Abstract
Alternative splicing generates a diversity of messenger RNA (mRNA) transcripts from a single mRNA precursor and contributes to the complexity of our proteome. Splicing is perturbed by a variety of mechanisms in cancer. Recurrent mutations in splicing factors have emerged as a hallmark of several hematologic malignancies. Splicing factor mutations tend to occur in the founding clone of myeloid cancers, and these mutations have recently been identified in blood cells from normal, healthy elderly individuals with clonal hematopoiesis who are at increased risk of subsequently developing a hematopoietic malignancy, suggesting that these mutations contribute to disease initiation. Splicing factor mutations change the pattern of splicing in primary patient and mouse hematopoietic cells and alter hematopoietic differentiation and maturation in animal models. Recent developments in this field are reviewed here, with an emphasis on the clinical consequences of splicing factor mutations, mechanistic insights from animal models, and implications for development of novel therapies targeting the precursor mRNA splicing pathway.
Introduction
The control of messenger RNA (mRNA) processing is a crucial component of gene regulation. The precursor messenger RNA (pre-mRNA) sequence contains exons that flank introns, which are removed during splicing.1 Human genes are remarkably complex, containing an average of 8 exons, with introns composing up to 90% of the pre-mRNA sequence.2,3 Up to 94% of human genes generate multiple mRNA isoforms.4-6 The spliceosome is a multiprotein complex consisting of 5 small nuclear ribonucleoproteins (snRNPs) and more than 150 proteins that recognizes the elements near intron-exon boundaries and the branch site that catalyzes the excision of intronic regions. Additional trans-acting factors including SR proteins bind the cis-regulatory sequences (exonic and intronic silencers and enhancers) to promote or prevent spliceosome assembly.7-9 The complexity of regulatory elements that control proper splicing makes this process not only critical to maintain tissue homeostasis, but also highly susceptible to hereditary and somatic mutations that are involved in disease. This review will discuss the implications of aberrant splicing on human disease, with a focus on the impact of somatic mutations in trans-acting splicing factors in hematologic malignancies.
Splicing and disease
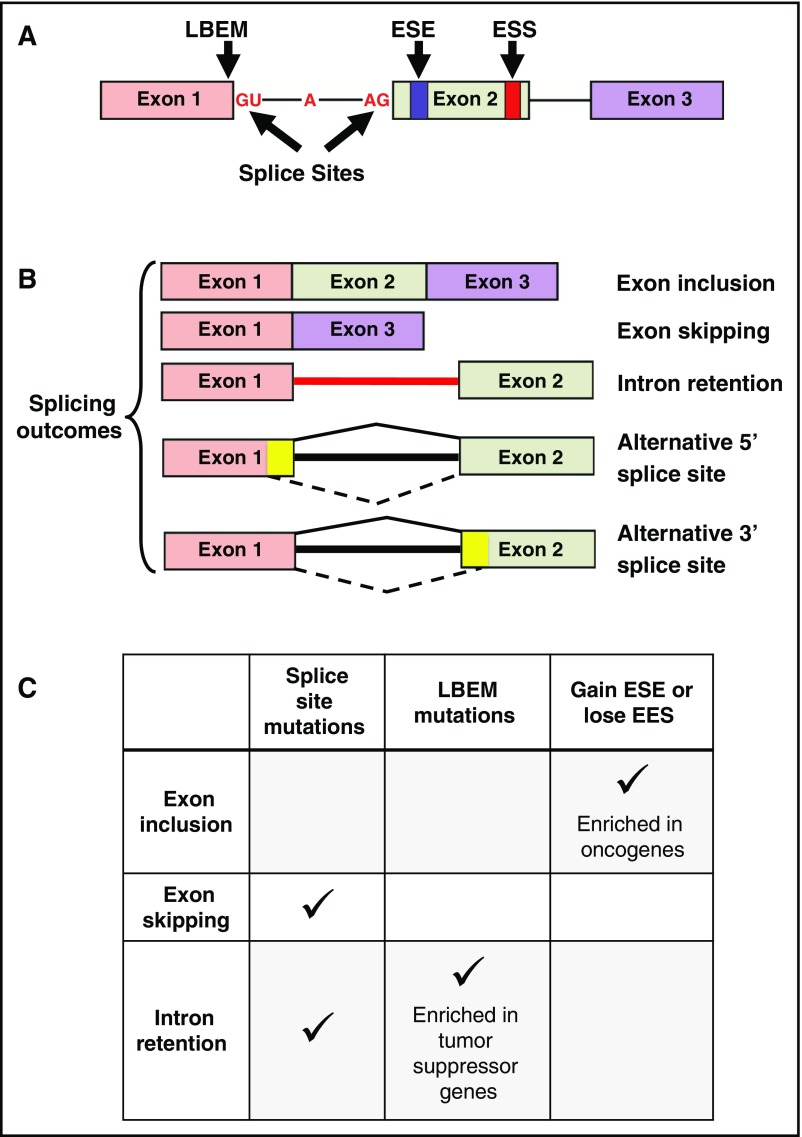
A comprehensive analysis of The Cancer Genome Atlas (TCGA) project RNA sequencing (RNA-seq) data from 16 tumor types and matched normal tissue revealed evidence of global alternative splicing involving a variety of splicing junctions, including cassette exons, retrained introns, and competing 5′ and 3′ splice sites (SSs).10 In particular, acute myeloid leukemia (AML) cells showed the largest number of alternative spliced events among tumor types.10 Recent evidence from analysis of paired DNA and RNA-seq data from TCGA has shown that somatic mutations affecting splicing regulatory sequences (splice donor/acceptor sites, last-base exon sites, and exonic splicing enhancer and silencer sites) can alter pre-mRNA splicing in tumor cells (Figure 1A-B).11-13
Figure 1.
Consequences of somatic mutations affecting cis-acting pre-mRNA sequences. (A) A representative 3-exon pre-mRNA model is depicted with annotation of the location of common cis-acting sequences affected by somatic mutations. (B) Common splicing outcomes that are caused by somatic mutations in cis-acting sequences or splicing factors include exon inclusion, exon skipping, and intron retention. Other splicing outcomes include alternative 5′ or 3′ splice sites (mutually exclusive exons, coordinate cassette exons, and alternative first and last exons are not shown). The yellow color in exons 1 and 2 represents exonic sequence that is excluded when splicing into an alternative cryptic splice site occurs, as indicated by the dashed lines at the bottom of each cartoon. (C) The predominant type of pre-mRNA splicing alteration induced by mutations in various cis-acting sequences.
Somatic mutations in the noncoding intronic 5′ splice donor (eg, GT dinucleotide) and 3′ splice acceptor (eg, AG dinucleotide) sites account for ∼1.6% of mutations in 12 TCGA tumor types, including AML.12 These mutations predominantly result in alterations of cassette exon splicing (ie, exon skipping) and intron retention (Figure 1C). Jung et al analyzed paired DNA and RNA-seq results from 6 TCGA tumor types and identified coding somatic single nucleotide variants that affected RNA splicing.11 Seventy percent of these single nucleotide variants occurred on the last base of exons, termed last-base exonic mutations (LBEMs). Collectively, 38% of LBEMs were associated with abnormal splicing. In contrast, only 4% of the penultimate position relative to the last base of exons, and only 3% of the first base of exons, caused abnormal splicing when mutated. Many of the LBEMs were synonymous and often involved tumor suppressor genes, suggesting that they may contribute to tumor pathogenesis (Figure 1C). Supek et al evaluated 11 TCGA tumor types using paired DNA and RNA-seq data and identified that ∼24% of all mutations were synonymous.13 Synonymous mutations were enriched within 30 base pairs of exon-intron junctions and often resulted in the gain of an exonic splice enhancer (ESE) or loss of an exonic splice silencer (ESS), both resulting in exon inclusion events. Somatic mutations affecting ESEs and ESSs were enriched in oncogenes, suggesting that they too may be important in disease pathogenesis (Figure 1C). In contrast, tumor suppressors such as p53 are also enriched in synonymous mutations. Some of these mutations affect sequences adjacent to SSs, which can lead to tumor suppressor inactivation.13 Collectively, somatic mutations in cis-acting regulatory sequences are likely to play a role in tumor pathogenesis by altering specific genes that have functional consequences.
Recurrent splicing mutations in hematologic malignancies
Mutation frequencies and clinical associations
In 2011, next-generation sequencing studies first revealed recurrent, somatic mutations in core components of the spliceosome.14-18 The initial studies focused on myelodysplastic syndromes (MDS) and chronic lymphocytic leukemia (CLL), where these mutations are most prevalent. Subsequent studies have clarified the frequency of these mutations across hematologic malignancies and solid tumors and begun to define their associations with disease subtypes and natural history. This information has been reviewed in detail elsewhere.19-23 The major clinical associations relevant to hematologic malignancies are summarized and updated here (Table 1). Mutations in more than 30 factors with established or putative roles in splicing have been reported in hematologic malignancies. The focus here will be on factors with the highest mutation frequency, because much larger datasets will be required to derive consistent and robust clinical associations for rare mutations.
Table 1.
Clinical features of splicing factor mutations in hematologic malignancies
| Gene | Disease | Frequency (%) | Co-occurring mutations | Clinical association(s) |
|---|---|---|---|---|
| SF3B1 | CLL | 5-1716,17,24,25,110 | del(11q)16 | Reduced PFS16 (clonal only),110 reduced OS,25 reduced PFS/OS17,24 |
| MDS | 7-2818,26,38,45,49,111-113 | DNMT3A38 | Improved LFS/OS18,26 | |
| RARS/RCMD-RS | 54-8315,18,38,114 | Improved OS,29 no impact on LFS/OS26,28,111 | ||
| RARS-T | 81-8845,114 | |||
| RA | 5-1218,38,45 | |||
| RAEB | 5-1118,45 | |||
| RCMD | 4-618,45 | |||
| Secondary AML | 5-1126,35,43,114 | |||
| De novo AML | 3-518,35,41,114 | |||
| tAML | 343 | |||
| CMML | 5-618,32 | |||
| MDS/MPN | 1826 | |||
| MPN | ||||
| PMF | 4-818,27,35,42 | No impact on OS27 | ||
| ET | 318 | |||
| Secondary AML | 435 | |||
| SRSF2 | MDS | 1-1215,33,37,38,45,49,112,113 | RUNX1,33,37,45 ASXL1,33,45 IDH2,33,45 IDH1,37 TET2,38,45 STAG245 | Reduced PFS,112 reduced LFS,45 reduced OS (lower risk IPSS),33 reduced OS (multivariate),113 reduced LFS/OS37 |
| Secondary AML | 5-2035,43 | |||
| CMML | 30-4728,31,33,39 | TET231 | No impact on OS,31,33 reduced LFS/OS39 | |
| MPN | ||||
| PMF | 3-1734,35,42 | IDH1/2 34 | Reduced LFS/OS34 | |
| Secondary AML | 1935 | Reduced OS35 | ||
| De novo AML | 5-635,41 | Reduced OS (univariate)41 | ||
| tAML | 1143 | |||
| U2AF1 | MDS | 5-1114,36-38,45,49,63,112,113,115 | del(20q),14,36,38 ASXL1,37,38,45 DNMT3A37 | Reduced PFS,112 reduced LFS,45 reduced OS (univariate),113 inferior OS, inferior LFS (lower risk)36 |
| Secondary AML | 9-1635,43 | |||
| MDS/MPN | 1463 | |||
| CMML | 5-932,39 | |||
| MPN | 863 | |||
| PMF | 3-1635,42 | Reduced OS (univariate)42 | ||
| Secondary AML | 5.735 | |||
| De novo AML | 2-715,35,41,63,115 | |||
| tAML | 543 | |||
| ZRSR2 | MDS | 1-1115,37,38,45,49 | TET238 | |
| Secondary AML | 2-835,43 | |||
| MPN | ||||
| PMF | 6-1135,42 | |||
| Secondary AML | 1.835 | |||
| De novo AML | 5.635 | |||
| tAML | 143 |
CMML, chronic myelomonocytic leukemia; ET, essential thrombocythemia; LFS, leukemia-free survival; IPSS, International Prognostic Scoring System; MPN, myeloproliferative neoplasm; OS, overall survival; PFS, progression-free survival; PMF, primary myelofibrosis; RA, refractory anemia; RAEB, refractory anemia with excess blasts; RARS, refractory anemia with ringed sideroblasts; RARS-T, refractory anemia with ringed sideroblasts and thrombocytosis; RCMD-RS, refractory cytopenia with multilineage dysplasia and ringed sideroblasts; tAML, therapy-related AML.
SF3B1 is mutated in ∼5% to 15% of CLL cases at initial diagnosis and is associated with shorter time to initial treatment and reduced overall survival.16,17 The frequency of mutations is higher in fludarabine-refractory cases.24 In the United Kingdom Leukaemia Research Fund Chronic Lymphocytic Leukemia 4 trial, SF3B1 mutations were associated with inferior overall survival in both treatment arms, but were not associated with progression-free survival in multivariate analysis.25 In myeloid malignancies, the frequency of SF3B1 mutations is highest in MDS, but recurrent mutations are also present in AML, myeloproliferative neoplasms (MPNs), and MDS/MPN overlap syndromes.18,26,27 Within MDS, the mutations are strongly enriched in cases with ring sideroblasts and are present at lower frequencies in refractory anemia, refractory cytopenia with multilineage dysplasia (RCMD), and refractory anemia with excess blasts.18,26,28 The prognostic significance of SF3B1 mutations in MDS has been controversial, but several studies have shown that they are associated with longer overall survival in multivariate analysis controlling for World Health Organization (WHO) category.26,29 The similarity in disease phenotype and clinical behavior for SF3B1-mutated cases with ≥15% or <15% ring sideroblasts provided the justification for lowering the threshold in the revised WHO classification to 5% ring sideroblasts for the diagnosis of MDS-RS if an SF3B1 mutation is present.30
SRSF2 is mutated in ∼5% to 15% of MDS cases overall, but is highly enriched in chronic myelomonocytic leukemia (CMML).15,31,32 CMML-1 and CMML-2 phenotypes are equally represented. SRSF2 mutations are associated with older age at CMML diagnosis, but have no significant consistent impact on survival, whereas overall survival (OS) is reduced in lower risk MDS cases with SRSF2 mutations.31,33 In primary myelofibrosis, SRSF2 is mutated in ∼17% of cases and is associated with worse OS, independent of other prognostic features.34 Although SRSF2 mutations are rare in de novo AML, they are more frequent in secondary AML derived from MDS or MPNs.35
U2AF1 is mutated in 5% to 10% of MDS cases, across all WHO and International Prognostic Scoring System subtypes.15 U2AF1 mutations are generally associated with adverse prognosis in MDS, including reduced OS and increased risk of progression to secondary AML reported in some studies.14,36
Other splicing factors are mutated at lower frequencies in hematologic malignancies, so there is less certainty about their associations with disease phenotype and clinical behavior. ZRSR2 is mutated in ∼5% of MDS patients.15,37 These mutations are associated with a higher risk of AML transformation and appear to be an independent unfavorable prognostic factor for overall survival when cooccurring with TET2 mutations.38 Recurrent mutations in SF3A1, U2AF2, SF1, and PRPF40B have also been reported in hematologic malignancies, but at insufficient frequency to have confidence in their clinical associations.15
Collectively, the frequency of splicing factor mutations is highest in MDS (>50% of cases), followed by MDS/MPN and CMML (∼50% to 75%), primary myelofibrosis (∼50%), secondary AML derived from MDS or MPN (∼10% to 55%), MPN (∼10% to 30%), and de novo AML (∼10%).15,39-43 The relative enrichment of splicing factor mutations in antecedent hematologic diseases with a propensity to evolve to AML (ie, MDS and MPN) and their paucity in de novo AML provided an opportunity to develop a genetic classifier than can objectively distinguish secondary AML arising from an known antecedent MDS from de novo AML, agnostic of clinical history.43 In this study, the presence of a mutation in 1 of 8 genes (4 of which are splicing factors) could differentiate secondary AML from de novo AML with >95% specificity (ie, secondary-type mutations).43 The authors demonstrate that patients classified as having de novo AML that harbor secondary-type mutations share many clinical and pathological features with secondary AML, suggesting that they may have had an unrecognized period of antecedent MDS before AML diagnosis. Furthermore, myeloid malignancies with 5% to 20% blasts and >20% to 30% blasts (defined as MDS-excess blasts and AML, respectively, in the WHO classification) have a similar frequency of splicing factor mutations and can be further stratified using gene expression and DNA methylation profiling.44 These studies provide additional evidence that splicing factor mutations define myeloid malignancies with a consistent phenotype and clinical behavior that may not be apparent using conventional clinicopathologic information.
Characteristics of splicing factor mutations.
Several principles emerged from the initial descriptions of splicing factor mutations and now have considerable support in the literature. First, the mutations tend to be acquired early in the disease course in MDS (ie, in the “founding” clone) and less often as subclonal mutations acquired at progression or relapse.45,46 In fact, splicing factor mutations have been found in the peripheral blood of aging individuals with clonally skewed hematopoiesis who do not have MDS or AML, implying that these mutations are sufficient to confer a selective growth advantage.47 Conversely, splicing factor mutations in CLL (principally, in SF3B1) are more often subclonal.48 Second, the mutations are mutually exclusive. That is, more than 1 splicing factor mutation is rarely seen in a single tumor (and rare exceptions may represent independent subclones harboring a single splicing factor mutation).15,45,46,49 Third, mutations in the 3 most frequently mutated genes (SF3B1, SRSF2, U2AF1) are heterozygous and are enriched at “hotspot” codons.14,15,18 Fourth, the spectrum of mutant alleles for these genes is consistent across different hematologic malignancy histologies, although there are notable exceptions in solid tumors (for example, distinct SF3B1 alleles in uveal melanoma).50,51 Finally, mutations in SF3B1 and U2AF1 affect 3′ SS usage and SRSF2 mutations affect exon recognition. Why the 5′ complex is largely spared, although it also comprises a large number of ubiquitously expressed proteins, is not understood. The main lessons will be reviewed and updated here because this information has also been the subject of other recent reviews.19,20,22,23
SF3B1 mutations in all hematologic malignancies are clustered in exons 14 through 16, which encode the fourth through sixth HEAT domains, with a hotspot (∼50% of mutations) at codon 700 (K700E).18 In CLL, SF3B1-mutated cases are less likely to co-occur with trisomy 12 and are largely mutually exclusive with mutations in NOTCH1.24,25 In MDS, SF3B1 mutations frequently co-occur with mutations in factors regulating DNA methylation (eg, DNMT3A, TET2).29
SRSF2 mutations invariably affect codon 95, most often as missense mutations (P95H>P95L>P95R>P95A/T) and less commonly as insertion/deletions at this location.15,31 In MDS, SRSF2 mutations co-occur with mutant IDH1/2, TET2, RUNX1, ASXL1, and STAG2.33,37,38,45 In CMML, SRSF2 mutations are largely mutually exclusive with EZH2 mutations, but frequently co-occur with mutations in TET2.31 In myelofibrosis, SRSF2 mutations frequently co-occur with mutant IDH1/2.34
U2AF1 mutations are almost exclusively missense substitutions at 2 codons—34 (S34F>S34Y) and 157 (Q157P/R/G) —and rare biallelic mutations in cis.14,15 U2AF1-mutated cases are enriched for co-occurring ASXL1 mutations and del(20q).38,45,49
Less frequently mutated splicing factors have distinct mutation spectra. ZRSR2 mutations are distributed across the coding region and include missense, nonsense, frameshift, and SS mutations, consistent with a putative loss-of-function role for this X-linked factor.15
Functional consequences of splicing factor mutations.
To investigate the functional consequences of splicing factor mutations, several studies have been performed using isogenic human cell lines and genetically engineered mouse models (summarized in Table 2).
Table 2.
Summary of hematopoietic changes observed in mouse models of mutant splicing factors
| Gene | Mutation | Mouse model | WBC | RBC | PLT | HSPCs | Competitive repopulation | MDS/AML | Pre-mRNA splicing | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Sf3b1 | K700E | Knockin (activated by cre) | = | ↓ Hg ↑ MCV | = | ↑54 ↓55 (LT-HSCs) = (MPPs)54,55 | ↓ | MDS-like (erythroid dysplasia) | Alternative 3′ SS | 54, 55 |
| Srsf2 | P95H | Knockin (activated by cre) | ↓ | ↓ Hg ↑ MCV | = | ↑ (LSK) | ↓ | MDS-like (multilineage dysplasia) | Altered affinity for ESE motif SSNG | 53 |
| U2AF1 | S34F | Transgenic (DOX inducible) | ↓ | = | = | ↑ (LSK) | ↓ | No | Altered 3′ consensus sequence at −3 bp | 52 |
=, no change; cre, cre-recombinase; DOX, doxycycline; Hg, hemoglobin; LKS: Lin<low>, Kit+, Sca1+; LT-HSC, Lin<low>, Kit+, Sca1+, CD48−, CD150+ or Lin<low>, Kit+, Sca1+, CD34−, CD135−; MCV, mean corpuscular volume; PLT, platelet; RBC, red blood cell; WBC, white blood cell.
Despite the predicted oncogenic role of U2AF1 mutations, Yoshida and colleagues first demonstrated that they have growth suppressive properties.15 Expression of the mutant U2AF1 (S34F) allele in HeLa cells led to growth delay and increased apoptosis. Furthermore, expression of U2AF1 mutations in highly purified hematopoietic stem and progenitor cells (HSPCs) reduced repopulation capacity in transplantation assays compared with WT-U2AF1 transduced cells.15 In a first-generation mouse model, a single copy of human U2AF1 complementary DNA (wild-type or S34F) was inserted in the collagen 1a1 locus downstream of a tetracycline-inducible promoter. This system allows for the temporal expression of the S34F mutation while maintaining its ratio similar to the endogenous U2af1 protein levels. Expression of the S34F mutation in the hematopoietic compartment led to a persistent reduction of the total white blood cell count without changes in the red blood cell parameters and platelet counts in the peripheral blood.52 Concomitantly, mutant mice showed a reduction in the numbers of monocytes and B lymphocytes and an accumulation of myeloid progenitors in the bone marrow. Nevertheless, this accumulation of HSPCs did not correlate with a functional advantage because competitive transplantation assays demonstrated a repopulation disadvantage of mutant cells in primary and secondary recipients. Furthermore, expression of the S34F mutation for up to a year was not sufficient to induce transformation to AML.52
A floxed SRSF2-P95H allele inserted in the endogenous Srsf2 locus in mice provided a conditional model to study the functional impact of this mutation on hematopoiesis.53 Srsf2-P95H mice developed a myelodysplastic phenotype characterized by leukopenia, macrocytic anemia, myeloid and erythroid dysplasia, and an accumulation of stem and progenitor cells in the bone marrow. Similar to the mutant U2AF1 mouse, Srsf2-P95H hematopoietic stem cells (HSCs) show limited repopulation ability in transplantation assays compared with their wild-type counterpart.53
A conditional murine knock-in of the Sf3b1-K700E mutation showed similar features to those observed in humans. Heterozygous expression of the mutation led to macrocytic anemia associated with elevated levels of erythropoietin, a block in erythroid differentiation, and no differences in the relative abundance of multipotent progenitor cells (although HSC quantification differs between both studied, most likely because of the use of alternative immunophenotypic definition for HSCs).54,55 However, these animals did not exhibit ring sideroblasts, the defining characteristic of SF3B1 mutant cells in human MDS. In competitive transplantation settings, mutant cells showed an engraftment disadvantage compared with wild-type stem cells.54,55 Limiting Sf3b1-K700E expression to the B-cell compartment led to global splicing changes, defects in B lymphopoiesis, and cell senescence but did not result in transformation to overt CLL (at least with the reported follow-up times).56
Molecular consequences of splicing factor mutations.
Most splicing factors mutations found in hematologic malignancies affect proteins implicated in the early exon recognition steps of pre-mRNA processing. These mutations universally lead to splicing defects affecting a specific subset of genes rather than globally affecting multiexonic genes.52-55,57,58
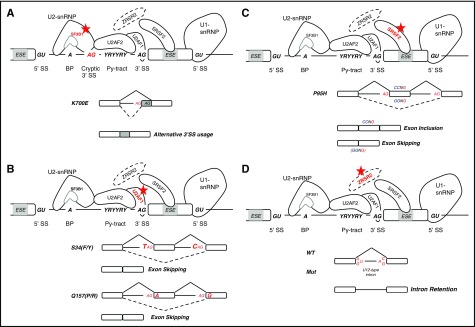
SF3B1 mutations cause alternative 3′ SS usage because of the increased recognition of a cryptic SS between the branch point and the canonical 3′ SS (Figure 2A).59 How this 3′ alternative SS is selected is being actively studied. Recent findings have suggested that the cryptic 3′ SS is associated with a shorter pyrimidine tract and enrichment in adenines upstream of the canonical 3′ SS, leading to the use of different branch sites.60 In addition, mutant SF3B1 may potentially induce a conformational change that would facilitate the use of the cryptic 3′ SS by modifying the ability to select SSs immediately downstream of the branch site, normally protected by steric hindrance upon binding of the U2 snRNP to the branch site.59,61 Analysis of a crystal structure for the SF3b core complex suggests that the hotspot mutations may induce conformational changes that alter interactions with pre-mRNA and other splicing factors.62
Figure 2.
Splicing factor (trans-acting) mutations and their impact on splicing in hematologic malignancies. (A) SF3B1 mutations lead to alternative 3′ splicing site (SS) usage because of the increased recognition of cryptic splice sites between the branch point and the canonical 3′ splice site. (B) U2AF1 mutations affect 3′ splice site recognition, leading to an increase in exon skipping because of use of an alternative splice sites. S34F-mutant-U2AF1 losses affinity for the 3′ SS motif when a T(U) is present in position −3. Conversely, mutations at the Q157 residue promote the recognition of 3′ SS when a G is present in position +1 and repress those bearing an A. (C) SRSF2 mutations affect its normal RNA-binding activity to the consensus ESE motif (SSNG). Mutant SRSF2 recognizes with higher affinity the CCNG motif vs the GGNG, promoting or repressing the inclusion of exons containing C- or G-rich motifs. (D) ZRSR2 loss-of-function mutations specifically affect splicing of U12-type introns leading to intron retention. BP, branch point; Py-tract, polypyrimidine-tract; snRNP, small nuclear ribonucleoproteins; Y, pyrimidine.
U2AF1 gene mutations also alter 3′ SS recognition of a subset of AG-dependent 3′ SSs leading predominantly to exon skipping and alternative 3′ acceptor site use (Figure 2B).52,57,58,63,64 These alterations are allele-specific, because S34 and Q157 mutations show alternative sequence specificity at position −3 and +1 in the 3′ SS, respectively.58 U2AF1 recognizes a pyrimidine-AG-purine motif. As a consequence of the S34F mutation, U2AF1 losses affinity for the motifs when a T (U) is present in position −3, leading to an increase in exon skipping and decreased recognition of the canonical junction resulting in the use of an alternative SS.57,58,63 Conversely, mutations at the Q157 residue promote the recognition of 3′ SS when a G is present in position +1 and repress those bearing an A at this position.58 Changes in RNA-binding affinity for mutant U2AF1 can, in general, account for observed changes in splicing preference.57,65 Furthermore, these sequence specific effects are distinct from those observed in U2AF1-deficient cells, reinforcing the concept that they do not behave as loss of function but rather have neomorphic activity.58 In addition, a recent study demonstrated that ectopic expression of S34F U2AF1 in Ba/F3 cells resulted in increased use of distal mRNA cleavage and polyadenylation sites.66 One of the affected transcripts was Atg7, which led to reduced protein levels and a functional autophagy defect.
Deep sequencing analysis of U2AF1 mutant patient samples, transgenic mice, and isogenic cell lines have demonstrated the presence of global changes that affect 5% to 10% of the transcriptome.52,57 Some of these recurrent alternatively spliced genes are implicated in essential cellular processes, such us RNA processing and splicing, protein translation, DNA methylation, and DNA damage response and apoptosis, and can also occur in genes that are recurrently mutated in MDS.52,57,58,63,64,67 However, the functional contribution to the disease phenotype of alternatively spliced genes remains an active area of research.
SRSF2 binds to ESEs to promote exon recognition in a sequence-specific manner. SRSF2 binds a consensus ESE motif (SSNG, where S represents a C or G) through its RNA recognition motif domain.68-70 The P95 mutation affects SRSF2 normal sequence-specific RNA binding activity.70 Mutant SRSF2 recognizes with higher affinity the CCNG motif vs GGNG, whereas the wild-type protein recognizes both with similar affinity (Figure 2C).70 As a result, mutant SRSF2 promotes or represses the inclusion of exons containing C- or G-rich motifs causing splicing changes in hundreds of genes.53,71 Some of these alternative splicing events affect genes commonly mutated in MDS, such as EZH2 and BCOR, suggesting that SRSF2 may contribute to MDS pathogenesis by, at least in part, controlling the expression of these genes.53 Similar to U2AF1 mutations, P95 mutant SRSF2 induces distinct splicing changes compared with those induced by the depletion of SRSF2, again suggesting that SRSF2 mutations are not loss of function, but rather have neomorphic activity.53,72
Less is known about the molecular consequences of ZRSR2 mutations. Because of its critical role in the minor spliceosome, ZRSR2 mutations are characterized by the accumulation of alternatively spliced U12-type introns (Figure 2D).73 These defects are mainly retention of U12-type introns. Furthermore, splicing defects resulting from ZSRS2 mutations are enriched for pathways including MAPK, ErbB signaling, and focal adhesion kinases, among others.73
Although the splicing junctions that are affected by these mutations are typically different, it is not known whether a shared set of downstream genes are affected. The discovery of common target genes has been limited by differences in the cell types that were sequenced and analytic approaches used in current studies; therefore, this remains an active area of research.
Therapeutic opportunities
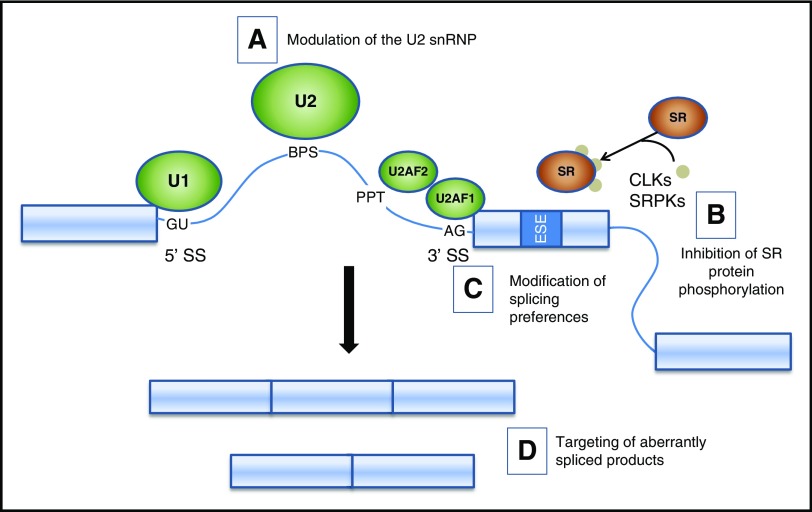
The strong selective pressure favoring acquisition of mutations in splicing factors suggests not only that these alterations are important for the biology of hematologic malignancies, but also that they may create vulnerabilities that could be exploited therapeutically. Preclinical work in this area has focused on 3 potential entry points (Figure 3): (1) perturbation of splicing, (2) modifying signaling pathways upstream of splicing, and (3) correcting downstream aberrant splicing. Recent developments and initial steps toward clinical translation will be reviewed here. Readers are referred to other reviews and primary sources for additional details on the medicinal chemistry, pharmacology, and biochemical properties of these compounds.74-76
Figure 3.
Potential therapeutic strategies targeting splicing factor mutations and splicing changes. A schematic of the major spliceosome components illustrates potential strategies to modulate splicing. (A) Small molecules (eg, PB and FR derivatives and analogs) alter the function of the U2 snRNP, leading to the accumulation of unspliced products. (B) Inhibition of SR protein phosphorylation alters intracellular trafficking and binding of SR proteins to exonic splice enhancers, leading to altered utilization of cassette exons. (C) Oligonucleotides and transplicing modulators can change splicing preference at specific targets. (D) Depletion or replacement of spliced products. BPS, branch point sequence; CLK, CDC-like kinase; PPT, polypyrimidine tract; SRPK, SR protein kinase.
Splicing modulators
A variety of natural plant and bacterial products have been isolated that have antitumor and splicing modulator properties (Figure 3A). Several of these molecules have been modified to improve potency and druglike properties, either by generating derivatives of the parent compounds or by de novo synthesis of structural analogs. FR901464 (FR) was isolated from Pseudomonas sp. Its methyl ketal derivative, spliceostatin A (SSA), physically interacts with components of the SF3b subcomplex and induces accumulation of unspliced products.77 Further study indicated that SSA alters splicing fidelity, rather than acting as a general splicing inhibitor, resulting in accumulation of alternatively spliced products containing premature termination codons, leading to their depletion by nonsense-mediated decay.78 In CLL cells, SSA potently depletes MCL-1 via alternative splicing, leading to induction of apoptosis at low nanomolar concentrations in vitro, with relative sparing of normal T and B cells.79
Pladienolide B (PB), FD-895, and herboxidiene are natural products of Streptomyces sp. that share a common pharmacophore with FR. The elaboration of structurally related splicing modulators by genetically diverse prokaryote species is a fascinating and unexplained example of convergent evolution. An elegant genetic study demonstrated unequivocally that SF3B1 is a direct target of PB.80 E7107 is a urethane derivative of PB that interferes with spliceosome assembly.81 In preclinical studies, E7107 reduced the leukemic burden in the MLL/AF9 retroviral transduction/transplantation model on a Srsf2P95H/+ background and impaired the growth of primary human AML samples with splicing factor mutations in a xenograft model.82 Similarly, in a competitive repopulation model in mice, Sf3b1K700E/+ cells were selectively depleted in vivo by E7107.54
Parallel phase 1 studies of E7107 with slightly different administration schedules were initiated in Europe and the United States for patients with advanced nonhematologic malignancies.83,84 Although the adverse effects were generally mild to moderate and reversible in 66 enrolled patients, both studies were terminated when 2 patients on the US study (NCT00499499) experienced impaired visual acuity.84 One patient on the European study (NCT00459823) developed visual disturbance at the time of treatment discontinuation, felt to be consistent with optic neuritis that partially improved with corticosteroid treatment.83 A phase 1 study of H3B-8800, an orally bioavailable splicing modulator, recently opened to accrual for patients with myeloid malignancies (NCT02841540). Whether the ocular toxicity in the E7107 studies is unique and attributable to this compound, the pharmacokinetics at these schedules, a class effect of PB derivatives, sensitivity of optic neuronal cells to splicing modulators in general, or other factors is unknown, but certainly merits close attention during clinical investigation with this class of compounds.
The sudemycins are synthetic analogs of PB and FR that share a common pharmacophore, but have fewer stereocenters and have improved solubility, chemical stability, and plasma half-life.85-87 Primary human CLL cells undergo apoptosis in a dose-dependent fashion after exposure to sudemycin D1 (SD1) in vitro.88 SF3B1 mutant samples were more sensitive in vitro than CLL cells lacking splicing factor mutations.88 SD1 and the BTK inhibitor, ibrutinib, act synergistically to induce apoptosis in CLL cells, possibly from inactivation of the endogenous BTK inhibitor, IBTK, by SD1 via alternative splicing.88 Primary human MDS/AML cells with mutant U2AF1 had impaired proliferation after treatment with SD1 in vitro compared with cells lacking splicing factor mutations and in vivo treatment of mice with SD6 reverted aberrant hematopoietic progenitor expansion in an inducible mouse model of mutant U2AF1.89
FD-895 caused induction of apoptosis in CLL cells in vitro (independent of SF3B1 genotype) at nanomolar concentrations, similar to PB.90 A semisynthetic analog (17S-FD-895) with improved pharmacokinetic properties91 administered to immunodeficient mice engrafted with primary human AML samples reduced their repopulating activity in secondary transplants and reverted the splicing signature in leukemia stems cells from “secondary AML-associated” to “healthy aging.”92
The genetic data from patients provided a priori support for the hypothesis that splicing factor mutant hematologic malignancies might be uniquely sensitive to splicing modulators. As discussed previously, the wild-type allele is retained and expressed in cells with mutations in the 3 most frequently mutated splicing factors, suggesting that some level of normal splicing is required for viability. Furthermore, the mutually exclusive nature of these mutations suggests that a cell with a splicing factor mutation cannot tolerate further perturbation of splicing induced by a second mutation in this pathway. Further support comes from the Srsf2P95H/+ mouse model, in which deletion of the wild-type allele caused rapid loss of hemizygous mutant cells.82 These preclinical studies suggest a synthetic interaction of splicing factor mutations and splicing modulator drugs that is not tolerated by mutant cells (ie, 2 perturbations in the spliceosome are not tolerated by cells). The results provide support for the initial hypothesis and lead to the following predictions that will be tested in planned and ongoing clinical trials: (1) patients with splicing factor mutant malignancies may derive the largest clinical benefit from splicing modulators and (2) a general splicing modulator may have activity across of broad range of splicing factor mutant genotypes. In fact, sensitivity to splicing modulators may extend to malignancies that lack splicing factor mutations. Myc-driven breast cancers show sensitivity to pharmacologic splicing modulation that has been attributed to the increased burden on the spliceosome to process Myc-induced pre-mRNA production.93 In addition, there is emerging evidence that some B-cell lymphomas (mantle cell and Myc-driven lymphomas) are sensitive to pharmacologic modulation of splicing.94,95 Future studies will be needed to explore the potential use of splicing modulator therapy in these diseases.
Targeting signaling inputs to splicing
A family of SR factors influences SS selection by the spliceosome (Figure 3B). The intracellular trafficking and protein-protein interactions of SR factors are regulated by the phosphorylation status of their arginine/serine (RS) domains. Hence, the upstream serine kinases (eg, SRPK1, SRPK2, CLK1) are potential targets for small molecules that could modulate splicing. Several compounds with these properties have been identified, including chlorhexidine, TG003, diospyrin, and indolocarbazole.96-99
Reverting aberrant splicing
Considerable progress has been made toward pharmaceutical approaches for diseases in which a single aberrant splice isoform is pathogenic. These include antisense oligonucleotides that target SSs or regulatory sequences and trans-splicing molecules that mediate exchange of a target sequence of an endogenous sequence containing a deleterious variant (Figure 3C) (reviewed in Daguenet et al100). These approaches may have value for splicing factor mutant hematologic malignancies in which 1 or a few aberrant splice isoforms prove to be pathogenic. Correction of a larger number of alternative splicing events will be challenging and may require the development of novel strategies and technologies (Figure 3D).
The tight coordination of splicing with other cellular processes (eg, transcription initiation and elongation, histone modification and DNA methylation, mRNA export and translation, DNA damage response)101-109 suggests that splicing factor mutations may create vulnerabilities in other pathways. Unbiased small molecule and genetic screens may uncover additional potentially druggable targets, some of which could be candidates for drug development.
Conclusion
Rapid progress has been made in understanding spliceosome gene mutations since their first description and their clinical associations in 2011. This was followed by studies focusing on the impact of mutations on splicing and then the development of genetically engineered mouse models to characterize their contribution to cellular and molecular phenotypes. Preclinical work on splicing modulators in cellular and mouse models, leading to a first in human study of an oral agent for patients with myeloid malignancies in 2016, just 5 years after discovery of the mutations, is evidence that the field is moving rapidly. This accelerated pace builds on preceding decades of work in the basic biology of splicing and the isolation and modification of naturally occurring splice modulator products. Finally, a highly collaborative research environment has been instrumental in moving this field forward with the goal of translating this research for the benefit of patients. Current and future research directions should clarify the mechanistic consequences of splicing factor mutations, define the role of cooperating mutations in mediating transformation to overt malignancy, and uncover vulnerabilities that can be exploited for novel therapeutic strategies.
Acknowledgments
This work was support by a National Institutes of Health/National Cancer Institute SPORE in Leukemia (P50CA171963) (T.A.G. and M.J.W.), the Edward P. Evans Foundation (M.J.W. and T.A.G.), the Lottie Caroline Hardy Trust (T.A.G. and M.J.W.), and a Leukemia and Lymphoma Society Scholar Award (M.J.W.).
Authorship
Contribution: All authors wrote, edited, reviewed, and approved the submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timothy A. Graubert, Massachusetts General Hospital Cancer Center, 149 13th St, Charlestown, MA 02129; e-mail: tgraubert@partners.org; and Matthew J. Walter, Washington University School of Medicine, 660 S. Euclid Ave, Campus Box 8007, St. Louis, MO 63110; e-mail: mjwalter@wustl.edu.
References
- 1.Gilbert W. Why genes in pieces? Nature. 1978;271(5645):501. [DOI] [PubMed] [Google Scholar]
- 2.Sakharkar MK, Chow VT, Kangueane P. Distributions of exons and introns in the human genome. In Silico Biol. 2004;4(4):387-393. [PubMed] [Google Scholar]
- 3.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8(10):749-761. [DOI] [PubMed] [Google Scholar]
- 4.Gerstein MB, Rozowsky J, Yan KK, et al. . Comparative analysis of the transcriptome across distant species. Nature. 2014;512(7515):445-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413-1415. [DOI] [PubMed] [Google Scholar]
- 6.Wang ET, Sandberg R, Luo S, et al. . Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn CN, Venugopal P, Scott HS, Hiwase DK. Splice factor mutations and alternative splicing as drivers of hematopoietic malignancy. Immunol Rev. 2015;263(1):257-278. [DOI] [PubMed] [Google Scholar]
- 8.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12(1):5-14. [DOI] [PubMed] [Google Scholar]
- 9.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dvinge H, Bradley RK. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung H, Lee D, Lee J, et al. . Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet. 2015;47(11):1242-1248. [DOI] [PubMed] [Google Scholar]
- 12.Kandoth C, McLellan MD, Vandin F, et al. . Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156(6):1324-1335. [DOI] [PubMed] [Google Scholar]
- 14.Graubert TA, Shen D, Ding L, et al. . Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44(1):53-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida K, Sanada M, Shiraishi Y, et al. . Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64-69. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Lawrence MS, Wan Y, et al. . SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365(26):2497-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quesada V, Conde L, Villamor N, et al. . Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44(1):47-52. [DOI] [PubMed] [Google Scholar]
- 18.Papaemmanuil E, Cazzola M, Boultwood J, et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anczuków O, Krainer AR. Splicing-factor alterations in cancers. RNA. 2016;22(9):1285-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida K, Ogawa S. Splicing factor mutations and cancer. Wiley Interdiscip Rev RNA. 2014;5(4):445-459. [DOI] [PubMed] [Google Scholar]
- 21.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17(1):19-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16(7):413-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue D, Bradley RK, Abdel-Wahab O. Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev. 2016;30(9):989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossi D, Bruscaggin A, Spina V, et al. . Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011;118(26):6904-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oscier DG, Rose-Zerilli MJ, Winkelmann N, et al. . The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood. 2013;121(3):468-475. [DOI] [PubMed] [Google Scholar]
- 26.Malcovati L, Papaemmanuil E, Bowen DT, et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium and of the Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118(24):6239-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lasho TL, Finke CM, Hanson CA, et al. . SF3B1 mutations in primary myelofibrosis: clinical, histopathology and genetic correlates among 155 patients. Leukemia. 2012;26(5):1135-1137. [DOI] [PubMed] [Google Scholar]
- 28.Patnaik MM, Lasho TL, Hodnefield JM, et al. . SF3B1 mutations are prevalent in myelodysplastic syndromes with ring sideroblasts but do not hold independent prognostic value. Blood. 2012;119(2):569-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malcovati L, Karimi M, Papaemmanuil E, et al. . SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126(2):233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arber DA, Orazi A, Hasserjian R, et al. . The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391-2405. [DOI] [PubMed] [Google Scholar]
- 31.Meggendorfer M, Roller A, Haferlach T, et al. . SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML). Blood. 2012;120(15):3080-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patnaik MM, Lasho TL, Finke CM, et al. . Spliceosome mutations involving SRSF2, SF3B1, and U2AF35 in chronic myelomonocytic leukemia: prevalence, clinical correlates, and prognostic relevance. Am J Hematol. 2013;88(3):201-206. [DOI] [PubMed] [Google Scholar]
- 33.Wu SJ, Kuo YY, Hou HA, et al. . The clinical implication of SRSF2 mutation in patients with myelodysplastic syndrome and its stability during disease evolution. Blood. 2012;120(15):3106-3111. [DOI] [PubMed] [Google Scholar]
- 34.Lasho TL, Jimma T, Finke CM, et al. . SRSF2 mutations in primary myelofibrosis: significant clustering with IDH mutations and independent association with inferior overall and leukemia-free survival. Blood. 2012;120(20):4168-4171. [DOI] [PubMed] [Google Scholar]
- 35.Zhang SJ, Rampal R, Manshouri T, et al. . Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119(19):4480-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu SJ, Tang JL, Lin CT, et al. . Clinical implications of U2AF1 mutation in patients with myelodysplastic syndrome and its stability during disease progression. Am J Hematol. 2013;88(11):E277-E282. [DOI] [PubMed] [Google Scholar]
- 37.Thol F, Kade S, Schlarmann C, et al. . Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119(15):3578-3584. [DOI] [PubMed] [Google Scholar]
- 38.Damm F, Kosmider O, Gelsi-Boyer V, et al. ; Groupe Francophone des Myélodysplasies. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood. 2012;119(14):3211-3218. [DOI] [PubMed] [Google Scholar]
- 39.Itzykson R, Kosmider O, Renneville A, et al. . Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31(19):2428-2436. [DOI] [PubMed] [Google Scholar]
- 40.Malcovati L, Papaemmanuil E, Ambaglio I, et al. . Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood. 2014;124(9):1513-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papaemmanuil E, Gerstung M, Bullinger L, et al. . Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barraco D, Elala YC, Lasho TL, et al. . Molecular correlates of anemia in primary myelofibrosis: a significant and independent association with U2AF1 mutations. Blood Cancer J. 2016;6:e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindsley RC, Mar BG, Mazzola E, et al. . Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125(9):1367-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taskesen E, Havermans M, van Lom K, et al. . Two splice-factor mutant leukemia subgroups uncovered at the boundaries of MDS and AML using combined gene expression and DNA-methylation profiling. Blood. 2014;123(21):3327-3335. [DOI] [PubMed] [Google Scholar]
- 45.Papaemmanuil E, Gerstung M, Malcovati L, et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haferlach T, Nagata Y, Grossmann V, et al. . Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steensma DP, Bejar R, Jaiswal S, et al. . Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landau DA, Carter SL, Stojanov P, et al. . Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walter MJ, Shen D, Shao J, et al. . Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia. 2013;27(6):1275-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45(2):133-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin M, Maßhöfer L, Temming P, et al. . Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45(8):933-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shirai CL, Ley JN, White BS, et al. . Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell. 2015;27(5):631-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim E, Ilagan JO, Liang Y, et al. . SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27(5):617-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Obeng EA, Chappell RJ, Seiler M, et al. . Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016;30(3):404-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mupo A, Seiler M, Sathiaseelan V, et al. . Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts [published online ahead of print 21 October 2016]. Leukemia. doi:10.1038/leu.2016.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L, Gambe R, Fan J, et al. . Expression of Sf3b1-K700E in murine B cells causes pre-mRNA splicing and altered B cell differentiation and function. Blood. 2015;126(23):366. [Google Scholar]
- 57.Okeyo-Owuor T, White BS, Chatrikhi R, et al. . U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia. 2015;29(4):909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ilagan JO, Ramakrishnan A, Hayes B, et al. . U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015;25(1):14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeBoever C, Ghia EM, Shepard PJ, et al. . Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLOS Comput Biol. 2015;11(3):e1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darman RB, Seiler M, Agrawal AA, et al. . Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Reports. 2015;13(5):1033-1045. [DOI] [PubMed] [Google Scholar]
- 61.Alsafadi S, Houy A, Battistella A, et al. . Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun. 2016;7:10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cretu C, Schmitzová J, Ponce-Salvatierra A, et al. . Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol Cell. 2016;64(2):307-319. [DOI] [PubMed] [Google Scholar]
- 63.Przychodzen B, Jerez A, Guinta K, et al. . Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood. 2013;122(6):999-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brooks AN, Choi PS, de Waal L, et al. . A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events [published correction appears online at http://dx.doi.org/10.1371/journal.pone.0096437]. PLoS One. 2014;9(1):e87361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fei DL, Motowski H, Chatrikhi R, et al. . Wild-type U2AF1 antagonizes the splicing program characteristic of U2AF1-mutant tumors and is required for cell survival. PLoS Genet. 2016;12(10):e1006384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park SM, Ou J, Chamberlain L, et al. . U2AF35(S34F) promotes transformation by directing aberrant ATG7 Pre-mRNA 3′ end formation. Mol Cell. 2016;62(4):479-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Damm F, Chesnais V, Nagata Y, et al. . BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood. 2013;122(18):3169-3177. [DOI] [PubMed] [Google Scholar]
- 68.Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell. 1998;1(5):765-771. [DOI] [PubMed] [Google Scholar]
- 69.Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol Cell Biol. 2000;20(3):1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daubner GM, Cléry A, Jayne S, Stevenin J, Allain FH. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2012;31(1):162-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J, Lieu YK, Ali AM, et al. . Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci USA. 2015;112(34):E4726-E4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Komeno Y, Huang YJ, Qiu J, et al. . SRSF2 is essential for hematopoiesis, and its myelodysplastic syndrome-related mutations dysregulate alternative pre-mRNA splicing. Mol Cell Biol. 2015;35(17):3071-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Madan V, Kanojia D, Li J, et al. . Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun. 2015;6:6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Webb TR, Joyner AS, Potter PM. The development and application of small molecule modulators of SF3b as therapeutic agents for cancer. Drug Discov Today. 2013;18(1-2):43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salton M, Misteli T. Small molecule modulators of pre-mRNA splicing in cancer therapy. Trends Mol Med. 2016;22(1):28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22(9):976-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaida D, Motoyoshi H, Tashiro E, et al. . Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3(9):576-583. [DOI] [PubMed] [Google Scholar]
- 78.Corrionero A, Miñana B, Valcárcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011;25(5):445-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Larrayoz M, Blakemore SJ, Dobson RC, et al. . The SF3B1 inhibitor spliceostatin A (SSA) elicits apoptosis in chronic lymphocytic leukaemia cells through downregulation of Mcl-1. Leukemia. 2016;30(2):351-360. [DOI] [PubMed] [Google Scholar]
- 80.Yokoi A, Kotake Y, Takahashi K, et al. . Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 2011;278(24):4870-4880. [DOI] [PubMed] [Google Scholar]
- 81.Folco EG, Coil KE, Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 2011;25(5):440-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee SC, Dvinge H, Kim E, et al. . Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med. 2016;22(6):672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eskens FA, Ramos FJ, Burger H, et al. . Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin Cancer Res. 2013;19(22):6296-6304. [DOI] [PubMed]
- 84.Hong DS, Kurzrock R, Naing A, et al. . A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest New Drugs. 2014;32(3):436-444. [DOI] [PubMed] [Google Scholar]
- 85.Lagisetti C, Pourpak A, Jiang Q, et al. . Antitumor compounds based on a natural product consensus pharmacophore. J Med Chem. 2008;51(19):6220-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lagisetti C, Pourpak A, Goronga T, et al. . Synthetic mRNA splicing modulator compounds with in vivo antitumor activity. J Med Chem. 2009;52(22):6979-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lagisetti C, Palacios G, Goronga T, Freeman B, Caufield W, Webb TR. Optimization of antitumor modulators of pre-mRNA splicing. J Med Chem. 2013;56(24):10033-10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xargay-Torrent S, López-Guerra M, Rosich L, et al. . The splicing modulator sudemycin induces a specific antitumor response and cooperates with ibrutinib in chronic lymphocytic leukemia. Oncotarget. 2015;6(26):22734-22749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shirai CL, White BS, Tripathi M, et al. . Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat Commun. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kashyap MK, Kumar D, Villa R, et al. . Targeting the spliceosome in chronic lymphocytic leukemia with the macrolides FD-895 and pladienolide-B. Haematologica. 2015;100(7):945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Villa R, Kashyap MK, Kumar D, et al. . Stabilized cyclopropane analogs of the splicing inhibitor FD-895. J Med Chem. 2013;56(17):6576-6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crews LA, Balaian L, Delos Santos NP, et al. . RNA splicing modulation selectively impairs leukemia stem cell maintenance in secondary human AML. Cell Stem Cell. 2016;19(5):599-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hsu TY, Simon LM, Neill NJ, et al. . The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525(7569):384-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koh CM, Bezzi M, Low DH, et al. . MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523(7558):96-100. [DOI] [PubMed] [Google Scholar]
- 95.Chan-Penebre E, Kuplast KG, Majer CR, et al. . A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11(6):432-437. [DOI] [PubMed] [Google Scholar]
- 96.Younis I, Berg M, Kaida D, Dittmar K, Wang C, Dreyfuss G. Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol Cell Biol. 2010;30(7):1718-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tazi J, Bakkour N, Soret J, et al. . Selective inhibition of topoisomerase I and various steps of spliceosome assembly by diospyrin derivatives. Mol Pharmacol. 2005;67(4):1186-1194. [DOI] [PubMed] [Google Scholar]
- 98.Pilch B, Allemand E, Facompré M, et al. . Specific inhibition of serine- and arginine-rich splicing factors phosphorylation, spliceosome assembly, and splicing by the antitumor drug NB-506. Cancer Res. 2001;61(18):6876-6884. [PubMed] [Google Scholar]
- 99.Muraki M, Ohkawara B, Hosoya T, et al. . Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem. 2004;279(23):24246-24254. [DOI] [PubMed] [Google Scholar]
- 100.Daguenet E, Dujardin G, Valcárcel J. The pathogenicity of splicing defects: mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO Rep. 2015;16(12):1640-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122(3):365-378. [DOI] [PubMed] [Google Scholar]
- 102.Xiao R, Sun Y, Ding JH, et al. . Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol. 2007;27(15):5393-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Savage KI, Gorski JJ, Barros EM, et al. . Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol Cell. 2014;54(3):445-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tresini M, Warmerdam DO, Kolovos P, et al. . The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 2015;523(7558):53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kfir N, Lev-Maor G, Glaich O, et al. . SF3B1 association with chromatin determines splicing outcomes. Cell Reports. 2015;11(4):618-629. [DOI] [PubMed] [Google Scholar]
- 106.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41(3):376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ji X, Zhou Y, Pandit S, et al. . SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153(4):855-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang Y, Gattoni R, Stévenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11(3):837-843. [DOI] [PubMed] [Google Scholar]
- 109.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18(2):210-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nadeu F, Delgado J, Royo C, et al. . Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127(17):2122-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Damm F, Thol F, Kosmider O, et al. . SF3B1 mutations in myelodysplastic syndromes: clinical associations and prognostic implications. Leukemia. 2012;26(5):1137-1140. [DOI] [PubMed] [Google Scholar]
- 112.Kang MG, Kim HR, Seo BY, et al. . The prognostic impact of mutations in spliceosomal genes for myelodysplastic syndrome patients without ring sideroblasts. BMC Cancer. 2015;15:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu L, Song L, Xu L, et al. . Genetic landscape of recurrent ASXL1, U2AF1, SF3B1, SRSF2, and EZH2 mutations in 304 Chinese patients with myelodysplastic syndromes. Tumour Biol. 2016;37(4):4633-4640. [DOI] [PubMed]
- 114.Visconte V, Rogers HJ, Singh J, et al. . SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood. 2012;120(16):3173-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Qian J, Yao DM, Lin J, et al. . U2AF1 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2012;7(9):e45760. [DOI] [PMC free article] [PubMed] [Google Scholar]