Abstract
The death of larval salivary gland cells during metamorphosis in Drosophila melanogaster has been a key system for studying steroid controlled programmed cell death. This death is induced by a pulse of the steroid hormone ecdysone that takes place at the end of the prepupal period. For many years, it has been thought that the ecdysone direct response gene Eip93F (E93) plays a critical role in initiating salivary gland cell death. This conclusion was based largely on the finding that the three “type” alleles of E93 cause a near-complete block in salivary gland cell death. Here, we show that these three mutations are in fact allelic to Idh3b, a nearby gene that encodes the β subunit of isocitrate dehydrogenase 3, a mitochondrial enzyme of the tricarboxylic acid (TCA) cycle. The strongest of the Idh3b alleles appears to cause a near-complete block in oxidative phosphorylation, as mitochondria are depolarized in mutant larvae, and development arrests early during cleavage in embryos from homozygous-mutant germline mothers. Idh3b-mutant larval salivary gland cells fail to undergo mitochondrial fragmentation, which normally precedes the death of these cells, and do not initiate autophagy, an early step in the cell death program. These observations suggest a close relationship between the TCA cycle and the initiation of larval cell death. In normal development, tagged Idh3b is released from salivary gland mitochondria during their fragmentation, suggesting that Idh3b may be an apoptogenic factor that functions much like released cytochrome c in mammalian cells.
Keywords: Idh3b, isocitrate dehydrogenase, E93, apoptosis, mitochondria, autophagy
The death of larval salivary glands during metamorphosis in Drosophila is an important model system for understanding the control of programmed cell death by steroid hormones [for review, see Yin and Thummel (2005)]. Salivary gland cell death shares many features with apoptosis, including hallmark events such as DNA fragmentation, mitochondrial fragmentation (Goyal et al. 2007), and exposure of phosphatidylserine on the outer leaflet of the plasma membrane (Lee and Baehrecke 2001). However, unlike apoptosis, in which dead or dying cells are engulfed and degraded by phagocytes, salivary gland cell death is autophagic, and involves degradation of cellular components within autolysosomes [Lee and Baehrecke (2001); reviewed in Baehrecke (2003); Tracy and Baehrecke (2013)]. The key initiating event in salivary gland death is expression of the cell death activators reaper (rpr) and head involution defective (hid). These activators interfere with the activity of the apoptosis inhibitor, diap1, which allows dimerization and activation of the initiator caspase dronc, which then cleaves and activates the downstream execution caspase drice, which, in turn, cleaves additional substrates in the cell death cascade.
The histolysis of larval salivary glands is triggered by the prepupal pulse of the steroid hormone ecdysone. Some early events in the death process are clear: transcription of the cell death activator rpr is activated directly by the ecdysone-receptor complex, with this activation being enhanced by the direct response gene Broad-Complex (BR-C) (Jiang et al. 2000). The ecdysone-receptor complex also directly induces expression of the initiator caspase dronc (Cakouros et al. 2004). In contrast, hid is not regulated directly by the ecdysone-receptor complex, but is activated by the immediate response genes BR-C and E74A (Jiang et al. 2000). This activation of rpr, dronc, and hid, as well as the partial repression of the apoptosis inhibitor diap1 by CBP (Yin et al. 2007), are key events in the initiation of salivary gland histolysis. To understand better the events of salivary gland cell death, large-scale genetic (Wang et al. 2008) and molecular (Gorski et al. 2003; Lee et al. 2003; Martin et al. 2007; McPhee et al. 2013) screens have been conducted to identify additional genes that mediate the death response. The genes identified encode functionally diverse products, including an RNA helicase (Ihry et al. 2012), chromatin remodelers (Ihry and Bashirullah 2014), an enzyme of the TCA cycle (Wang et al. 2010), subunits of the mediator complex (Wang et al. 2010), subunits of the cop9 signalsome (McPhee et al. 2013), several autophagy (Atg) proteins (Berry and Baehrecke 2007), and several proteins of unknown function. The roles of these proteins in salivary gland cell death, and how these roles are coordinated, are, for the most part, poorly understood.
The ecdysone primary response gene Eip93F (E93) has been thought to play a key role in initiating salivary gland cell death. In their original characterization of E93, Lee et al. (2000) set out to identify loss-of-function alleles of the locus by screening for new lethal mutations that fail to complement a deficiency that includes the E93 locus. One of the complementation groups identified became the focus of attention, as its three alleles cause lethality early in metamorphosis, coinciding with the beginning of E93 transcription. This group was assigned to E93 when it was found that one of its alleles (“E931”) is associated with a nonsense mutation at codon 995 of the E93 A isoform, near the 3′ end of the E93 coding sequence. The other two alleles in this complementation group (“E932” and “E933”) do not show changes in the E93 coding sequence, but were assumed to have changes elsewhere in the gene. Since this early work, “E931,” “E932,” and “E933” have been considered the “type” alleles of the gene, and have been utilized in almost all subsequent studies of E93 function. These mutations cause an almost complete block in the death of salivary gland cells during metamorphosis. The “E93” mutants eliminate, or reduce, expression of several apoptotic genes in the pupal salivary gland, including rpr, hid, crq, ark, and dronc (Lee et al. 2000), and show low levels of activated Drice (Martin and Baehrecke 2004). In addition, these mutants do not initiate autophagic cell death changes in the salivary glands (Lee and Baehrecke 2001). These phenotypes led to the view that the transcription factor encoded by E93 is a critical regulator of salivary gland cell death during metamorphosis (Lee et al. 2000).
This view of E93 function has been challenged recently by the finding that a separate complementation group, identified by Lee et al. (2000) in their lethal screens of the E93 region, is allelic to E93. The alleles of this group (the E934 through E936 alleles) are nonsense changes at codons 360, 545, and 783 of the 1165 codon E93 A coding sequence (Mou et al. 2012). These mutations cause lethality at a much later pupal stage than do alleles of the “E931-3” group, producing pharate adults that show numerous defects in structures normally patterned at the pupal stage (Lee et al. 2000; Mou et al. 2012). Indeed, essentially all cuticular patterning events that normally occur during the pupal stage fail in mutants of this group, suggesting that E93 functions as a key temporal determinant of adult development during metamorphosis (Mou et al. 2012). Recent work in other insects strongly supports this conclusion (Ureña et al. 2014).
The relationship between the E934-6 and “E931-3” groups of alleles has been puzzling. Alleles of the two groups complement, and, in cell clones alleles of the “E931-3” group, show no defects in imaginal patterning, unlike alleles of the E934-6 group (Mou et al. 2012). Complementation of the E934-6 alleles and “E931” has been particularly hard to understand, as the E934-6 nonsense alleles truncate the coding region at positions upstream of the E931 nonsense allele. In an attempt to resolve this paradoxical behavior, we localized the “E931-3” alleles by deletion mapping and sequencing. Here, we show that the causative mutations in this group are not in fact allelic to E93; rather, they are allelic to a nearby gene (CG6439) that encodes the β subunit of isocitrate dehydrogenase-3 (Idh3b), a key enzyme of the citric acid cycle. This finding shifts the focus from E93 to Idh3b and/or mitochondrial function in the initiation of salivary gland cell death. We show that Idh3b is required for mitochondrial polarization and the generation of normal ATP levels. For the strongest of the Idh3b alleles, embryos from homozygous mutant germline mothers arrest development during early cleavage, consistent with a near-complete block in oxidative phosphorylation. We show that Idh3b is required for mitochondrial fragmentation, an early step in the death program of larval salivary gland cells (Goyal et al. 2007), and that, in normal development, tagged Idh3b is expelled from mitochondria during this fragmentation. This observation suggests that Idh3b may be an apoptogenic factor functionally similar to cytochrome c and other factors released by mammalian mitochondria in the initiation of cell death. Idh3b is the second TCA cycle enzyme (after malate dehydrogenase-2) shown to be required for the death of larval salivary gland cells (Wang et al. 2010), suggesting a key role for the TCA cycle in initiating larval cell death during metamorphosis.
Materials and Methods
Unless indicated otherwise, all fly stocks used were obtained from the Bloomington Drosophila Stock Center (BDSC).
Generation of E93 region deletions
Defined deletions within the E93 region were produced by FLP-catalyzed recombination, basically as described by Parks et al. (2004). Deletions were generated between the following pairs of insertions (Thibault et al. 2004): P{XP}Eip93Fd07598 and PBac{WH}Eip93Ff01771, P{XP}d06373 and PBac{WH}Eip93Ff01771, and P{XP}Eip93Fd07598 and PBac{WH}CG6439f07670. Stocks of each insertion were obtained from the Exelixis Harvard Medical School Collection. Males and females heterozygous for a pair of insertions, carrying an X chromosome marked by w, and containing a heat-shock inducible FLP transgene (hs-FLP122), were heat-shocked as larvae three times for 30 min each at 37° in plastic vials in a water bath. Emerging adults were then mated to w/Y; TM3, Sb/TM6B, Tb or w/w; TM3, Sb/TM6B, Tb flies. Deletion recombinants were identified among the progeny by their white eye color, and recombinant chromosomes placed into balanced stock with TM6B, Tb.
Germline clones
Germline clones were generated as described by Chou et al. (1993). y w hs-FLP122/w; FRT82B Idh3b/FRT82B P{ovoD1}18 female larvae were heat-shocked at 37° three times during larval development. Control females not subject to heat shock produced no eggs, confirming the presence and efficacy of P{ovoD1}18. However, heat-shocked females containing Idh3b-homozygous clones in their germlines produced numerous, normal-appearing eggs. A major concern in these experiments was the elimination of extraneous lethals from the Idh3b mutant chromosomes. Each of the three Idh3b alleles was subject to multiple rounds of recombination to generate chromosomes for each that survive to pupariation when homozygous. These chromosomes were then recombined with a homozygous viable FRT82B chromosome to generate FRT82B Idh3b chromosomes. All recombinant chromosomes used for clone production survive to pupariation when homozygous.
Amplification and sequencing of mutants
For each of the “E931-3” alleles, the region between the PBac{WH}Eip93Ff01771 and PBac{WH}CG6439f07670 insertions was amplified and cloned as three ∼3 kb subfragments into pJET 2.1, which were then sequenced with primers spaced at 800 bp intervals (primer sequences available upon request). A single nucleotide difference within the Idh3b gene was found for each mutation.
Fly transformation
All transgenes were generated by standard molecular methods. DNA was prepared with PureYield Midi preps (Promega), and resuspended in injection buffer (5 mM potassium chloride, 0.1 mM sodium phosphate, pH 7.8) at a concentration of 1 µg/µl. Staged embryos of appropriate genotype were dechorionated in bleach, and injected under oil with pulled glass capillary needles using a micromanipulator (Narishige) controlled with a N2 driven Picospritzer (General Valve Corporation). All constructs were injected into one or more of the following ϕc31 integrase and attP site strains: y1 M{vas-int.DM}ZH-2A w; M{3XP3-RFP.attP′}ZH-22A (BDSC line 24481), y1 M{vas-int.DM}ZH-2A w; M{3XP3-RFP.attP′}ZH-68E (BDSC line 24485), and/or y1 M{vas-int.DM}ZH-2A w; M{3XP3-RFP.attP′}ZH-51D (BDSC line 24483).
Transgene constructions
The Idh3b rescue construct was derived by amplifying the Idh3b region using wild-type genomic DNA as template, followed by cloning as a NotI–Acc65I fragment into pUASTattB (Bischof et al. 2007). The genomic fragment tested includes the entire Idh3b A form transcribed region, as well as 198 bp of upstream sequence. The HA/FLAG tagged Idh3b construct was ordered directly from the Drosophila Genome Resource Center Tagged ORF collection (accession number UFO02744).
ATP assays
Assays were performed with the Enlighten ATP Bioluminescence Kit (Promega) according to the manufacturer’s protocols. Briefly, pupae aged 18 hr after puparium formation (APF) were collected, and ground in 100 µl of homogenization buffer (five animals per sample); 20 µl was removed for Bradford Protein Assays (Sigma), and the rest of each sample was boiled 5 min and spun 3 min at 14,000 rpm to remove debris. The supernatant was diluted 1:1000× with ATP-free water, and 10 µl aliquots were mixed with 100 µl rL/L reagent in white 96-well plates, and read immediately on a Varioskan Luminometer (Thermo Scientific), along with a freshly prepared ATP standard series. Three replicates were assayed for each sample, and three readings were conducted for each experiment. ATP concentration was determined by comparison to the ATP standard curve, and normalized to total protein.
Antibody/mitotracker/lysotracker staining
For MitoTracker and LysoTracker staining, tissues were dissected at room temperature in Phosphate Buffered Saline (PBS), rinsed briefly in PBS, and incubated for 5 min in 100 nM MitoTracker RedCMXRos, 100 nM MitoTracker Deep Red, or 50 nM LysoTracker Red (all reagents from Life Technologies). The samples were quickly rinsed three times in PBS, and fixed in 4% Paraformaldehyde in PBS for 1 min at room temperature. The samples were then rinsed in PBS with 0.05% Triton X (PBSTx), stained for 5 min with 2 µg/ml DAPI, rinsed twice with PBSTx, and mounted in 50% glycerol/50% PBS. Antibody staining was performed as described previously (Kankel et al. 2004). Antibodies used in this study were monoclonal mouse anti-Engrailed (cell line gift from N. Patel, Univ. California, Berkeley), monoclonal rat anti FLAG (BioLegend), Cy3 Donkey anti mouse, and Cy3 Donkey anti rat (Jackson Immunolabs). Images were collected with a Nikon A1 confocal microscope and a Zeiss SV11 fluorescent stereomicroscope with GFP filters.
Data availability
All Drosophila strains used or generated in this study are available on request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
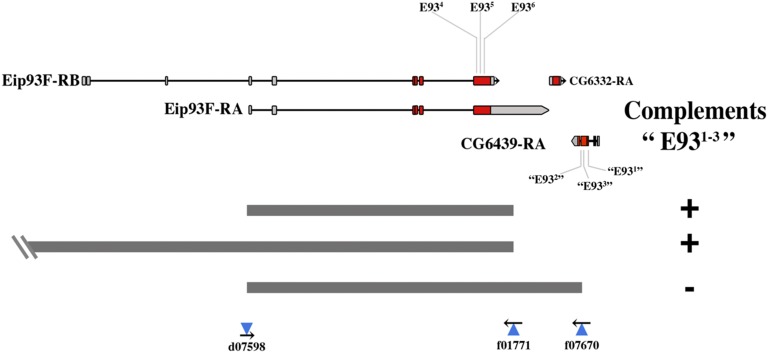
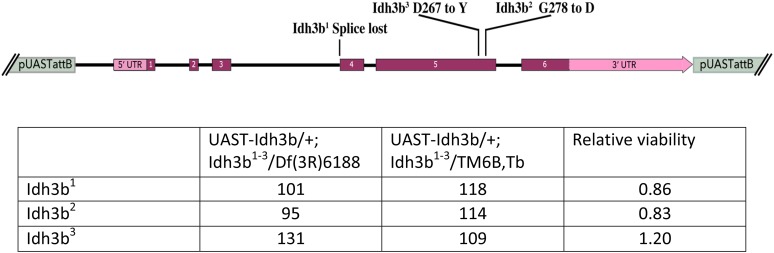
To explore the relationship between the E934-6 and “E931-3” complementation groups, our first goal was to generate a true null allele. This was accomplished by FLP-mediated recombination between FRT elements (Parks et al. 2004) carried by existing transposon insertions that lie at either end of the major coding region of the gene [insertions d07598 and f01771 (Thibault et al. 2004)] (Figure 1). The deletion generated, E93Δ1, lacks the entire E93A coding region, and all but the 5′-most 32 codons of the E93B isoform coding sequence. Homozygotes for E93Δ1 survive to the pharate adult stage, and show cuticular defects essentially identical to those shown by homozygotes for alleles of the E934-6 group. As expected, E93Δ1 fails to complement alleles of this group. However, E93Δ1 fully complements the three alleles of the “E931-3” group. This result strongly suggests that alleles of the latter group affect some product other than E93. To identify this product, we generated two additional deletions: one includes the E93Δ1 deletion and extends to the left (produced by FLP catalyzed recombination between the insertions d06373 and f01771), while the other includes E93Δ1 and extends to the right (produced by recombination between the insertions d07508 and f07670) (Figure 1). The former deletion complements the “E931-3” group, whereas the latter does not, indicating that the “E931-3” alleles lie close to the right of the E93 coding sequence. Amplification and sequencing of this region from each of the mutant alleles revealed that all three carry mutations within a nearby gene (CG6439), which encodes a 370 amino acid protein orthologous to the β subunit of human isocitrate dehydrogenase 3. “E931” is an A to T change in the AG splice acceptor at the 5′ boundary of exon 4 of this gene; “E932” is a G to A missense allele causing the change of G278 to D; and “E933” is a G to A missense allele causing the change of D267 to Y (Figure 2). Allelism of the E931-3 mutations with CG6439 was confirmed by essentially complete rescue of their lethality by a 3.0 kb genomic fragment containing the CG6439 gene (see Figure 2 for details of the construct used, and data on its rescue ability). Henceforth, these alleles will be called Idh3b1, Idh3b2, and Idh3b3, respectively. The full complementation of E93Δ1 and “E931” indicates that the E93 C-terminal nonsense mutation present in the latter is incidental, and does not compromise any essential function of E93.
Figure 1.
Deletion mapping of the “E931-3” alleles. At the top are shown the transcripts of the E93, CG6332, and CG6439 genes, and at the bottom are shown the extents of deletions generated by recombination between FRT elements carried by the indicated insertions. At the right, the ability of each deletion to complement the “E931-3” alleles is indicated. Coding regions of each transcript are indicated in red.
Figure 2.
Rescue of the Idh3b mutants by an Idh3b+ transgene. (A) The Idh3b A form transcribed region, showing the locations of exons, coding sequences, untranslated regions, the Idh3b alleles, and the extent of the Idh3b rescue fragment. The indicated fragment was cloned into pUASTattB and transformants generated. These transformants rescue the Idh3b mutants in the absence of a Gal4 driver. (B) Ability of the Idh3b rescue construct to rescue the lethality of Idh3b mutant hemizygotes. Crosses to test rescue were of the form w; UAST-Idh3b/SM6; Idh3b1-3/TM6B, Tb males x w/w; Df(3R)Exel6188/TM6B females. Adult progeny numbers are shown only for Idh3b1-3/Df(3R)Exel6188 and Idh3b1-3/TM6B, Tb progeny that carry the UAST-Idh3b transgene. Almost complete rescue of survival to adulthood is seen for all three alleles. For the Idh3b2 mutant, rescue of salivary gland death was also examined. Of nine UAST-Idh3b/+; Idh3b2/Df(3R)Exel6188 pupae dissected at 24–30 hr after puparium formation, none contained salivary glands.
A second reason that the Idh3b alleles were thought to be allelic to E93 is that E93 transcripts were reported to be absent in Idh3b3 mutants, and strongly reduced in Idh3b2 mutants (Lee et al. 2000). However, we find that antibody staining for E93 protein is normal in salivary glands from heterozygotes for Idh3b3 and a deficiency that removes the Idh3b and E93 genes [Df(3R)Exel6188] (Supplemental Material, Figure S1). E93 staining is also seen Idh3b2/Df(3R)Exel6188 salivary glands, although the level of staining is reduced compared to wild type. This reduction is likely due to developmental arrest, as E93 staining is normal in salivary glands from Idh3b2/E93Δ1 heterozygotes. These observations indicate that the Idh3b mutants are not transcript null alleles of E93.
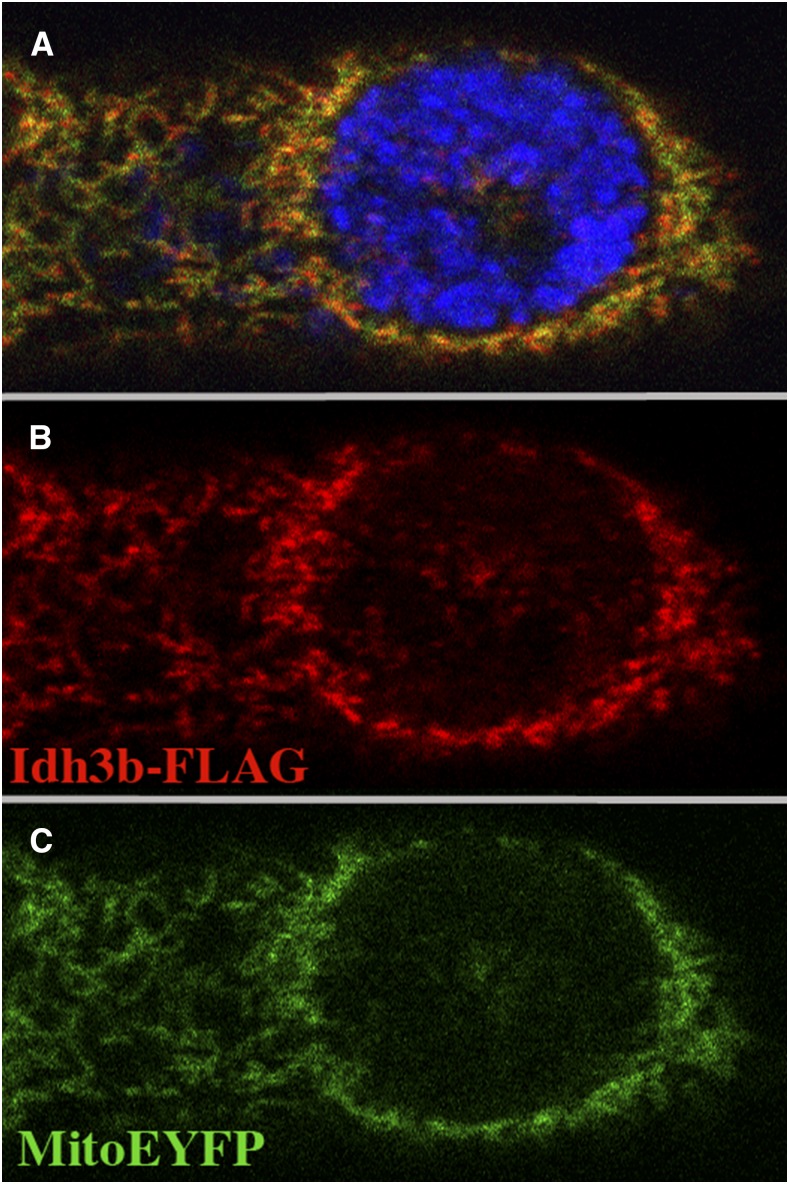
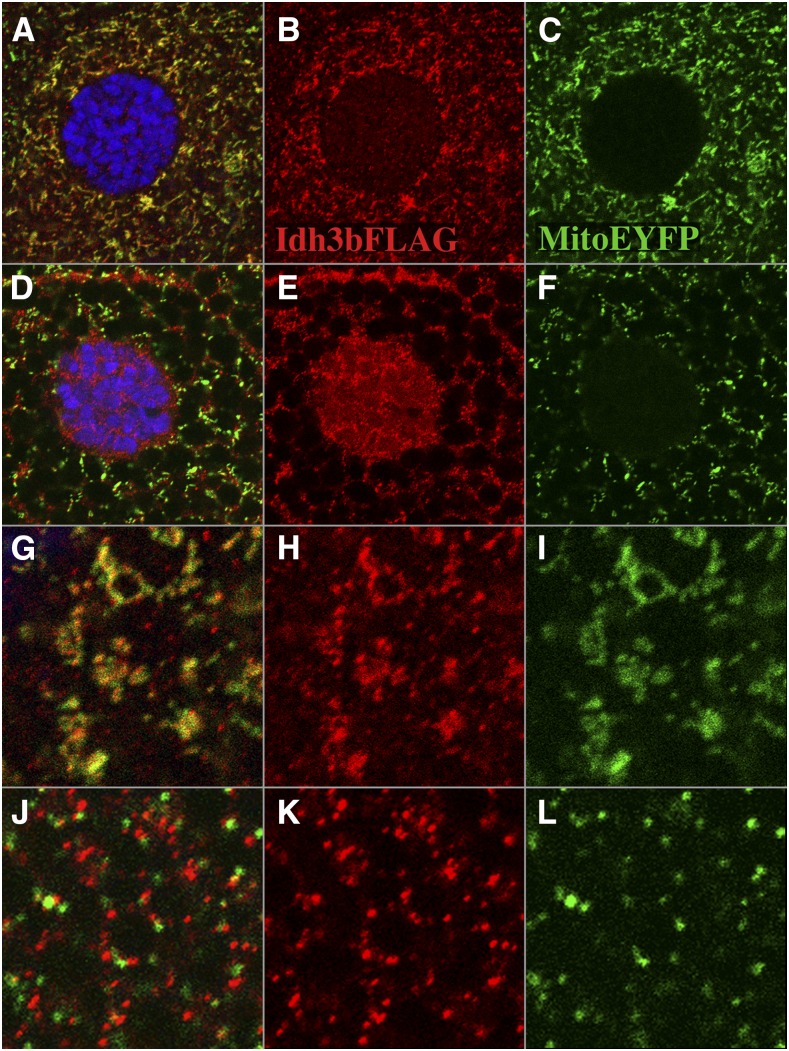
Three forms of isocitrate dehydrogenase are present in humans: IDH1 and IDH2 are NADP-dependent enzymes, whereas IDH3 is NAD dependent. IDH1 is found within the cytosol and in peroxisomes, while IDH2 and IDH3 localize to mitochondria. IDH2 is known to function in the elimination of reactive oxygen species generated by electron transport (Jo et al. 2001), while IDH3 is thought to function within the tricarboxylic acid (TCA) cycle to catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate. IDH3 is a heterotetramer, consisting of two α subunits, one β subunit, and one γ subunit. Drosophila CG6439 is most similar in sequence to the β subunit of IDH3 (61% identity and 77% similarity), with the next highest similarity being to the IDH3γ subunit (51% identity and 68% similarity). To localize Drosophila Idh3b, we drove expression of FLAG-tagged Idh3b (Drosophila Genome Resource Center) using the tubulin-Gal4 driver, and compared its localization to that of mito-EYFP, which localizes to mitochondria (LaJeunesse et al. 2004). Tagged Idh3b and mito-EYFP colocalize in all cell types examined (imaginal disc, tracheal, and salivary gland cells) (Figure 3), consistent with the presence of a predicted mitochondrial targeting peptide at the N-terminus of the protein (TargetP prediction; Emanuelsson et al. 2000), and the identification of Idh3b within the proteome of the Drosophila mitochondrial matrix (Chen et al. 2015).
Figure 3.
FLAG-tagged Idh3b localizes to mitochondria. A tracheal cell from a mid third-instar larva of the genotype UAS-HA/FLAG-Idh3b/+; tub-Gal4/mito-EYFP that has been stained with anti-FLAG (red) and the DNA dye DAPI (blue) is shown. FLAG-Idh3b (red) (B) and mito-EYFP (green) (C) colocalize (see merged image in A).
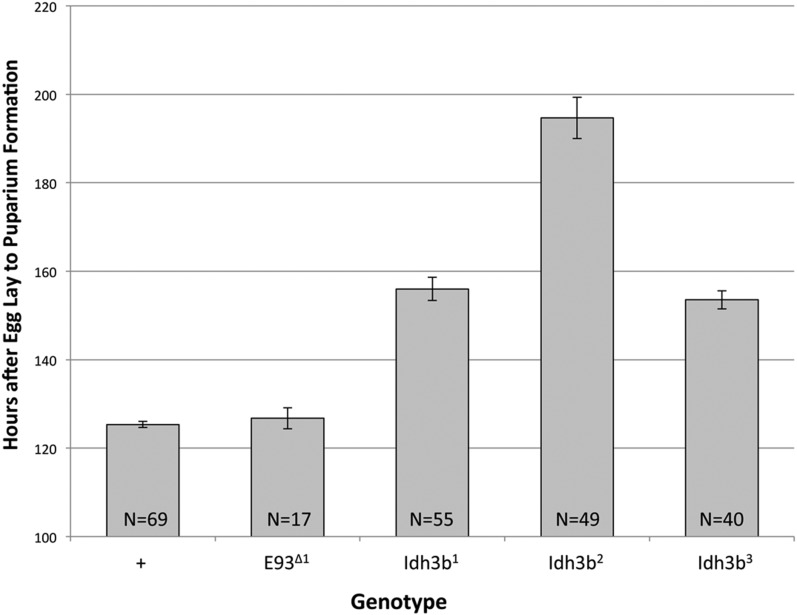
As would be expected for impairment of mitochondrial function, Idh3b mutants develop more slowly than wild type. Egg to puparium development time was determined for each Idh3b allele when heterozygous with Df(3R)Exel6188. Compared to wild-type hemizygotes, Idh3b1, Idh3b2, and Idh3b3 hemizygotes show increases in average egg to puparium development of 24, 55, and 22%, respectively (Figure 4). Despite this developmental delay, the egg to puparium survival of the Idh3b mutant hemizygotes (∼80%) does not differ significantly from wild-type control hemizygotes. For all three mutant alleles, anterior spiracle eversion is often abnormal at pupariation, and, for Idh3b2 mutants, many puparia resemble elongate sclerotized larvae. Head eversion, which marks the transition between the prepupal and pupal periods, occurs in about half of Idh3b1 and Idh3b2 hemizygotes (22/39 and 19/38 animals, respectively), but in essentially all Idh3b3 hemizygotes (37/38 animals). Based on developmental delay, morphological abnormalities at pupariation, and head eversion failure, the Idh3b alleles can be ordered in severity as Idh3b2 > Idh3b1 > Idh3b3.
Figure 4.
The Idh3b alleles cause slowed development. Average egg to puparium development times are indicated for heterozygotes of +, E93Δ1, and each of the Idh3b alleles with Df(3R)Exel6188. The Idh3b alleles cause increases of 24–55% in egg to puparium development time, whereas the E93Δ1 allele has no effect. N = total number of puparia scored.
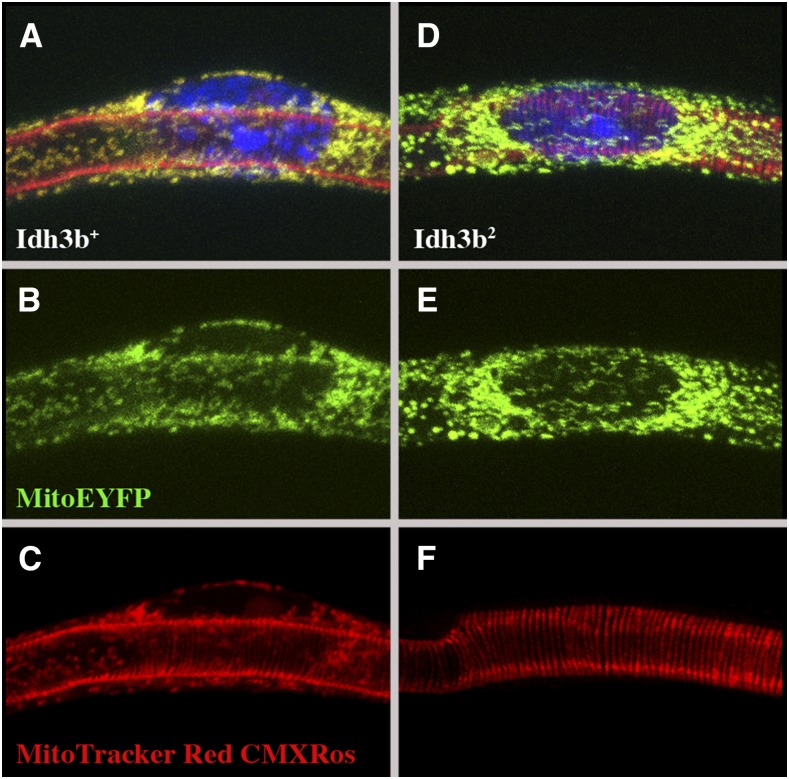
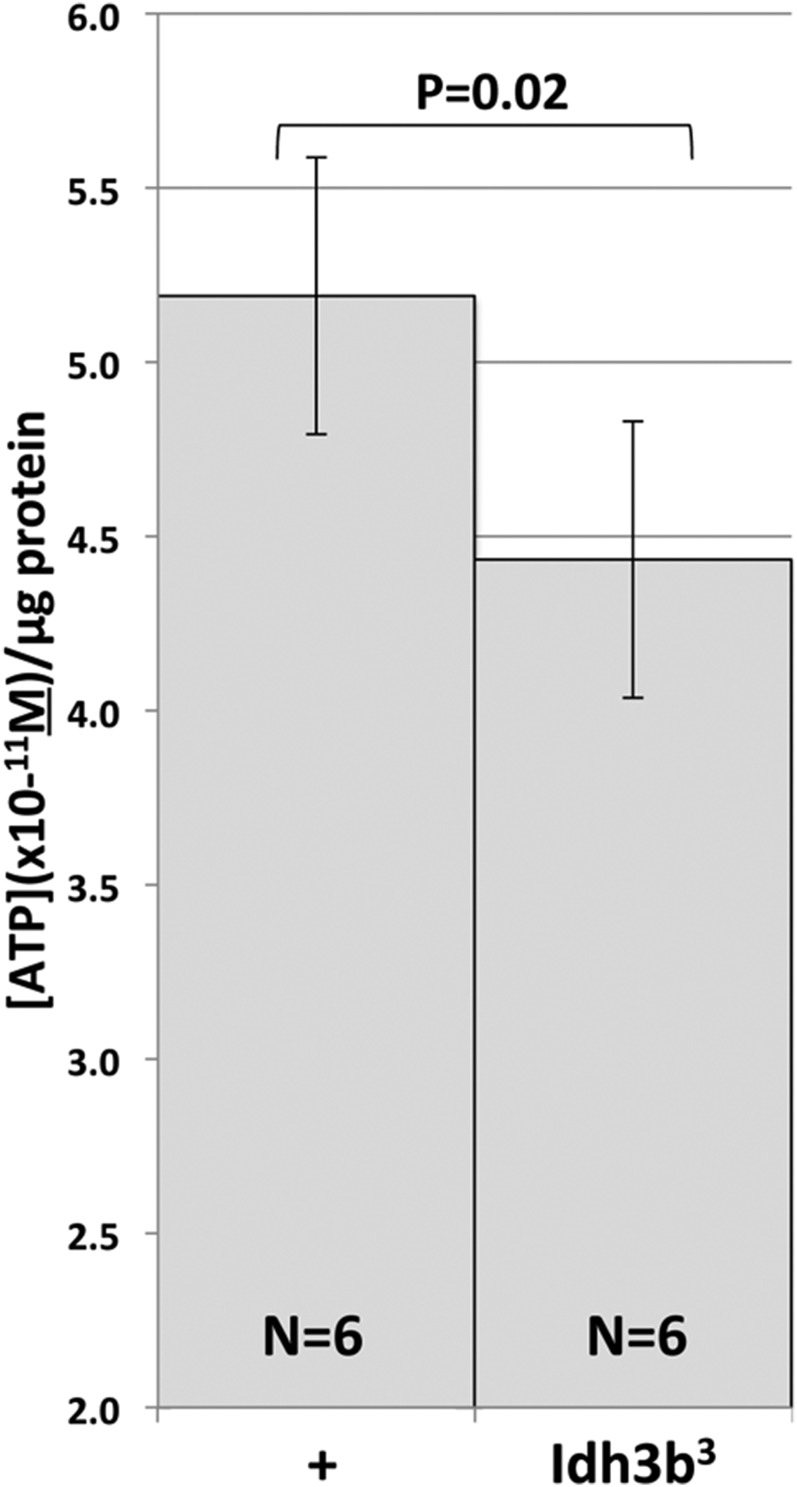
Disruption of the TCA cycle in the Idh3b mutants would be expected to result in mitochondrial dysfunction. To test this possibility, we stained living cells with the dye MitoTracker Red CMXRos (ThermoFisher), which is taken up only by polarized mitochondria. Tracheal cells from late third-instar larvae were examined, as these cells have prominent mitochondria, and are readily accessible to the MitoTracker dye. In Idh3b+ mito-EYFP/Df(3R)Exel6188 control animals, essentially all mitochondria in these cells stain with the MitoTracker dye, whereas in Idh3b2 mito-EYFP/Df(3R)Exel6188 cells, MitoTracker staining is absent (Figure 5). MitoTracker staining is also reduced, although to a lesser extent, in Idh3b1 mito-EYFP/Df(3R)Exel6188 and Idh3b3 mito-EYFP/Df(3R)Exel6188 cells (not shown). The dye MitoTracker Deep Red, which is largely, but not completely, dependent on mitochondrial polarization, gave similar results (not shown). To test whether this mitochondrial dysfunction is associated with reduced ATP production, we compared ATP levels in Idh3b mutant and wild-type animals at 18 hr APF, a time at which oxidative phosphorylation, rather than glycolysis, has become an important source of ATP (Wang et al. 2010; Tennessen et al. 2011, 2014). The weakest allele (Idh3b3) was selected for examination because, unlike Idh3b1 and Idh3b2, Idh3b3-mutant animals do not arrest development until well after 18 hr APF, allowing accurate staging in the prepupal/early pupal periods. We find that ATP levels in Idh3b3/Df(3R)Exel6188 animals are ∼15% lower (P = 2%) than in +/Df(3R)Exel6188 controls at 18 hr APF (Figure 6). Since Idh3b3 is the weakest of the alleles, this reduction in ATP is a minimal estimate of the requirement for Idh3b.
Figure 5.
Loss of mitochondrial polarization in Idh3b2 mutants. (A–C) a tracheal cell from a wandering third instar mito-EYFP Idh3b+/Df(3R)Exel6188 larva stained with the dye MitoTracker Red CMXRos, which is specific for polarized (functional) mitochondria. MitoTracker staining (C) is coincident with mito-EYFP (B) (see merged image in A). (D–F) a tracheal cell from a wandering third instar mito-EYFP Idh3b2/Df(3R)Exel6188 larva. MitoTracker Red CMXRos staining (F) is absent from mitochondria (labeled in E by mito-EYFP; merged image in D). The striated tubes showing red fluorescence in A, C, D and F are the tracheae; this fluorescence results from the combined effects of cuticle autofluorescence and affinity of the dye for tracheal cuticle. In A and D, DNA is stained by DAPI (blue).
Figure 6.
ATP levels are reduced in Idh3b3 mutants. ATP levels are shown for +/Df(3R)Exel6188 and Idh3b3/Df(3R)Exel6188 18 hr pupae. The Idh3b3 mutant shows a decline in ATP level of ∼15% relative to wild type (P = 0.02). Six independent samples were tested for each genotype.
In previous work, the Idh3b mutations (then thought to be E93 alleles) were found to cause strong persistence of larval salivary glands in the pupa (Lee et al. 2000). We have confirmed this observation. Loss of glands was monitored in intact pupae in which GFP expression was driven specifically within salivary glands by the Sg-Gal4 driver (Wang et al. 2008). In control +/Df(3R)Exel6188 pupae, salivary gland loss is complete at between 21 and 23 hr APF at room temperature (Figure S2 and Figure S3). However, salivary glands persist beyond 30 hr APF in the three Idh3b mutant hemizygotes. In contrast, E93Δ1 hemizygotes cause only slightly increased persistence of salivary glands relative to wild type (Figure S2 and Figure S3). The nonsense alleles E934, E935, and E936 behave similarly. E93Δ1 hemizygotes also show no developmental delay during larval development (Figure 4), unlike the Idh3b mutants.
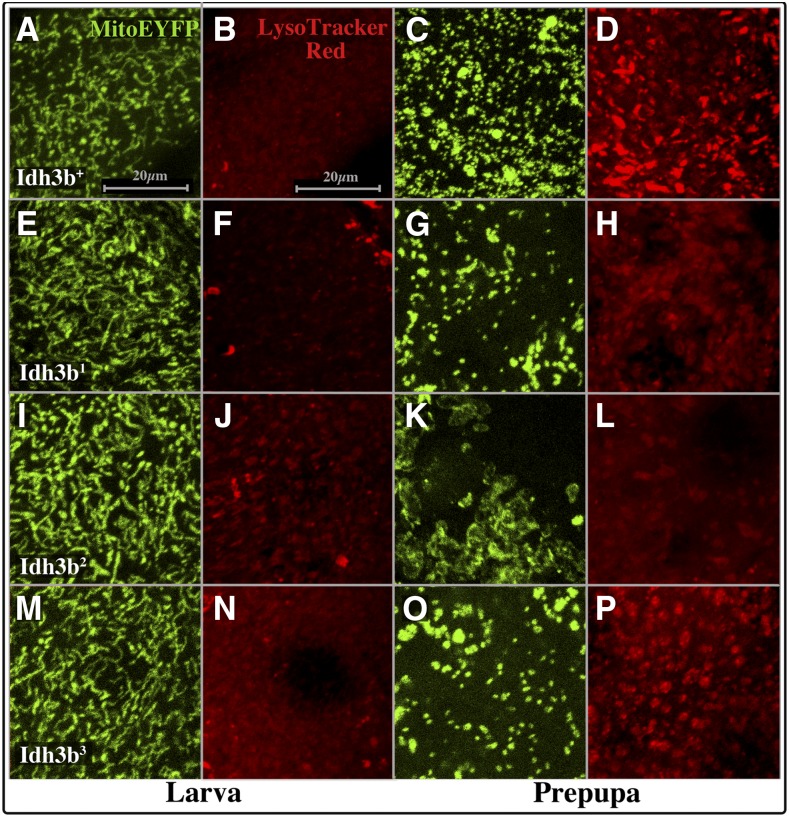
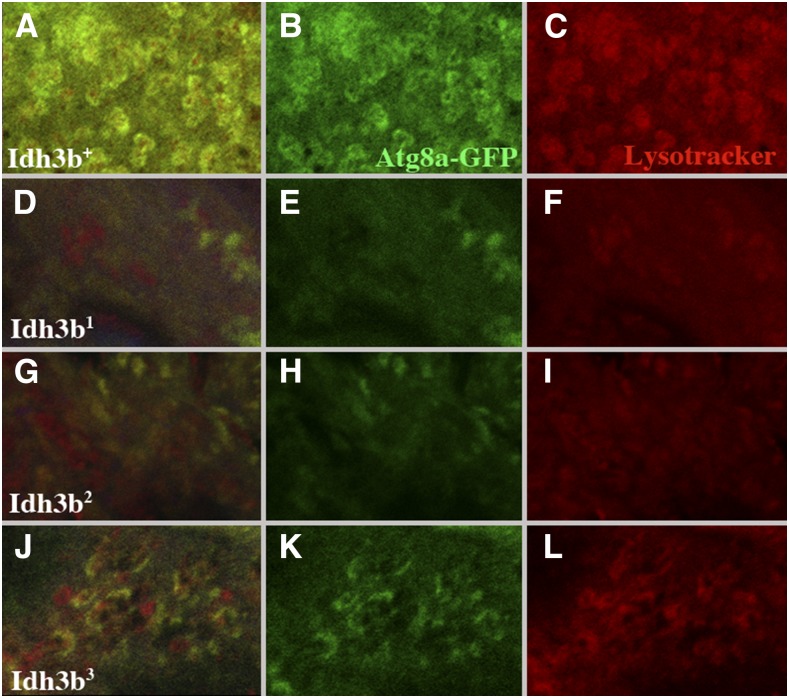
The death of larval salivary gland cells is known to involve autophagy (Lee and Baehrecke 2001). To monitor autophagy we used the dye LysoTracker (Thermo Fisher), which labels acidic cellular compartments, including lysosomes and autolysosomes, and Atg8a-GFP, which labels the membranes of autophagosomes and autolysosomes (Scott et al. 2004). In wild type, LysoTracker staining in salivary gland cells increases dramatically in the prepupal stage (Figure 7, B and D). When Atg8a-GFP is driven by Sg-Gal4, very little fluorescence is seen at 0 hr APF, but, by 13 hr APF salivary gland cytoplasm contains densely packed GFP-labeled vesicles (Figure 8, A–C). These vesicles colabel with LysoTracker, identifying them as autolysosomes. The high density of these autolysosomes accounts for the rapid degradation of the salivary gland cytoplasm at this stage in wild type. In Idh3b1 and Idh3b2 mutants, LysoTracker staining increases only slightly, or not at all, after pupariation, and the number of Atg8a-GFP-labeled vesicles at 13 hr APF is dramatically reduced (Figure 7, H and L and Figure 8, D–I). Idh3b3 mutants have an intermediate phenotype: LysoTracker increases after pupariation, but less so than wild type (Figure 7P), and at 13 hr APF many partially formed Atg8a-GFP-labeled vesicles are present (Figure 8, J and K) These partially formed vesicles are associated with LysoTracker staining, but the association is not as complete as in wild type. Taken together, these observations are consistent with cytological observations of Lee and Baehrecke (2001) that demonstrate a failure to initiate autophagy in Idh3b mutant salivary glands.
Figure 7.
Idh3b is required for mitochondrial fragmentation and increase in acidic compartments after pupariation in salivary gland cells. Panels show salivary gland cells of the genotypes mito-EYFP Idh3b+/Df(3R)Exel6188 (A–D), mito-EYFP Idh3b1/Df(3R)Exel6188 (E–H), mito-EYFP Idh3b2/Df(3R)Exel6188 (I–L), and mito-EYFP Idh3b3/Df(3R)Exel6188 (M–P). In mid third-instar larvae, mitochondria are unfragmented (A, E, I, and M) and LysoTracker staining is very weak (B, F, J, and N) in all genotypes shown. However, in 8–12 hr APF prepupal glands (C, D, G, H, K, L, O, and P), mitochondria have undergone extensive fragmentation in mito-EYFP Idh3b+/Df(3R)Exel6188 animals (C), and LysoTracker staining has increased dramatically (D), consistent with the initiation of autophagic cell death. In glands from mito-EYFP Idh3b1/Df(3R)Exel6188 and mito-EYFP Idh3b3/Df(3R)Exel6188 prepupae, mitochondrial fragmentation is not as extensive (G and O), and LysoTracker staining increases to a lesser extent (H and P). In glands from mito-EYFP Idh3b2/Df(3R)Exel6188 prepupae, mitochondria do not fragment, but appear clumped (K), and LysoTracker staining fails to increase (L). All panels are at the same magnification (see scale bars in A and B).
Figure 8.
Autophagy is blocked in Idh3b-mutant salivary gland cells. All images show salivary gland cells from 8 to 12 APF prepupae, in which expression of UAS-Atg8a-GFP is driven by Sg-Gal4. Atg8a-GFP is shown in green, and LysoTracker staining is in red. (A–C) In Idh3b+/Df(3R)Exel6188 cells, Atg8a-GFP labeled vesicles are densely packed, and these vesicles colabel with LysoTracker, identifying them as autolysosomes. (D–I) In Idh3b1/Df(3R)Exel6188 and Idh3b2/Df(3R)Exel6188 cells, Atg8a-GFP-labeled vesicles are dramatically reduced in number, and LysoTracker staining is much reduced, indicating an almost complete block in autophagy. (J–L) Idh3b3/Df(3R)Exel6188 cells show an intermediate phenotype, in which Atg8a-GFP vesicles are partially formed, and LysoTracker staining is weakened, and incompletely associated with Atg8a-GFP fluorescence.
To explore further the role of Idh3b in salivary gland elimination, we examined the behavior of mitochondria in mutant larval salivary glands. In wild type, salivary gland mitochondria undergo extensive fragmentation in late larvae (Goyal et al. 2007) (Figure 7, A–D). This fragmentation is thought to be an early initiating step in the death of the glands. We find that mitochondrial fragmentation is absent or very much reduced in Idh3b2 mutants compared to wild type (Figure 7, I–L). Mutant mitochondria remain elongated, and appear clumped together. Weaker reduction in fragmentation is seen in Idh3b1 (Figure 7, E–H) and Idh3b3 (Figure 7, M–P) mutants. Surprisingly, mitochondrial localization of Idh3b is stage-specific in salivary gland cells. In glands from Idh3b+ mid third-instar larvae, FLAG-Idh3b, and mito-EYFP colocalize (Figure 9, A–C and G–I). However, in prepupae, after mitochondrial fragmentation, FLAG-Idh3b and mito-EYFP fluorescence show near mutually exclusive punctate localization (Figure 9, D–F and J–L). Strikingly, after mitochondrial fragmentation, FLAG-Idh3b puncta are present within the nucleus, while mitochondria (i.e., mito-EYFP) are not (Figure 9, D–F). Other mitochondrial proteins, including the TCA cycle enzyme fumarase, are known to become localized to the nucleus under certain conditions [see Tang (2015) for review]. Whether the nuclear localization of Idh3b seen here after mitochondrial fragmentation is of functional significance is not clear.
Figure 9.
Expulsion of Idh3b from mitochondria after fragmentation. All images are from larvae of the genotype UAS-HA/FLAG-Idh3b/+; tub-Gal4/mito-EYFP. (A–C) Salivary gland cell from a mid third-instar larva. FLAG-Idh3b and mito-EYFP colocalize. The nucleus (marked by DAPI staining in blue) shows no mito-EYFP fluorescence, and is very weakly labeled for FLAG-Idh3b. (D–F) Salivary gland cell from a late (wandering) third-instar larva. Mitochondrial fragmentation has taken place. FLAG-Idh3b staining and mito-EYFP are no longer coincident (see merged image in D). Note that FLAG-Idh3b staining within the nucleus has become more intense (E), but that mito-EYFP staining continues to be excluded from the nucleus (F). The dark lacunae in these images are secretory vacuoles, which are prominent at this time. (G–I) High magnification of part of a mid third-instar salivary gland cell. Note that FLAG-Idh3b staining (H) and mito-EYFP (I) largely colocalize (G). (J–L) High magnification of part of a late third-instar salivary gland cell. FLAG-Idh3b puncta (K) and mito-EYFP (L) are now almost mutually exclusive (merged image in J).
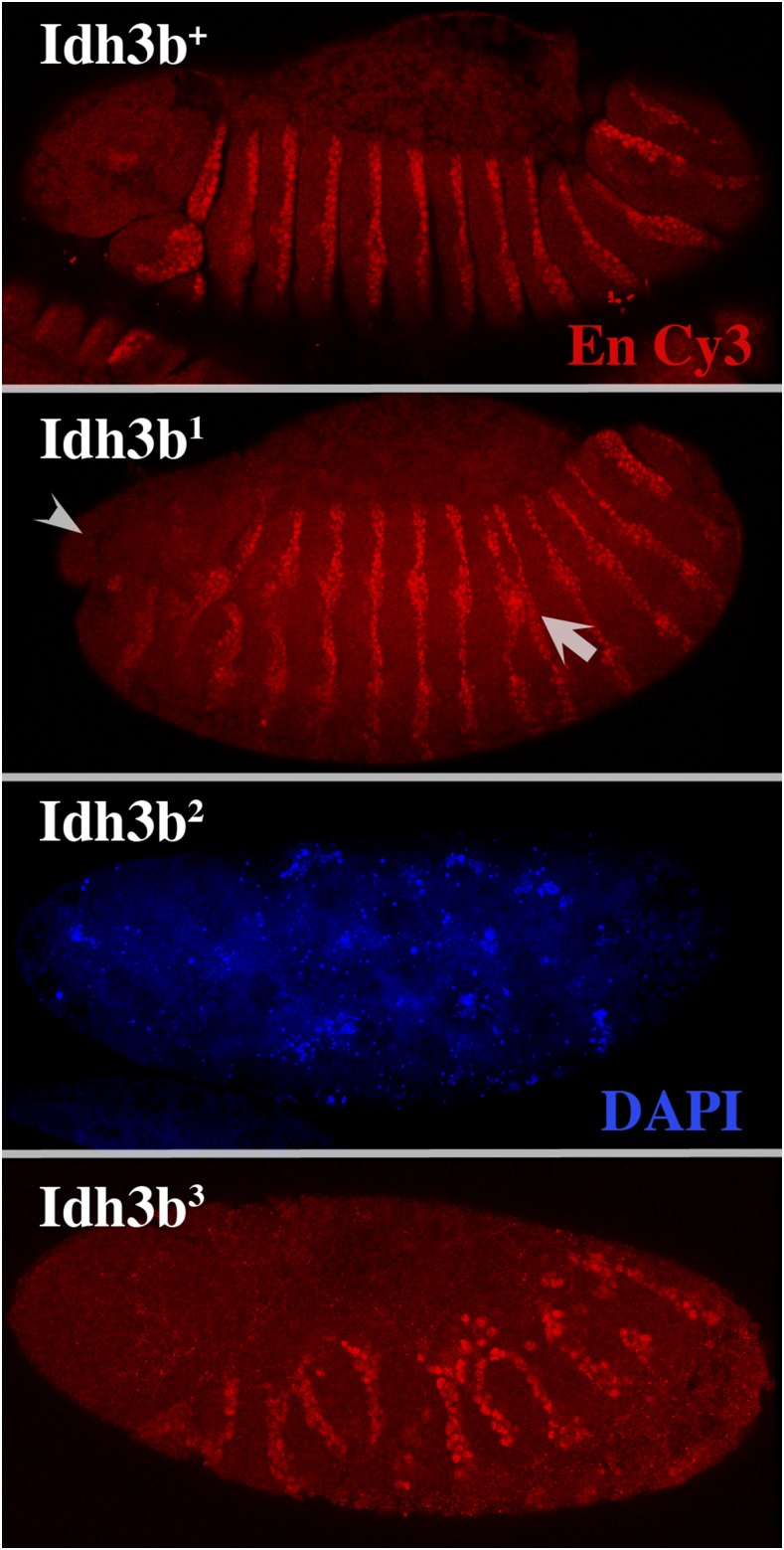
To test whether maternally contributed Idh3b is important for normal development, we used the ovoD dominant female sterile method (Chou et al. 1993) to generate mothers whose germlines were homozygous mutant for each of the Idh3b alleles. These females were crossed to males heterozygous for Df(3R)Exel6188, which is deficient for Idh3b. Remarkably, females whose germlines are homozygous for the Idh3b alleles produce large numbers of normal-appearing eggs, indicating that the female germline does not require autonomous Idh3b function. However, the embryos that develop from these eggs are highly abnormal. For Idh3b2-mutant germline mothers, all embryos arrest early during cleavage (Figure 10). Nuclei in arrested embryos are highly variable in size, and often appear incompletely separated, suggesting extensive mitotic abnormalities. These defects are similar to those caused by anoxia at this stage (DiGregorio et al. 2001; Foe and Alberts 1985), consistent with an almost complete block in oxidative phosphorylation. Almost all embryos from Idh3b3-mutant germline mothers also fail to complete embryogenesis. However, these embryos fare much better, with the majority surviving to the blastoderm stage or beyond. About half show some degree of segmentation, as indicated by the development of engrailed-expressing stripes. For most such embryos, these stripes are highly abnormal, being fragmented and showing spacing irregularities (Figure 10), and the head region is strongly reduced. Surprisingly, the Idh3b1 allele has much weaker effects than Idh3b3 in maternal germline clones. This is despite Idh3b1 being significantly stronger than Idh3b3 in its effects on homozygotes from heterozygous mothers. Almost all embryos from Idh3b1 mutant germline mothers survive to segmented stages, although segmentation defects are frequent (Figure 10); ∼15% of such embryos hatch, with a few surviving to pupariation and adulthood.
Figure 10.
Absence of maternally supplied Idh3b causes developmental arrest. Embryos shown are from mothers whose germlines were homozygous for the indicated Idh3b alleles crossed to Df(3R)Exel6188/TM6B fathers. For embryos from Idh3b+, Idh3b1, and Idh3b3 homozygous germline mothers, the segmentation protein engrailed (En) is stained using Cy3-labeled anti-engrailed (red). Since embryos from Idh3b2 mutant germline mothers all arrest prior to segmentation, the Idh3b2 mutant embryo shown is stained instead with DAPI (blue), which stains nuclei. Most embryos from Idh3b1 germline mothers segment, but engrailed stripes usually show spacing defects or are fused with one another (arrow). The head region (arrowhead) is almost always reduced. Embryos from Idh3b2 germline mothers all arrest during cleavage, and contain nuclei of variable size. Embryos from Idh3b3 germline mothers arrest at stages ranging from cleavage to cuticle formation; almost all of those that express engrailed have grossly abnormal striping patterns, like the embryo shown here.
Discussion
We report here that the three mutations initially described as the “type alleles” of E93 are in fact alleles of Idh3b, which encodes the β subunit of NAD-dependent isocitrate dehydrogenase—a key mitochondrial enzyme of the TCA cycle. This finding is important because almost all studies aimed at understanding E93 function have relied upon these mutations. Reports of the interaction of E93 with other genes involved in the ecdysone response hierarchy, as well as the regulation by E93 of genes involved in cell death and autophagy, will all have to be revisited. Most importantly, the view that E93 serves as a dedicated regulator of cell death will need to be revised. We generate an allele of E93 (E93Δ1) that is deleted for almost the entire E93 coding sequence. This allele behaves essentially the same as the E934-6 alleles described previously (Mou et al. 2012), consistent with E93 functioning as a temporal identity determinant for pupal development (Mou et al. 2012; Ureña et al. 2014).
The three Idh3b mutations were misidentified as E93 alleles primarily because one of them (Idh3b1, called initially E931) was found to be associated with a nonsense change near the 3′ end of the E93 coding sequence (at codon 995 of the E93 A isoform). Allelism with E93 seemed to make good sense, as the three alleles had the expected phenotype (death after pupariation). However, we find that hemizygotes for Idh3b1 are rescued to adulthood by an Idh3b+ transgene, indicating that the nonsense change in E93 in this mutant is not responsible for its phenotypic effects. Indeed, this nonsense change has no apparent phenotypic effect, as E93Δ1/Idh3b1 heterozygotes are fully viable and appear wild type as adults. The E93 nonsense change associated with Idh3b1 is almost certainly an incidental EMS-induced change. At the doses of EMS used in the screen that produced the Idh3b alleles (25 and 37 mM) (Lee et al. 2000), the expected frequency of base changes induced is about one altered base pair in 300–400 kb (Blumenstiel et al. 2009; Cooper et al. 2008). Therefore, the expected frequency of such incidental changes in the E93 coding sequence (∼3300 bp) is ∼1% of treated chromosomes.
In their initial description of the “E931-3” alleles, Lee et al. (2000) performed a “rescue” experiment, in which they showed that driving expression of E93 in salivary glands at the larval stage by use of forkhead-Gal4 causes elimination of the glands in an “E931” (i.e., Idh3b1) mutant background. This result was taken as supporting evidence that “E931-3” are alleles of E93, and that E93 plays a key role in directing larval cell death. However, a significant weakness of this experiment is that E93 expression was driven in salivary glands at the larval stage, rather than at the pupal stage, when E93 expression and salivary gland death normally occur. In imaginal discs, ectopic expression of a wide variety of cell fate regulators causes apoptosis and/or segregation of cell clones [see, for example, Bielmeier et al. (2016)]. The elimination of salivary glands in larvae by ectopic E93 may occur for similar reasons; that is, E93 may interfere with the normal developmental program of the larval salivary gland, leading secondarily to gland elimination.
By all measures, the strongest of the Idh3b alleles is Idh3b2. This allele is a missense change of an invariant glycine at position 278 to aspartic acid. Idh3b2 may be a null allele. However, another possibility is that it encodes an interfering subunit. Mammalian IDH3 is a heterotetramer consisting of two α, one β, and one γ subunit. Biochemical reconstitution experiments show that the α and γ subunits can produce weakly active enzyme in the absence of β (Ehrlich and Colman 1983). The strength of the Idh3b2 allele relative to the other alleles suggests it may encode an inactive β subunit that associates with α and γ, and blocks their residual activity. Idh3b1 is an A to T change within the AG splice acceptor sequence at the 5′ boundary of exon 4. One might have expected Idh3b1 to behave as a null allele, as total failure of splicing of the intron between exons 3 and 4 would result in the production of a protein consisting of only the N-terminal 61 amino acids of the 370 amino acid Idh3b protein, followed by 29 unrelated residues encoded by the unspliced intron. The relative weakness of this allele, especially when homozygous in the female germline, suggests that a cryptic splice site within exon 4 can be utilized in the mutant, or that exon 4 is entirely skipped. Splicing at the first downstream inframe AG in exon 4 would generate a transcript deleted for the first nine codons of exon 4; skipping exon 4 entirely would result in the deletion of 39 codons. Idh3b3 is a missense change of an aspartic acid at position 267 to tyrosine. This position tolerates several different residues among known Idh3b sequences, but is never occupied by tyrosine in a wild-type sequence. Curiously, the relative strengths of Idh3b1 and Idh3b3 depend on the phenotype considered. For homozygous mutant zygotes from mutant/+ mothers, Idh3b1 is stronger in its effects than Idh3b3. However, for embryos from homozygous mutant germline mothers, the reverse is true. This difference may reflect differing splicing specificities in germline- and zygotic-development, or differing Idh3b domain requirements at the two stages.
Strikingly, Idh3b is located within a cluster of six contiguous mitochondrial genes. These genes encode mitofilin, mitochondrial ribosomal protein L35, fumarylacetoacetase, cytochrome c heme lyase, and NADH dehydrogenase (ubiquinone) 42-kDa subunit, as well as Idh3b. In situ hybridization results reported by Baehrecke and Thummel (1995) suggest that all of these genes are contained within the 93F pupal ecdysone puff, consistent with their coregulation.
Given the central importance of the TCA cycle in metabolism, it seemed at first surprising that Idh3b mutants survive until the pupal stage. In humans, an IDH3B allele has been described that also has a surprisingly weak phenotype (Hartong et al. 2008). This allele causes a frameshift at codon 197, and is therefore likely a null allele. Individuals homozygous for this allele develop retinitis pigmentosa, but are otherwise normal, arguing that IDH3β is dispensable in all human tissues outside the retina. Residual IDH3 enzyme activity (presumably due to activity of IDH3α and IDH3γ) in mutant cells is only 2–5% of normal. The authors suggest that, in humans, a second mitochondrial IDH, IDH2, can fully substitute for IDH3 outside of the retina. Unlike IDH3, which is NAD dependent, IDH2 is NADP dependent, and is located within the mitochondrial intermembrane space, rather than the matrix, where the TCA cycle takes place (Chen et al. 2015). IDH2 is known to play an important role in the regeneration of reduced glutathione, which is utilized in the elimination of reactive oxygen species produced by the electron transport chain (Jo et al. 2001). However, Hartong et al. (2008) raise the possibility that IDH2 may also be the primary enzyme utilized in the TCA cycle. Although this hypothesis may well be correct for human cells, two observations argue that it is not true for Drosophila. First, we find that mitochondria from Idh3b2 mutant larvae show no, or dramatically reduced, staining by dyes specific for polarized mitochondria (MitoTracker RedCMXRos and MitoTracker Deep Red), although mitochondria from wild-type control larvae are stained by these dyes. Second, we find that embryos from homozygous-mutant Idh3b2 germline mothers arrest development during cleavage, with arrested embryos showing nuclear abnormalities similar to those caused by anoxia at this stage. Both of these observations suggest that the Idh3b2 mutation causes an almost complete block in the TCA cycle and oxidative phosphorylation.
The survival of Idh3b mutants until pupariation is consistent with recent work showing that Drosophila larvae generate energy largely by aerobic glycolysis (Tennessen et al. 2011, 2014). In this mode of metabolism, which is characteristic of very rapid growth in many systems, sugars are largely converted to biomass rather than being totally oxidized, as occurs during oxidative phosphorylation [see Lunt and Vander Heiden (2011) for review]. Early embryos are dependent upon oxidative phosphorylation for energy production, and are very sensitive to anoxia (Foe and Alberts 1985; DiGregorio et al. 2001). However, midway through embryogenesis, genes encoding enzymes of the glycolytic pathway are globally upregulated by the Drosophila estrogen-related receptor (Tennessen et al. 2011). The glycolytic pathway then comes to dominate energy metabolism, and facilitates biomass increase during the larval stages. Consistent with early dependence on oxidative phosphorylation, embryos from homozygous-mutant Idh3b2 germline mothers arrest development during cleavage. Idh3b mutant homozygotes from heterozygous mothers survive the embryonic period due to the presence of maternally supplied Idh3b, and then survive larval development, likely because of the metabolic shift to aerobic glycolysis at this stage. Although Idh3b mutants survive as larvae, their development is slowed considerably, indicating that oxidative phosphorylation continues to play an important role during normal larval development. This conclusion is supported by the polarization of mitochondria in wild-type larvae revealed by MitoTracker RedCMXRos and MitoTracker Deep Red staining.
Although the shift to aerobic glycolysis during larval development is likely the major factor allowing the survival of Idh3b mutants until pupariation, residual Idh3 activity provided by the Idh3 α and γ subunits may also play a role. Bypass of the TCA cycle step blocked by loss of Idh3 might also be a factor. Such bypass could occur by the action of Idh2 in the intermembrane space. As suggested by Hartong et al. (2008), the NADPH produced by Idh2 could be converted to NADH by nicotinamide-nucleotide transhydrogenase and transferred into the matrix. Alternatively, α-ketoglutarate produced by Idh2 could be transported into the matrix. It is also possible that citrate exported from mitochondria (as normally occurs in lipid biosynthesis) is converted to α-ketoglutarate or glutamate by cytosolic enzymes, followed by reimport of these intermediates [see Lunt and Vander Heiden (2011)].
The behavior of mitotic recombination clones homozygous for the Idh3b alleles is striking. In this report, we find that female germline clones homozygous for each of the Idh3b alleles survive and produce large numbers of normal-appearing eggs. Previously, we showed that homozygous-mutant somatic clones show no obvious growth defect, and produce normal adult cuticular structures throughout the body (Mou et al. 2012). The simplest interpretation is that oxidative phosphorylation is of limited importance within these cell types. Alternatively, Idh2 may be able to substitute for Idh3 in these cells, or homozygous mutant cells within clones may be rescued by their wild-type neighbors by transfer of ATP or other metabolites through gap junctions (Goldberg et al. 1999).
Like Idh3b alleles, mutations affecting another TCA cycle enzyme, Malate dehydrogenase-2 (Mdh2), arrest development after pupariation, and are defective in salivary gland death (Wang et al. 2010). In addition, both Idh3b-mutant pupae (this report) and Mdh2-mutant pupae (Wang et al. 2010) show reduced ATP levels. However, in other ways, mutants for these genes behave quite differently. Mdh2 mutants show normal growth and produce normal-appearing puparia (Wang et al. 2010), whereas Idh3b mutations cause significant developmental delay, and show defects in puparium shape and spiracle eversion. In salivary gland cells, Mdh2 mutants have no effect on expression of the ecdysone-induced cell death regulators rpr, hid, and dronc, while it has been reported that Idh3b mutants fail to activate transcription of these genes (Lee et al. 2000). Finally, Mdh2 mutants show a normal increase in LysoTracker staining in pupal salivary glands (which signals the initiation of autophagy), while Idh3b2 mutants fail to show such an increase. We do not understand why Idh3b mutants are more severely affected than Mdh2 mutants. One possibility is that the TCA cycle block in Mdh2 mutants can be partially bypassed, possibly via enzymes of the malate-aspartate shuttle.
The block in salivary gland death in Idh3b and Mdh2 mutants indicates an important role for the TCA cycle and mitochondria in driving the death of larval cells during metamorphosis. As in many cell types, mitochondria in salivary gland cells undergo extensive fragmentation prior to cell death (Goyal et al. 2007). This fragmentation fails to occur in salivary gland cells of the Idh3b2 mutant. In mammalian cells, mitochondrial fragmentation is associated with the release of apoptogenic factors such as cytochrome c from the mitochondrial intermembrane space [for review see Arnoult (2006)]. Although release of cytochrome c does not appear to promote cell death in Drosophila [for review see Clavier et al. (2016)], the failure of mitochondrial fragmentation in Idh3b mutants may block the release of other mitochondrial factors that promote larval cell death. Idh3b itself is a candidate for such a factor, as we find that it is released from the mitochondrial matrix after fragmentation, becoming concentrated in puncta that localize to both the cytoplasm and the nucleus. In addition, Idh3b has been reported to interact physically with the effector caspase DrICE (Guruharsha et al. 2011). Experiments are underway to test whether Idh3b, or other factors released from the mitochondrial matrix, play a role in promoting larval cell death.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.037366/-/DC1.
Acknowledgments
We thank Eric Baehrecke for supplying the E93 and Idh3b mutants used in this study, suggesting the deletion approach used, and many discussions. We also thank Jennifer Duncan (Department of Pediatrics, Washington University School of Medicine) for the gift of the dyes MitoTracker Red CMXRos and MitoTracker Deep Red, Dr. Yehuda Ben-Shahar (Department of Biology, Washington University) for discussions, and providing the mito-EYFP line, and Carl Thummel for helpful comments on the manuscript. The research described in this report was supported by award 1257579 from the National Science Foundation.
Footnotes
Communicating editor: A. Bashirullah
Literature Cited
- Arnoult D., 2006. Mitochondrial fragmentation in apoptosis. Trends Cell Biol. 17: 6–12. [DOI] [PubMed] [Google Scholar]
- Baehrecke E. H., 2003. Autophagic programmed cell death in Drosophila. Cell Death Differ. 10: 940–945. [DOI] [PubMed] [Google Scholar]
- Baehrecke E. H., Thummel C. S., 1995. The Drosophila E93 gene from the 93F early puff displays stage- and tissue-specific regulation by 20-hydroxyecdysone. Dev. Biol. 171: 85–97. [DOI] [PubMed] [Google Scholar]
- Berry D. L., Baehrecke E. H., 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131: 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielmeier C., Alt S., Weichselberger V., Fortezza M. L., Harz H., et al. , 2016. Interface contractility between differently fated cells drives cell elimination and cyst formation. Curr. Biol. 26: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific ϕc31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel J. P., Noll A. C., Griffiths J. A., Perera A. G., Walton K. N., et al. , 2009. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakouros D., Daish T. J., Kumar S., 2004. Ecdysone receptor directly binds the promoter of the Drosophila caspase dronc, regulating its expression in specific tissues. J. Cell Biol. 165: 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-L., Hu Y., Udeshi N. D., Lau T. Y., Wirtz-Peitz F., et al. , 2015. Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc. Natl. Acad. Sci. USA 112: 12093–12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. B., Noll E., Perrimon N., 1993. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development 119: 1359–1369. [DOI] [PubMed] [Google Scholar]
- Clavier A., Rincheval-Arnold A., Colin J., Mignotte B., Guénal I., 2016. Apoptosis in Drosophila: which role for mitochondria? Apoptosis 21: 239–251. [DOI] [PubMed] [Google Scholar]
- Cooper J. L., Greene E. A., Till B. J., Codomo C. A., Wakimoto B. T., et al. , 2008. Retention of induced mutations in a Drosophila reverse-genetic resource. Genetics 180: 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGregorio P. J., Ubersax J. A., O’Farrell P. H., 2001. Hypoxia and nitric oxide induce a rapid, reversible cell cycle arrest of the Drosophila syncytial divisions. J. Biol. Chem. 276: 1930–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich R. S., Colman R. F., 1983. Separation, recombination, and characterization of dissimilar subunits of the DPN-dependent isocitrate dehydrogenase from pig heart. J. Biol. Chem. 258: 7079–7086. [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G., 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Foe V. E., Alberts B. M., 1985. Reversible chromosome condensation induced in Drosophila embryos by anoxia: visualization of interphase nuclear organization. J. Cell Biol. 100: 1623–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg G. S., Lampe P. D., Nicholson B. J., 1999. Selective transfer of endogenous metabolites through gap junctions composed of different connexins. Nat. Cell Biol. 1: 457–459. [DOI] [PubMed] [Google Scholar]
- Gorski S. M., Chittaranjan S., Pleasance E. D., Freeman J. D., Anderson C. L., et al. , 2003. A SAGE approach to discovery of genes involved in autophagic cell death. Curr. Biol. 13: 358–363. [DOI] [PubMed] [Google Scholar]
- Goyal G., Fell B., Sarin A., Youle R. J., Sriram V., 2007. Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster. Dev. Cell 12: 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha K. G., Rual J. F., Zhai B., Mintseris J., Vaidya P., et al. , 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartong D. T., Dange M., McGee T. L., Berson E. L., Dryja T. P., et al. , 2008. Insights from retinitis pigmentosa into the roles of isocitrate dehydrogenases in the Krebs cycle. Nat. Genet. 40: 1230–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihry R. J., Bashirullah A., 2014. Genetic control of specificity to steroid-triggered responses in Drosophila. Genetics 196: 767–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihry R. J., Sapiro A. L., Bashirullah A., 2012. Translational control by the DEAD box RNA helicase belle regulates ecdysone-triggered transcriptional cascades. PLoS Genet. 8: e1003085 10.1371/journal.pgen.1003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Lamblin A.-F. J., Steller H., Thummel C. S., 2000. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol. Cell 5: 445–455. [DOI] [PubMed] [Google Scholar]
- Jo S.-H., Son M.-K., Koh H.-J., Lee S.-M., Song I.-H., et al. , 2001. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 276: 16168–16176. [DOI] [PubMed] [Google Scholar]
- Kankel M. W., Duncan D. M., Duncan I., 2004. A screen for genes that interact with the Drosophila pair-rule segmentation gene fushi tarazu. Genetics 168: 161–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse D. R., Buckner S. M., Lake J., Na C., Pirt A., et al. , 2004. Three new Drosophila markers of intracellular membranes. Biotechniques 36: 784–788. [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Baehrecke E. H., 2001. Steroid regulation of autophagic programmed cell death during development. Development 128: 1443–1455. [DOI] [PubMed] [Google Scholar]
- Lee C.-Y., Wendel D. P., Reid P., Lam G., Thummel C. S., et al. , 2000. E93 directs steroid-triggered programmed cell death in Drosophila. Mol. Cell 6: 433–443. [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Clough E. A., Yellon P., Teslovich T. M., Stephan D. A., et al. , 2003. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr. Biol. 13: 350–357. [DOI] [PubMed] [Google Scholar]
- Lunt S. Y., Vander Heiden M. G., 2011. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27: 441–464. [DOI] [PubMed] [Google Scholar]
- Martin D. N., Baehrecke E. H., 2004. Caspases function in autophagic programmed cell death in Drosophila. Development 131: 275–284. [DOI] [PubMed] [Google Scholar]
- Martin D. N., Baigley B., Dutta S., Chen J., Rudnick P., et al. , 2007. Proteomic analysis of steroid-triggered autophagic programmed cell death during Drosophila development. Cell Death Differ. 14: 916–923. [DOI] [PubMed] [Google Scholar]
- McPhee C. K., Baigley B. M., Nelson C., Hill J. H., Batlevi Y., et al. , 2013. Identification of factors that function in Drosophila salivary gland cell death during development using proteomics. Cell Death Differ. 20: 218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou X., Duncan D. M., Baehrecke E. H., Duncan I., 2012. Control of target gene specificity during metamorphosis by the steroid response gene E93. Proc. Natl. Acad. Sci. USA 109: 2949–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A. L., Cook K. R., Belvin M., Dompe N. A., Fawcett R., et al. , 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- Scott R. C., Schuldiner O., Neufeld T. P., 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7: 167–178. [DOI] [PubMed] [Google Scholar]
- Tang B. L., 2015. Mitochondrial protein in the nucleus. Cellbio (Irvine Calif.) 4: 23–29. [Google Scholar]
- Tennessen J. M., Baker K. D., Lam G., Evans J., Thummel C. S., 2011. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 13: 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. M., Bertagnolli N. M., Evans J., Sieber M. H., Cox J., et al. , 2014. Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3 (Bethesda) 4: 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., et al. , 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. [DOI] [PubMed] [Google Scholar]
- Tracy K., Baehrecke E. H., 2013. The role of autophagy in Drosophila metamorphosis. Curr. Top. Dev. Biol. 103: 101–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ureña E., Manjón C., Franch-Marro X., Martin D., 2014. Transcription factor E93 specifies adult metamorphosis in hemimetabolous and holometabolous insects. Proc. Natl. Acad. Sci. USA 111: 7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Lam G., Thummel C. S., 2010. Med24 and Mdh2 are required for Drosophila larval salivary gland death. Dev. Dyn. 239: 954–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Evans J., Andrews H. K., Beckstead R. B., Thummel C. S., et al. , 2008. A genetic screen identifies new regulators of steroid-triggered programmed cell death in Drosophila. Genetics 180: 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin V. P., Thummel C. S., 2005. Mechanisms of steroid-triggered programmed cell death in Drosophila. Semin. Cell Dev. Biol. 16: 237–243. [DOI] [PubMed] [Google Scholar]
- Yin V. P., Thummel C. S., Bashirullah A., 2007. Down-regulation of inhibitor of apoptosis levels provides competence for steroid-triggered cell death. J. Cell Biol. 178: 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All Drosophila strains used or generated in this study are available on request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.