, albeit fragmented due to changes in gene order. This was largely the case (Figure 1), where each contig aligned to a single D. melanogaster chromosome arm but with minor sections of alignment to other chromosome arms toward the contig edges where repetitive elements were more likely to be found. The one major exception to this general pattern of conservation was found in the longest contig in the assembly, contig 3208, which aligned mainly to D. melanogaster 3R but contained an ∼600 kbp segment that aligned to D. melanogaster 3L. To test whether this was likely to be a misassembly, we searched the contig for previously published SNP markers that have been placed on the D. serrata linkage map. The marker m25 (Stocker et al. 2012), which maps to 3L, was located in the suspected misassembled region (contig 3208 and position 3,537,591) indicating that a misassembly rather than a genomic translocation rearrangement between 3R and 3L was most likely.
Figure 1.
Alignment of the six longest contigs from the D. serrata assembly to D. melanogaster genome version 6.05. Red dots indicate a MUMmer alignment that matches to the D. melanogaster genome in the forward orientation; blue dots indicate a MUMmer alignment that matches to the D. melanogaster genome in the reverse orientation. M, million.
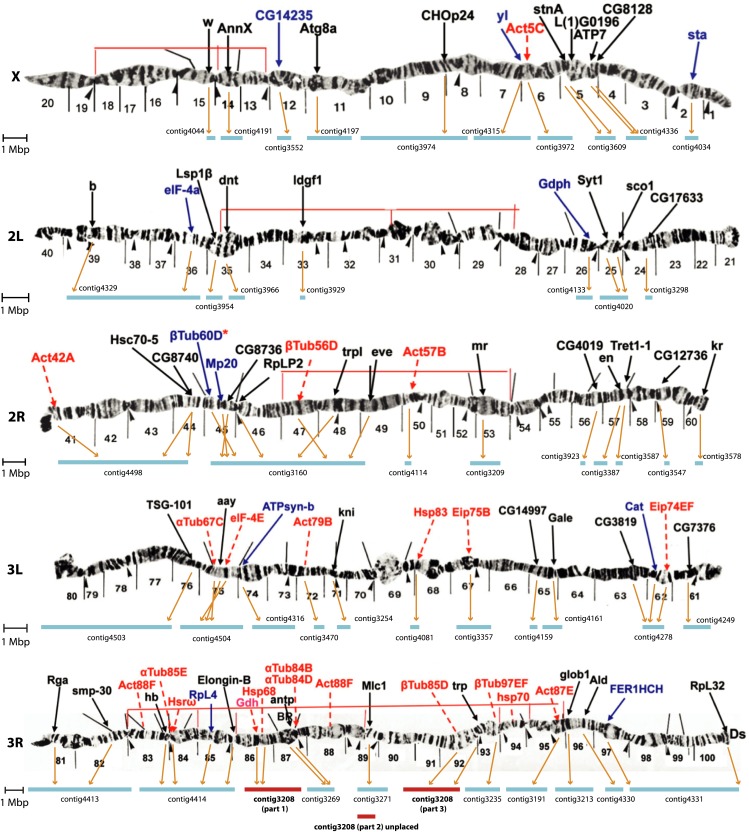
To further examine assembly quality, we compared our assembly to the entire physical genome map of D. serrata (Stocker et al. 2012), where in situ hybridization was used to physically locate 78 genes. We were able to assess possible misassemblies when a contig contained multiple physically mapped genes (11 contigs ranging in size from ∼1 to ∼6 Mbp). Using this approach, we observed no apparent chromosome arm-level assignment errors beyond that seen for contig 3208 (Figure 2). Furthermore, when contigs contained three or more physically mapped genes, gene order could be examined. We saw three cases of apparent gene order reversal (two on 2R and one on 3L). Interestingly, two of these regions map to the positions of known chromosomal inversions (Mavragani-Tsipidou et al. 1990), which is perhaps not unexpected given that different inbred lines were used for the physical map and genome sequencing. After considering these probable inversions, gene order and location appears to be largely correct for these 11 contigs at least. For contig 3208, each section that aligned to 3R could be placed on the physical map only after splitting the contig into three pieces based on the previously identified misassembly.
Figure 2.
Comparison between the draft genome assembly and the physical D. serrata genome map, image is adapted from Stocker et al. (2012). Genes in red were mapped by Drosopoulou and Scouras (1995, 1998), Drosopoulou et al. (1996, 1997, 2002), and Pardali et al. (1996). Genes in blue are also included in the linkage map produced by Stocker et al. (2012). Thin red lines are inversions found by Stocker et al. (2004) and thin black lines are inversions found by Mavragani-Tsipidou et al. (1990). Contig3208 (shown in red), was split into three parts based on the misassembly; parts 1 and 3 aligned with D. melanogaster 3R and part 2 aligned with 3L (Figure 1). Markers Act88F and hsp70 were not mapped to contigs because the former appears twice and nomenclature changes meant we could not be certain exactly which gene hsp70 was referring to.
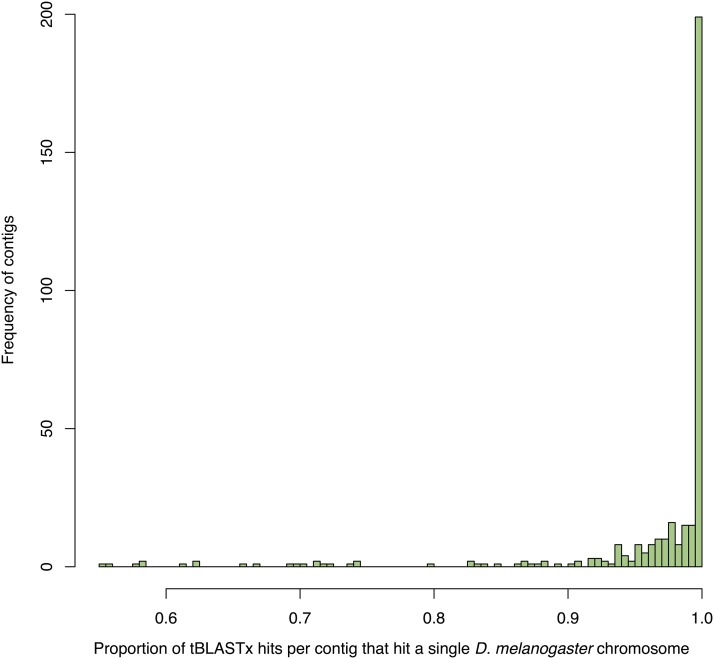
The conservation of chromosome arm-level gene content was a common feature of the remaining contigs as well. For example, while only 354 contigs contained significant tBLASTx hits to at least one D. melanogaster gene (genome version 6.05), these contigs spanned 167 Mbp, and the vast majority had > 90% tBLASTx hits to a single D. melanogaster chromosomal arm (mean = 96.35% and median = 100%) (Figure 3). Furthermore, only 34 contigs displayed < 90% similarity to D. melanogaster and a linear regression where contig size predicted percent similarity indicated no significant relationship (F(1,32) = 0.4003, P = 0.5314), suggesting that very large contigs were no more likely to be misassembled than short contigs.
Figure 3.
Comparison of D. serrata gene locations relative to D. melanogaster. On average, > 95% of tBLASTx hits to D. melanogaster genes (version 6.05) in each contig map to a single D. melanogaster arm.
Annotation
To facilitate annotation of the D. serrata genome, we sequenced mRNA from male and female adult flies. The in silico gene predictors SNAP (Johnson et al. 2008) and Augustus (Stanke and Morgenstern 2005) found 22,718 and 15,984 genes, respectively. Of these in silico predicted genes, a total of 14,271 protein coding genes were sufficiently supported by RNA-seq and/or protein sequence data to be annotated by Maker2 (Holt and Yandell 2011). Maker scores annotations using the annotation edit distance (AED), a zero-to-one score where a value of zero indicates that the in silico annotation and the empirical evidence are in perfect agreement and a value of one indicates that the in silico annotation has no support from empirical data (Eilbeck et al. 2009). The AED for the D. serrata genome had a mean score of 0.18 and median of 0.13, suggesting that most annotations were of high quality with strong empirical support. While the number of genes we annotated in D. serrata is similar to the 13,929 protein coding genes that have currently been annotated in D. melanogaster (genome version 6.05), we annotated far fewer total transcripts (31,482 identified in D. melanogaster versus 16,202 in D. serrata) (Attrill et al. 2016); this is likely due to the larger number of tissue types and life stages for which D. melanogaster gene expression has been characterized with RNA-seq. For instance, considering that in Drosophila appreciable numbers of genes peak in expression during early life stages such as embryogenesis (Arbeitman et al. 2002), our use of adult fly RNA-seq data may mean that some such genes are yet to be annotated. Furthermore, as we used mRNA-seq, we have not yet annotated noncoding genes of which there are 3503 in the D. melanogaster genome (Attrill et al. 2016). Future RNA-seq datasets will be used to update the existing gene models.
We observed differences in gene, exon, and intron lengths between D. serrata and D. melanogaster. In D. serrata, there were on average 3.9 exons per protein coding gene and the gene, exon, and intron lengths were 4655, 451, and 699 bp respectively. Apart from average exon number, which does not differ between the two species, these values are lower than those for D. melanogaster protein coding genes (genome version 6.05), where the mean gene, exon, and intron lengths are 6962, 539, and 1704 bp, respectively (Attrill et al. 2016). The lower average intron length observed in D. serrata may be a consequence of annotating far fewer alternate splice variants. In total, coding sequence comprised 33.6% of the genome when including introns and 15.4% of the genome when considering only exons. Lower percentage intron content has been associated with overall longer genomes in Drosophilidae (Gregory and Johnston 2008), which is consistent with our observations here.
Many of the annotated genes in D. serrata were found to be putative orthologs of D. melanogaster genes (Table S1). In total, 10,995 (77%) were found to be orthologs via best reciprocal BLAST (Huynen and Bork 1998; Moreno-Hagelsieb and Latimer 2008; Tatusov et al. 1997) using tBLASTx with default settings (Camacho et al. 2009) and version 6.05 of the D. melanogaster genome (Drosophila 12 Genomes Consortium 2007; McQuilton et al. 2012). The median e-value of each reciprocal comparison was zero, indicating that most orthologs are very similar to one another. Furthermore, when comparing D. serrata genes to D. melanogaster, the largest e-value was 1.6 with only 85 orthologs having an e-value > 1e−10. Similarly, when comparing D. melanogaster genes to D. serrata, the largest e-value was 0.18 with only 78 orthologs having an e-value > 1e−10. The correlation between e-values for the reciprocal BLAST was 0.88.
Conclusions
We have assembled a draft genome for a species with no existing genome using only 3GS data. Our study indicates the feasibility of long-read-only genome assembly for nonmodel species with modest sized genomes when using an inbred line. While either greater 3GS coverage or a hybrid merged assembly (Chakraborty et al. 2016) may be required to provide greater genome contiguity, it is clear that the genome has a high degree of completeness in terms of gene content and that misassemblies at chromosome arm-level are rare. The genome and its initial annotation provide a useful resource of future population genomic and trait mapping studies in this species.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.037598/-/DC1.
Acknowledgments
We thank S. Koren for advice regarding the PBcR pipeline. Funding for this research was provided by The University of Queensland.
Footnotes
Communicating editor: B. Oliver
Literature Cited
- Alkan C., Sajjadian S., Eichler E. E., 2011. Limitations of next-generation genome sequence assembly. Nat. Methods 8(1): 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S., 2010. A Quality Control Tool for High Throughput Sequence Data. Reference Source, Fast, QC. [Google Scholar]
- Arbeitman M. N., Furlong E. E., Imam F., Johnson E., Null B. H., et al. , 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297(5590): 2270–2275. [DOI] [PubMed] [Google Scholar]
- Attrill H., Falls K., Goodman J. L., Millburn G. H., Antonazzo G., et al. , 2016. FlyBase: establishing a gene group resource for Drosophila melanogaster. Nucleic Acids Res. 44(D1): D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K., Koren S., Chin C.-S., Drake J. P., Landolin J. M., et al. , 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33: 623–630. [DOI] [PubMed] [Google Scholar]
- Blows M. W., Hoffman A. A., 1993. The genetics of central and marginal populations of Drosophila serrata. I. Genetic variation for stress resistance and species borders. Evolution 47: 1255–1270. [DOI] [PubMed] [Google Scholar]
- Blows M. W., Chenoweth S. F., Hine E., 2004. Orientation of the genetic variance-covariance matrix and the fitness surface for multiple male sexually selected traits. Am. Nat. 163(3): 329–340. [DOI] [PubMed] [Google Scholar]
- Brake, I., and G. Bächli, 2008 Drosophilidae (Diptera), pp. 1–412 in World Catalogue of Insects 9. Apollo Books Aps., Stenstrup, Denmark. [Google Scholar]
- Butler J., MacCallum I., Kleber M., Shlyakhter I. A., Belmonte M. K., et al. , 2008. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 18(5): 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10(1): 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M. S., Holt C., Moore B., Yandell M., 2014. Genome annotation and curation using MAKER and MAKER-P. Curr. Protoc. Bioinformatics. 48: 4.11.1– 4.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M. J., Brinza D., Pevzner P. A., 2009. De novo fragment assembly with short mate-paired reads: does the read length matter? Genome Res. 19(2): 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M., Baldwin-Brown J. G., Long A. D., Emerson J. J., 2016. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 44(19): e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. X., Sturgill D., Qu J., Jiang H., Park S., et al. , 2014. Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome Res. 24(7): 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenoweth S. F., Blows M. W., 2003. Signal trait sexual dimorphism and mutual sexual selection in Drosophila serrata. Evolution 57(10): 2326–2334. [DOI] [PubMed] [Google Scholar]
- Chenoweth S. F., Petfield D., Doughty P., Blows M. W., 2007. Male choice generates stabilizing sexual selection on a female fecundity correlate. J. Evol. Biol. 20(5): 1745–1750. [DOI] [PubMed] [Google Scholar]
- Chenoweth S. F., Rundle H. D., Blows M. W., 2008. Genetic constraints and the evolution of display trait sexual dimorphism by natural and sexual selection. Am. Nat. 171(1): 22–34. [DOI] [PubMed] [Google Scholar]
- Chenoweth S. F., Rundle H. D., Blows M. W., 2010a The contribution of selection and genetic constraints to phenotypic divergence. Am. Nat. 175(2): 186–196. [DOI] [PubMed] [Google Scholar]
- Chenoweth S. F., Rundle H. D., Blows M. W., 2010b Experimental evidence for the evolution of indirect genetic effects: changes in the interaction effect coefficient, psi (ψ), due to sexual selection. Evolution 64(6): 1849–1856. [DOI] [PubMed] [Google Scholar]
- Chenoweth S. F., Appleton N. C., Allen S. L., Rundle H. D., 2015. Genomic evidence that sexual selection impedes adaptation to a novel environment. Curr. Biol. 25(14): 1860–1866. [DOI] [PubMed] [Google Scholar]
- Chin C. S., Sorenson J., Harris J. B., Robins W. P., Charles R. C., et al. , 2011. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364(1): 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. S., Alexander D. H., Marks P., Klammer A. A., Drake J., et al. , 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10(6): 563–569. [DOI] [PubMed] [Google Scholar]
- Chung H., Loehlin D. W., Dufour H. D., Vaccarro K., Millar J. G., et al. , 2014. A single gene affects both ecological divergence and mate choice in Drosophila. Science 343(6175): 1148–1151. [DOI] [PubMed] [Google Scholar]
- Delcourt M., Blows M. W., Rundle H. D., 2009. Sexually antagonistic genetic variance for fitness in an ancestral and a novel environment. Proc. Biol. Sci. 276: 2009–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Clark A. G., Eisen M. B., Smith D. R., Bergman C. M., et al. , 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450(7167):203–218. [DOI] [PubMed] [Google Scholar]
- Drosopoulou E., Scouras Z. G., 1995. The β-tubulin gene family evolution in the Drosophila montium subgroup of the melanogaster species group. J. Mol. Evol. 41(3): 293–298. [PubMed] [Google Scholar]
- Drosopoulou E., Scouras Z. G., 1998. The organization of the alpha-tubulin gene family in the Drosophila montium subgroup of the melanogaster species group. Genome 41(4): 504–509. [DOI] [PubMed] [Google Scholar]
- Drosopoulou E., Konstantopoulou I., Scouras Z. G., 1996. The heat shock genes in the Drosophila montium subgroup: chromosomal localization and evolutionary implications. Chromosoma 105(2): 104–110. [DOI] [PubMed] [Google Scholar]
- Drosopoulou E., Tsiafouli M., Mavragani-Tsipidou P., Scouras Z. G., 1997. The glutamate dehydrogenase, E74 and putative actin gene loci in the Drosophila montium subgroup. Chromosomal homologies among the montium species and D. melanogaster. Chromosoma 106(1): 20–28. [DOI] [PubMed] [Google Scholar]
- Drosopoulou E., Wiebauer K., Yiangou M., Mavragani-Tsipidou P., Domdey H., et al. , 2002. Isolation, characterization, and localization of beta-tubulin genomic clones of three Drosophila montium subgroup species. Genome 45(3): 604–607. [DOI] [PubMed] [Google Scholar]
- Eilbeck K., Moore B., Holt C., Yandell M., 2009. Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics 10(1): 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu F. D., Chenoweth S. F., 2008. Polyandry and paternity skew in natural and experimental populations of Drosophila serrata. Mol. Ecol. 17(6): 1589–1596. [DOI] [PubMed] [Google Scholar]
- Frentiu F. D., Chenoweth S. F., 2010. Clines in cuticular hydrocarbons in two Drosophila species with independent population histories. Evolution 64(6): 1784–1794. [DOI] [PubMed] [Google Scholar]
- Frentiu F. D., Adamski M., McGraw E. A., Blows M. W., Chenoweth S. F., 2009. An expressed sequence tag (EST) library for Drosophila serrata, a model system for sexual selection and climatic adaptation studies. BMC Genomics 10: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosden T. P., Chenoweth S. F., 2011. On the evolution of heightened condition dependence of male sexual displays. J. Evol. Biol. 24(3): 685–692. [DOI] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., et al. , 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29(7): 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P., 1997. Against a whole-genome shotgun. Genome Res. 7(5): 410–417. [DOI] [PubMed] [Google Scholar]
- Gregory T. R., Johnston J. S., 2008. Genome size diversity in the family Drosophilidae. Heredity (Edinb) 101(3): 228–238. [DOI] [PubMed] [Google Scholar]
- Hallas R., Schiffer M., Hoffmann A. A., 2002. Clinal variation in Drosophila serrata for stress resistance and body size. Genet. Res. 79(2): 141–148. [DOI] [PubMed] [Google Scholar]
- Higgie M., Chenoweth S., Blows M. W., 2000. Natural selection and the reinforcement of mate recognition. Science 290(5491): 519–521. [DOI] [PubMed] [Google Scholar]
- Hine E., Lachish S., Higgie M., Blows M. W., 2002. Positive genetic correlation between female preference and offspring fitness. Proc. Biol. Sci. 269: 2215–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C., Yandell M., 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12(1): 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M. A., Bork P., 1998. Measuring genome evolution. Proc. Natl. Acad. Sci. USA 95(11): 5849–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Handsaker R. E., Pulit S. L., Nizzari M. M., O’Donnell C. J., et al. , 2008. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24(24): 2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann V., van Heerwaarden B., Sgro C. M., Hoffmann A. A., 2009. Fundamental evolutionary limits in ecological traits drive Drosophila species distributions. Science 325(5945): 1244–1246. [DOI] [PubMed] [Google Scholar]
- Kim K. E., Peluso P., Babayan P., Yeadon P. J., Yu C., et al. , 2014. Long-read, whole-genome shotgun sequence data for five model organisms. Sci. Data 1: 140045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Schatz M. C., Walenz B. P., Martin J., Howard J. T., et al. , 2012. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 30(7): 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A. L., Smoot M., Shumway M., et al. , 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2): R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latimer C. A., Wilson R. S., Chenoweth S. F., 2011. Quantitative genetic variation for thermal performance curves within and among natural populations of Drosophila serrata. J. Evol. Biol. 24(5): 965–975. [DOI] [PubMed] [Google Scholar]
- Latimer C. A., Foley B. R., Chenoweth S. F., 2015. Connecting thermal performance curve variation to the genotype: a multivariate QTL approach. J. Evol. Biol. 28(1): 155–168. [DOI] [PubMed] [Google Scholar]
- Lee H., Gurtowski J., Yoo S., Marcus S., McCombie W. R., et al. , 2014. Error correction and assembly complexity of single molecule sequencing reads. bioRxiv. DOI: https://doi.org/10.1101/006395. [Google Scholar]
- Li R., Zhu H., Ruan J., Qian W., Fang X., et al. , 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20(2): 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiafoglou A., Carew M., Hoffmann A., 2002. Shifting clinal patterns and microsatellite variation in Drosophila serrata populations: a comparison of populations near the southern border of the species range. J. Evol. Biol. 15(5): 763–774. [Google Scholar]
- Mavragani-Tsipidou P., Kyrpides N., Scouras Z. G., 1990. Evolutionary implications of duplications and balbiani rings in Drosophila. A study of Drosophila serrata. Genome 33(4): 478–485. [DOI] [PubMed] [Google Scholar]
- McGuigan K., Petfield D., Blows M. W., 2011a Reducing mutation load through sexual selection on males. Evolution 65(10): 2816–2829. [DOI] [PubMed] [Google Scholar]
- McGuigan K., Rowe L., Blows M. W., 2011b Pleiotropy, apparent stabilizing selection and uncovering fitness optima. Trends Ecol. Evol. 26(1): 22–29. [DOI] [PubMed] [Google Scholar]
- McGuigan K., Collet J. M., Allen S. L., Chenoweth S. F., Blows M. W., 2014a Pleiotropic mutations are subject to strong stabilizing selection. Genetics 197(3): 1051–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuigan K., Collet J. M., McGraw E. A., Ye Y. H., Allen S. L., et al. , 2014b The nature and extent of mutational pleiotropy in gene expression of male Drosophila serrata. Genetics 196(3): 911–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuilton P., St Pierre S. E., Thurmond J., FlyBase Consortium , 2012. FlyBase 101–the basics of navigating FlyBase. Nucleic Acids Res. 40(Database issue): D706–D714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. R., Delcher A. L., Koren S., Venter E., Walenz B. P., et al. , 2008. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24(24): 2818–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G., Latimer K., 2008. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24(3): 319–324. [DOI] [PubMed] [Google Scholar]
- Nagarajan N., Pop M., 2013. Sequence assembly demystified. Nat. Rev. Genet. 14(3): 157–167. [DOI] [PubMed] [Google Scholar]
- Pardali E., Feggou E., Drosopoulou E., Konstantopoulou I., Scouras Z. G., et al. , 1996. The Afrotropical Drosophila montium subgroup: Balbiani ring 1, polytene chromosomes, and heat shock response of Drosophila vulcana. Genome 39(3): 588–597. [DOI] [PubMed] [Google Scholar]
- Phillippy A. M., Schatz M. C., Pop M., 2008. Genome assembly forensics: finding the elusive mis-assembly. Genome Biol. 9(3): R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J., Quinlan A. R., Loman N. J., 2014. A reference bacterial genome dataset generated on the MinION portable single-molecule nanopore sequencer. Gigascience 3(22): 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A., Webster D. R., Sahl J. W., Bashir A., Boisen N., et al. , 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365(8): 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle H. D., Chenoweth S. F., Blows M. W., 2009. The diversification of mate preferences by natural and sexual selection. J. Evol. Biol. 22(8): 1608–1615. [DOI] [PubMed] [Google Scholar]
- Sahlin K., Chikhi R., Arvestad L., 2016. Assembly scaffolding with PE-contaminated mate-pair libraries. Bioinformatics 32(13): 1925–1932. [DOI] [PubMed] [Google Scholar]
- Schatz M. C., Delcher A. L., Salzberg S. L., 2010. Assembly of large genomes using second-generation sequencing. Genome Res. 20(9): 1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J., Ji H., 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26(10): 1135–1145. [DOI] [PubMed] [Google Scholar]
- Simao F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19): 3210–3212. [DOI] [PubMed] [Google Scholar]
- Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J., et al. , 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19(6): 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. F., R. Hubley, and P. Green, 1996 RepeatMasker Open-3.0. http://www.repeatmasker.org Accessed December 6th, 2015.
- Stanke M., Morgenstern B., 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33(Suppl. 2): W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker A. J., Foley B., Hoffmann A., 2004. Inversion frequencies in Drosophila serrata along an eastern Australian transect. Genome 47(6): 1144–1153. [DOI] [PubMed] [Google Scholar]
- Stocker A. J., Rusuwa B. B., Blacket M. J., Frentiu F. D., Sullivan M., et al. , 2012. Physical and linkage maps for Drosophila serrata, a model species for studies of clinal adaptation and sexual selection. G3 2(2): 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Subramanian S., Kumar S., 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21(1): 36–44. [DOI] [PubMed] [Google Scholar]
- Tatusov R. L., Koonin E. V., Lipman D. J., 1997. A genomic perspective on protein families. Science 278(5338): 631–637. [DOI] [PubMed] [Google Scholar]
- Treangen T. J., Salzberg S. L., 2012. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 13(1): 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukkonen E., 1992. Approximate string-matching with Q-grams and maximal matches. Theor. Comput. Sci. 92(1): 191–211. [Google Scholar]
- van Heesch S., Kloosterman W. P., Lansu N., Ruzius F.-P., Levandowsky E., et al. , 2013. Improving mammalian genome scaffolding using large insert mate-pair next-generation sequencing. BMC Genomics 14(1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel J., Kingsford C., Pop M., 2011. Assessing the benefits of using mate-pairs to resolve repeats in de novo short-read prokaryotic assemblies. BMC Bioinformatics 12(1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassin A., Delaney E. K., Reddiex A. J., Seher T. D., Bastide H., et al. , 2016. The pdm3 locus is a hotspot for recurrent evolution of female-limited color dimorphism in Drosophila. Curr. Biol. 26(18): 2412–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data including PacBio and RNA-seq reads have been submitted to public repositories and are available via the D. serrata genome NCBI project accession PRJNA355616. The genome assembly and annotation tracks are available from http://www.chenowethlab.org. We also supply a list of D. melanogaster orthologs in Table S1.