Key Points
TP53RK confers poor prognosis in MM patients.
TP53RK knockdown or inhibition by IMiDs triggers MM cell apoptosis, validating TP53RK as a novel therapeutic target in MM.
Abstract
p53-related protein kinase (TP53RK, also known as PRPK) is an upstream kinase that phosphorylates (serine residue Ser15) and mediates p53 activity. Here we show that TP53RK confers poor prognosis in multiple myeloma (MM) patients, and, conversely, that TP53RK knockdown inhibits p53 phosphorylation and triggers MM cell apoptosis, associated with downregulation of c-Myc and E2F-1–mediated upregulation of pro-apoptotic Bim. We further demonstrate that TP53RK downregulation also triggers growth inhibition in p53-deficient and p53-mutant MM cell lines and identify novel downstream targets of TP53RK including ribonucleotide reductase-1, telomerase reverse transcriptase, and cyclin-dependent kinase inhibitor 2C. Our previous studies showed that immunomodulatory drugs (IMiDs) downregulate p21 and trigger apoptosis in wild-type-p53 MM.1S cells, Importantly, we demonstrate by pull-down, nuclear magnetic resonance spectroscopy, differential scanning fluorimetry, and isothermal titration calorimetry that IMiDs bind and inhibit TP53RK, with biologic sequelae similar to TP53RK knockdown. Our studies therefore demonstrate that either genetic or pharmacological inhibition of TP53RK triggers MM cell apoptosis via both p53-Myc axis-dependent and axis-independent pathways, validating TP53RK as a novel therapeutic target in patients with poor-prognosis MM.
Introduction
There have been many recent therapeutic advances in multiple myeloma (MM) resulting from the development of proteasome inhibitors, immunomodulatory drugs (IMiDs), monoclonal antibodies (Abs), and histone deacetylase inhibitors.1 However, this disease is genetically heterogeneous, with ongoing DNA damage,2-4 and novel targeted therapies are urgently needed. In our clinically annotated gene expression profiling (GEP) data set, we identified increasing p53-related protein kinase (TP53RK, also known as PRPK) expression to be associated with progression from normal plasma cells to smoldering MM (SMM) to active MM, and, importantly, found its expression to be inversely correlated with MM patient survival. In addition, short hairpin RNA (shRNA) screening of cell lines in the Achilles Project at Broad Institute, Massachusetts Institute of Technology (http://www.broadinstitute.org/achilles), also showed that downregulation of TP53RK induces MM cell line cytotoxicity. Previous studies have also shown that TP53RK regulates p53 activity via phosphorylation of serine residue (Ser15),5 and that TP53RK inhibition sensitizes cancer cells to taxanes.6 However, the biologic sequelae of TP53RK in MM have not yet been elucidated.
Our early studies showed that IMiDs trigger direct MM cytotoxicity via caspase 8– mediated apoptosis; downregulate adhesion molecules, thereby abrogating MM cell binding in the bone marrow (BM) milieu; inhibit angiogenesis; and enhance immune effector antitumor response, while inhibiting T-regulatory cells.7-10 More recently, multiple groups have shown that IMiDs thalidomide (Thal),11 lenalidomide (Len), and pomalidomide (Pom)12 directly bind to cereblon (CRBN), forming an E3 ubiquitin ligase complex with damaged DNA binding protein 1, cullin-4A, and regulator of cullins1.11 The crystal structure of CRBN bound to DNA binding protein 1 and Len has been resolved, and the drug-binding residues critical for its antiproliferative effects have been defined.13 Upon Len treatment, CRBN triggers proteasomal degradation of Ikaros (IKZF1) and Aiolos (IKZF3), followed by inhibition of interferon regulatory factor 4 and MM cell growth.14,15 Conversely, downregulation of CRBN induces resistance to both Len and Pom in MM cell lines.12,16 Importantly, mutations in CRBN have recently been associated with clinical resistance to Len.17
Because P53RK expression increases with progression from SMM to active MM and because increased P53RK expression in a subset of MM patients portends poor prognosis, in the present study, we validated TP53RK as a novel therapeutic target in MM. Genetic downregulation of TP53RK inhibited phosphorylation (Ser15) and activity of p53, thereby triggering MM cell growth inhibition and apoptosis associated with both downregulation of c-Myc and E2F-1–mediated upregulation of pro-apoptotic Bim. We also identified novel TP53RK downstream target genes including ribonucleotide reductase M1 (RRM1) and cyclin-dependent kinase inhibitor 2C (CDKN2C, p18). Because our prior studies showed that IMiDs trigger downregulation of p21CIP1, growth inhibition, and apoptosis in MM.1S MM cells with wild-type (wt)-p53,7 we hypothesized that IMiDs may target TP53RK. Pull-down, nuclear magnetic resonance (NMR) spectroscopy, differential scanning fluorimetry (DSF), and isothermal titration calorimetry (ITC) indeed confirmed that IMiDs bind and inhibit TP53RK and p53, with biologic and molecular sequelae similar to TP53RK knockdown. These studies show that increased TP53RK expression confers poor prognosis in MM, and, importantly, that genetic or pharmacologic downregulation of TP53RK triggers MM cell apoptosis, validating TP53RK as a novel therapeutic target in an MM patient subset with poor prognosis.
Materials and methods
Cells
MM.1S, NCI-H929 (H929), RPMI8226, and p53-mutant (U266) MM cells were obtained from American Type Culture Collection (Manassas, VA). Doxorubicin-resistant RPMI-Dox40 cell line was provided by William Dalton (H Lee Moffitt Cancer Center, Tampa, FL). OPM1 and OPM2 were provided from P. Leif Bergsagel (Mayo Clinic, Tucson, AZ). KMS11 cell line was obtained from JCRB Cell Bank (Japan). All MM cells were cultured in RPMI1640 medium. HEK-293FT cells were obtained from Invitrogen (Grand Island, NY) and maintained in Dulbecco’s modified Eagle medium. Cell lines have been tested and authenticated by STR DNA fingerprinting analysis (Molecular Diagnostic Laboratory, Dana-Farber Cancer Institute) and used within 3 months after thawing. All media were supplemented with 10% fetal bovine serum, 100 U/mL of penicillin, and 100 μg/mL of streptomycin.
Patient MM cells were purified from BM aspirates by negative selection (RosetteSep Separation System, StemCell Technologies, Vancouver, Canada). The purity of MM cells (>85%) was confirmed by flow cytometric analysis using anti-CD138 Ab (BD Pharmingen, San Diego, CA), as in prior studies. All experiments with patient samples were performed under the auspices of a Dana-Farber Cancer Institute institutional review board–approved protocol.
Reagents and antibodies
Thal, Len, Pom, and MDM2 inhibitor Nutlin-3a were purchased from Selleck Chemicals (Houston, TX). E2F inhibitor HLM006474 was obtained from EMD Millipore (Darmstadt, Germany). TAS-117 was provided by TAIHO Pharmaceutical Co., Ltd. (Ibaraki, Japan). Anti-FLAG M2-agarose beads were purchased from Sigma. Anti-p53 (clone DO-1) and anti-nucleolin (C23), anti-actin, and anti-MDM2 Abs were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); anti-TP53RK (PRPK); anti-CRBN Abs were obtained from Sigma; and anti-TPRKB and anti-RRM1 Abs were from Abcam (Cambridge, MA). All other Abs used were purchased from Cell Signaling Technologies (Danvers, MA). For detection of TP53RK-Myc DDK and TPRKB-Myc-DDK, an infrared imaging system (LI-COR, Lincoln, NE) using anti-mouse IRDye 680RD/LiCor/926-68070 (red color) and anti-rabbit IRDye 800CW/LiCor/926-32211 (green color) secondary Abs was used.
Results
Clinical significance and biologic impact of TP53RK in MM
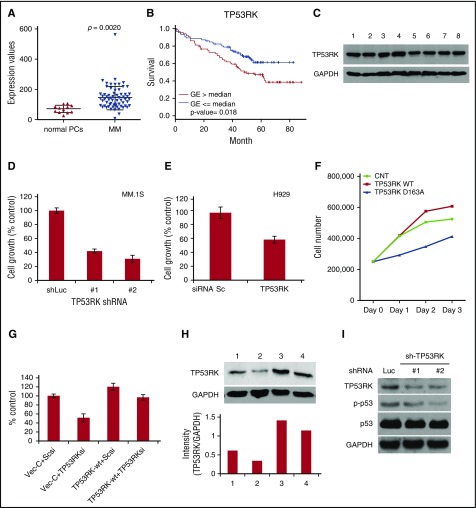
Our GEP analyses revealed that tumor cells from patients with active MM (Figure 1A) or SMM (see supplemental Figure 1, available on the Blood Web site) express higher levels of TP53RK than normal plasma cells, suggesting that TP53RK expression correlates with disease progression in MM. Importantly, newly diagnosed patients with MM expressing high TP53RK levels have significantly shorter survival than patients whose tumor cells express lower levels of TP53RK (P = .018) (Figure 1B), indicating that TP53RK identifies a poor-prognosis patient subset. TP53RK is also constitutively expressed in MM cell lines (Figure 1C). We next examined the sequelae of TP53RK downregulation using shRNA and small interfering RNA (siRNA) in MM.1S and H929 cells, respectively. TP53RK messenger RNA (mRNA) levels were markedly decreased in knockdown cells (supplemental Figure 2A-B) and were associated with growth inhibition in both MM.1S (Figure 1D) and H929 cells (Figure 1E). These results suggest that TP53RK is a growth and/or survival factor in MM cells.
Figure 1.
TP53RK is upregulated in MM patients with poor prognosis and MM cell lines. (A) Comparative GEP analysis of TP53RK between normal plasma cell (PC) and MM cells. (B) Overall survival relative to TP53RK expression in patients with newly diagnosed MM (log-rank test). (C) Whole cell lysates from MM cell lines (1: MM.1S, 2: H929, 3: OPM1, 4: KMS11, 5: RPMI8226, 6: U266, 7: doxorubicin-40 (Dox-40), 8: OPM2) were subjected to western blot. (D) MM.1S cells were infected with control (Luc) or TP53RK (1, 2) shRNAs. After puromycin selection, cells were cultured for 72 hours and growth was assessed by MTT assay. (E) H929 cells were transfected with scrambled (Sc) or TP53RK siRNAs. After transfection, cells were cultured for 72 hours and growth was assessed by MTT assay. (F) H929 cells were transfected with control, wt, or TP53RK kinase-dead mutant (D163A), and viable cells enumerated by trypan blue. (G, H) H929 cells were cotransfected with wt-TP53RK and/or TP53RK siRNA. 1: vector control + Sc siRNA, 2: vector control + TP53RK siRNA, 3: wt-TP53RK + Sc siRNA, 4: wt-TP53RK + TP53RK siRNA. Cell growth and TP53RK protein expression were assessed by MTT assay (G) and western blot (H), respectively. ImageJ was used for densitometric analysis. (I) TP53RK was knocked down in MM.1S cells. Whole cell lysates from MM cells were subjected to western blot with indicated Abs. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide.
To further confirm the impact of TP53RK on MM cell growth, we first overexpressed wt and kinase-dead mutant (D163A)18 TP53RK. Cell type wt-TP53RK modestly enhanced, whereas D163A decreased, growth of H929 (Figure 1F) and U266 cells (supplemental Figure 3). We next examined whether reexpression of TP53RK could rescue TP53TK knockdown-induced MM cell growth inhibition. Importantly, reexpression of wt-TP53RK in TP53RK knockdown H929 cells overcame cell growth inhibition (Figure 1G) and downregulation of TP53RK protein (Figure 1H). Similar results were observed in MM.1S cells (supplemental Figure 4). Because TP53RK regulates phosphorylation of p53 at Ser15,5 we next confirmed that TP53RK knockdown by shRNA downregulated p-p53 in MM.1S cells (Figure 1I). Taken together, these results show that TP53RK mediates MM cell growth, and, conversely, that TP53RK downregulation triggers MM cell growth inhibition.
p53 inhibition triggers MM cell apoptosis via E2F-Bim pathway and c-Myc
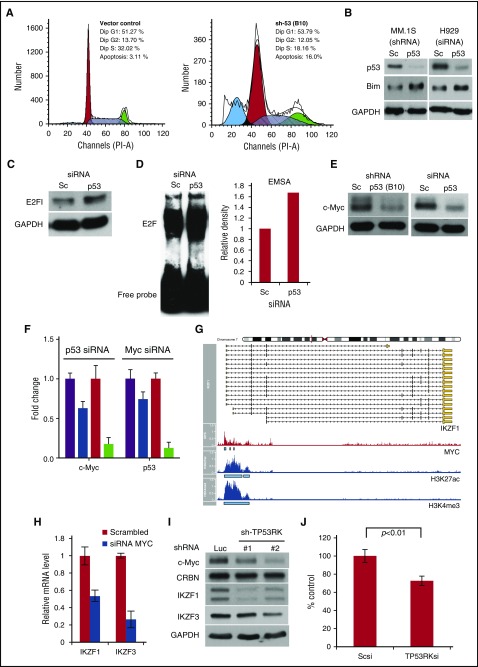
To date, p53 is the only known downstream target of TP53RK. Because our results show that TP53RK knockdown downregulates phosphorylation of p53 associated with MM growth inhibition, we next examined the biologic sequelae of p53 inhibition in MM cells. Importantly, p53 knockdown by shRNA (supplemental Figure 5A) decreased cell growth in MM.1S cells (supplemental Figure 5B). Cell-cycle profiling using propidium iodide staining showed increased sub-Go/G1 fraction (Figure 2A), indicating that downregulation of p53 triggered apoptotic cell death. Interestingly, upregulation of pro-apoptotic protein Bim19 was also observed in p53 knockdown cells (Figure 2B). Because Bim is transcriptionally regulated by E2F1,20 we examined transcriptional activity of E2F1 in p53 knockdown cells and found that p53 knockdown upregulated E2F1 level and its DNA binding activity, assessed by western blot (Figure 2C) and electrophoretic mobility shift assay (EMSA) (Figure 2D), respectively. Consistent with these results, p53 knockdown increased nuclear translocation of E2F1 in MM.1S cells (supplemental Figure 5C). These results suggest that p53 negatively regulates Bim via E2F1.
Figure 2.
p53 inhibition triggers MM cell growth inhibition. (A) MM.1S infected with control (left panel) or p53 shRNA (B10, right panel) was subjected to propidium iodide staining and cell-cycle analysis using flow cytometry. (B) MM.1S cells (left panel) and H929 cells (right panel) were infected with lentiviral Sc or p53 shRNA/siRNA, respectively. (C, D) H929 cells were transfected with Sc or p53 siRNA. Whole cell lysates and nuclear extracts were subjected to western blot (C) and EMSA (D), respectively. Densitometric analysis was performed on E2F EMSA. (E) p53 was knocked down in MM.1S cells (left panel) and H929 (right panel) using shRNA and siRNA, respectively. (F) MM.1S and H929 cells were transfected with p53 or Myc siRNA. mRNAs from transfectants were extracted and subjected to real-time qPCR for c-Myc or p53; purple, MM.1S Sc shRNA; blue, MM.1S c-Myc shRNA; red, H929 Sc siRNA; green, H929 c-Myc siRNA. (G) Normalized (average reads per million) c-MYC, ac-H3K27, or tri-methylated H3K4 chromatin immunoprecipitation-sequencing tag counts around (±5 kb) the transcription start site of IKZF1. (H) H929 cells were transfected with Sc or c-Myc siRNA. Extracted mRNA was subjected to qPCR for IKZF1/3. (I) MM.1S cells were infected with Luc or TP53RK shRNA. (J) p53-deficient (KMS-11) MM cells were transfected with Sc or TP53RK siRNA. Cell growth was assessed by MTT assay after 72-hour incubation.
c-Myc plays a crucial role in MM pathogenesis,21,22 and we unexpectedly observed downregulation of c-Myc protein (Figure 2E) and mRNA (Figure 2F, left panel) after p53 knockdown. Conversely, c-Myc knockdown downregulated p53 mRNA (Figure 2F, right panel), indicating crosstalk transcriptional regulation between p53 and c-Myc in MM cells. By analyzing previously reported chromatin immunoprecipitation sequence data,23 we found that c-Myc binds to promoter regions in both IKZF1 (Figure 2G) and IKZF3 (supplemental Figure 6A), novel therapeutic targets in MM.14,15 Regulation of IKZF1/3 by c-Myc was further confirmed in c-Myc knockdown MM cells using quantitative polymerase chain reaction (qPCR) (Figure 2H). We next showed that TP53RK knockdown markedly downregulated IKZF1/3 in both MM.1S (Figure 2I) and H929 cells (supplemental Figure 6B). Consistent with these results, GEP analyses showed that TP53RK, c-Myc, and IKZF1 expression were all significantly correlated in patient MM cells (supplemental Figure 7A). Taken together, these data indicate that TP53RK knockdown modulates IKZF1/3 expression via the p53-c-Myc axis.
Identification of novel TP53RK downstream proteins
Although p53 is the only known downstream target of TP53RK, we observed that TP53RK knockdown inhibited cell growth even in p53-deficient KMS11 cells (Figure 2J; supplemental Figure 7B) and in p53-mutant/c-Myc–deficient U266 cells (supplemental Figure 8). We did not observe alteration of L-Myc in U266 cells by TP53RK knockdown (data not shown). These results suggest that TP53RK mediates MM cell growth/survival via p53-cMyc axis-dependent and axis-independent pathways.
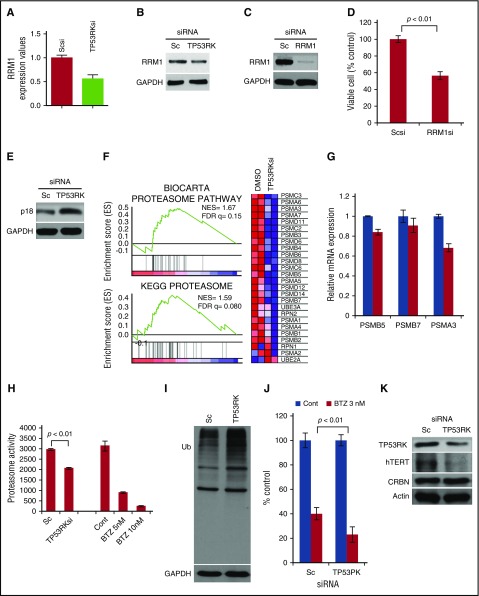
We therefore next carried out GEP analyses and validation studies before and after TP53RK knockdown in H929 cells. We identified genes downregulated (supplemental Table 1) or upregulated (supplemental Table 2) by TP53RK knockdown. We next validated MM-relevant genes including RRM124,25 and CDK4/6 inhibitor p18 (CDKN2C).26,27 Reverse transcription (RT)-qPCR (Figure 3A) and western blot (Figure 3B) confirmed downregulation of RRM1 after TP53RK knockdown. Importantly, downregulation of RRM1 by targeted siRNA (Figure 3C) triggered significant MM cell growth inhibition (Figure 3D). Although mRNA level did not show significant change, we also confirmed upregulation of p18 (CDKN2C) by western blot (Figure 3E) after TP53RK knockdown.
Figure 3.
Novel mechanism of actions whereby inhibition of TP53RK triggers anti-MM activities. H929 cells were transfected with Sc or TP53RK siRNA. Extracted mRNA was subjected to qPCR (A) and western blot (B) for RRM1. H929 cells were transfected with Sc or RRM1 siRNA. Transfectants were subjected to western blot for RRM1 (C) and viable cell count (D). H929 cells were transfected with Sc or TP53RK siRNA. Whole cell lysates were subjected to western blot (E) for p18 (CDKN2C). H929 cells were transfected with Sc or TP53RK siRNA. Extracted RNA/mRNA was subjected to GSEA for proteasome pathway (F) and qPCR for proteasome subunits (G). The cells were subjected to chymotrypsin-like proteasome activity assay (H) and western blot for ubiquitin (I). (J) H929 cells were transfected with Sc or TP53RK, followed by bortezomib (BTZ; 3 nM) treatment of 24 hours. Cell growth was assessed by MTT assay. Data represent mean ± standard deviation from triplicate cultures. Cont, control. (K) H929 cells were transfected with Sc or TP53RK siRNA and subjected to western blot. All western blots were carried out using whole cell lysates and indicated Abs.
Gene set enrichment analysis (GSEA) also identified the proteasome pathway to be downregulated by TP53RK knockdown (Figure 3F). RT-qPCR confirmed downregulation of proteasome subunits PSMB5, PSMB7, and PSMA3 (Figure 3G); chymotrypsin-like proteasome activity was also significantly inhibited (Figure 3H), resulting in accumulation of polyubiquitinated proteins (Figure 3I). Of note, TP53RK knockdown synergistically (P = .0064) enhanced cytotoxicity induced by proteasome inhibitor bortezomib (Figure 3J).
Previous studies have demonstrated that the “kinase endopeptidase and other proteins of small size” (KEOPS) complex promotes telomere uncapping and elongation.28 Because TP53RK is part of the KEOPS complex, we next determined whether TP53RK knockdown affects expression and/or activity of telomerase reverse transcriptase (TERT). Importantly, TP53RK knockdown downregulated human TERT (hTERT) expression in H929 cells (Figure 3K). Taken together, these studies show that TP53RK regulates not only p53, but also other targets including RRM1, CDKN2C (p18), proteasome subunits, and TERT, which may also contribute to TP53RK downregulation-induced MM cell growth inhibition.
IMiDs bind to TP53RK and inhibit its kinase activity
Currently, no small molecule inhibitors targeting TP53RK are available. However, we have previously shown that IMiDs (Pom more potently than Len) trigger MM cell growth inhibition associated with downregulated p21CIP1, in MM.1S cells with wt-p53.7 Other studies have shown that Len downregulates p53 by proteasomal degradation of p53 in Namalwa cells with a 5q deletion and in myeloblastic syndrome patient samples 29; therefore, we hypothesized that IMiDs, either directly or indirectly, inhibit the TP53RK-p53 axis and/or its interacting proteins in MM cells.
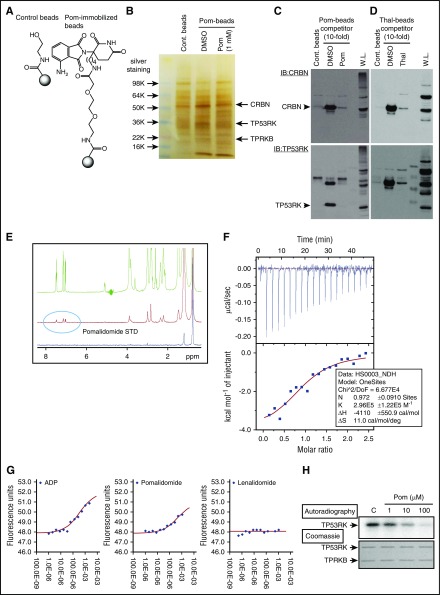
To test this possibility, we first generated Pom-immobilized beads (Pom-beads) (Figure 4A; supplemental Figure 9A), distinct from previously reported Thal-11 (supplemental Figure 9B) or Len-immobilized beads,12 for use in pull-down experiments. Importantly, Pom-beads pulled down not only CRBN as expected, but also TP53RK and its binding protein (TPRKB), confirmed by gel staining and mass spectrometry as well as western blot (Figure 4B,C; supplemental Figure 10). As a positive control, Thal-beads pulled down CRBN,11,12 but not TP53RK or TPRKB (Figure 4D).
Figure 4.
Pom binds to TP53RK. (A) A 3′-n-butylamine derivative of Pom was synthesized and attached via linker to generate Pom-based affinity reagent. (B) MM.1S whole cell lysates were incubated with Pom-beads in the presence or absence of competitor (1 mM free Pom) for 1 hour. After elution, samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining. DMSO, dimethyl sulfoxide. Specific bands pulled down by Pom-beads were subjected to mass spectometry analysis. Protein lysates from MM.1S pulled down by Pom-beads (C), Thal-beads (D), or whole cell lysate (W.L.) were subjected to western blotting using anti-CRBN Abs (top panel). Membranes were subsequently blotted with anti-TP53RK Abs without stripping. IB, immunoblot. (E) Binding of Pom and recombinant TP53RK was assessed by saturation-transfer difference NMR spectrometry. (F) ITC analysis of TP53RK/TPRKB complex and Pom. (G) DSF measurements showed that Pom, but not Len, stabilized the complex. ADP, adenosine 5′-diphosphate. (H) Recombinant C-terminal Myc-DDK-tagged TP53RK and TPRKB proteins from HEK-293FT cells were subjected to in vitro autophosphorylation assay.
We further confirmed Pom binding to the TP53RK/TPRKB complex (TP53RK complex) by saturation-transfer difference NMR spectroscopy, ITC, and DSF using TP53RK/TPRKB recombinant proteins. NMR demonstrated that Pom, with higher affinity than Len, binds recombinant TP53RK (Figure 4E; supplemental Figure 11). ITC showed that TP53RK complex bound to Pom in a 1:1 stoichiometry, with dissociation constant of 3.4 μM (Figure 4F). DSF showed that Pom stabilized the TP53RK complex by >2°C in melting temperature (Tm) (Figure 4G). We next examined the effect of Pom on phosphorylation of TP53RK, using previously reported methods.18 Importantly, Pom downregulated in vitro autophosphorylation of TP53RK in a dose-dependent fashion (Figure 4H). Taken together, our results show that IMiDs bind to TP53RK and inhibit its kinase activity.
TPRKB is a cofactor of TP53RK-IMiDs binding
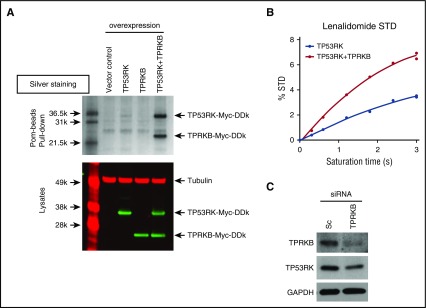
In pull-down experiments, Pom-beads showed weak binding activity to recombinant TP53RK and TPRKB alone, respectively; however, binding was strongly enhanced in the presence of TPRKB complex (Figure 5A). Binding of Len to TP53RK, assessed by NMR spectrometry, was also enhanced in the presence of TPRKB (Figure 5B). The initial slope of the percent saturation-transfer difference vs the saturation time curve is greater for the TP53RK + TPRKB case than would be expected for the difference in protein molecular weight, suggesting tighter Len binding to the complex than to TP53RK alone. Moreover, TPRKB knockdown markedly downregulates TP53RK protein expression (Figure 5C). These results suggest that TPRKB plays a crucial role in maintaining stability and binding activity of TP53RK.
Figure 5.
TPRKB enhances TP53RK binding to IMiDs. (A) TP53RK, TPRKB, or both were overexpressed in HEK293FT cells using c-Myc–tagged expression vectors. Whole cell lysates were subjected to Pom-bead pull-down assay and silver staining (top panel) or western blot using an infrared imaging system (bottom panel). (B) TPRKB was knocked down in H929 cells. STD, saturation-transfer difference. (C) Len binding to TP53RK in the presence of TPRKB. Binding of Len to TP53RK was assessed by NMR spectrometry in the presence or absence of TPRKB.
IMiDs inhibit TP53RK and its downstream targets
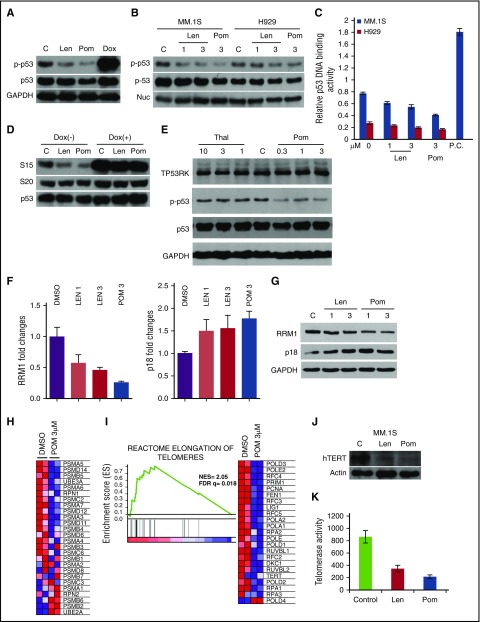
Because IMiDs inhibit TP53RK phosphorylation, we next examined whether IMiDs similarly inhibit downstream targets of TP53RK, as described previously in TP53RK knockdown cells. As with TP53RK knockdown, IMiDs (Pom more potently than Len) also triggered downregulation of phosphorylated (p-Ser15)-p53 (Figure 6A), indicating that IMiDs inhibit TP53RK. This downregulation is further confirmed in nuclear extracts from MM.1S and H929 cell lines (Figure 6B) and in patient MM cells (supplemental Figure 12A,B). Ser15 phosphorylation is required for p53 function,30 and both Len and Pom downregulated p53 activity in MM.1S and H929 cells (Figure 6C). Moreover, phosphorylation at Ser20 (Figure 6D), Ser6, and Ser37 (data not shown) was not altered by the treatment, suggesting that IMiDs selectively inhibit p53 phosphorylation at Ser15. In contrast, Thal does not bind to TP53RK (Figure 4D) or induce p-p53 inhibition (Figure 6E). Our results therefore show that IMiDs, Pom more potently than Len, bind and inhibit TP53RK and downstream p53 activity.
Figure 6.
Biologic significance of pharmacological inhibition of TP53RK by IMiDs in MM cells. (A) MM.1S cells were cultured with Len (2.5 μM) or Pom (2.5 μM) in the presence (40 nM) or absence of Dox for 48 hours. (B, C) MM.1S and H929 cells were treated with Len (1.3 μM) or Pom (3 μM) for 48 hours. Nuclear proteins were subjected to (B) western blot or (C) p53 DNA binding activity assay. Nucleolin served as a loading control. P.C., plasma cell. (D) MM.1S cells were treated with Len (2.5 μM) or Pom (2.5 μM) in the presence or absence of Dox (40 nM) for 48 hours. (E) MM.1S cells were treated with Thal or Pom for 48 hours. In each case, western blot was carried out using cell lysates and the indicated Abs. (F, G) H929 cells were transfected with Sc or TP53RK siRNA. Extracted mRNA was subjected to qPCR (F) and western blot (G) for RRM1 and p18. (H) H929 cells were treated with Pom for 48 hours and subjected to GSEA for proteasome pathway. Extracted RNA/mRNA and cell lysates were subjected to GSEA (I), western blot (J), or telomerase activity assay (K). All western blots were carried out using whole cell lysates and the indicated Abs.
Because previous studies show that phosphorylation of TP53RK at Ser250 can be modulated by Akt,18 we next determined whether downregulation of TP53RK phosphorylation triggered by Pom is mediated, at least in part, via Akt inhibition. MM.1S cells were treated with Akt inhibitor TAS-11731 or cytokines (interleukin-6, insulinlike growth factor 1) to inhibit or activate Akt, respectively; in neither case was p-p53 modulation observed (supplemental Figure 13). Moreover, neither Len nor Pom treatment modulated Akt phosphorylation (supplemental Figure 14). Taken together, these results indicate that phosphorylation and activity of TP53RK in MM cells are not regulated via Akt.
Previous studies have also shown that DNA damage triggers phosphorylation of p53 at different serine residues (activated ATM, ATR, or DNA-PK phosphorylate Ser15 and/or Ser37), whereas Chk1/2 phosphorylates Ser20.32 Because MM cells have ongoing constitutive DNA damage,33 we next asked whether IMiDs Pom and Len downregulate serine residues of p53 by inhibiting these kinases. Both Len and Pom treatment inhibited phosphorylation of Chk1 and p53, but not ATM, ATR, or Chk2 (supplemental Figure 14). Coupled with the lack of p-Ser20 modulation (Figure 6D), these results suggest that p-53 downregulation triggered by IMiDs is not induced via DNA damage-mediated signaling pathways.
We next examined the effects of IMiDs on novel TP53RK downstream targets, RRM1 and p18 (CDKN2C). Similar to TP53RK knockdown, Len and Pom also downregulate RRM1 and upregulate p18, evidenced by RT-qPCR (Figure 6F) and western blot (Figure 6G), respectively. Moreover, Pom-treated cells also showed downregulation of proteasome pathway (Figure 6H) and activity (supplemental Figure 15), indicating that Pom-induced proteasome inhibition is mediated via TP53RK.
Finally, GSEA identified reactome elongation of telomeres to be downregulated by Pom (Figure 6I). Moreover, both Len and Pom treatment of MM.1S and H929 cells significantly inhibited hTERT expression (Figure 6J) and its activity (Figure 6K; supplemental Figure 16A). Importantly, GEP analysis of MM patient samples showed lower hTERT levels after Len treatment (supplemental Figure 16B), indicating that IMiDs may impair telomere maintenance via inhibiting TP53RK.
Inhibition of TP53RK-p53 by IMiDs upregulates Bim
As described previously, p53 knockdown upregulates Bim expression associated with enhanced transcriptional activity of E2F1 (Figure 2B-D); we next showed that IMiDs similarly upregulate Bim (supplemental Figure 17A) resulting from enhanced transcription, confirmed by real-time qPCR (supplemental Figure 17B). IMiDs also enhanced DNA binding activity of E2F1, confirmed by EMSA (supplemental Figure 17C). Finally, IMiDs triggered increased levels of both E2F1 protein (supplemental Figure 18A) and mRNA (supplemental Figure 18B). Enhanced E2F1 DNA binding activity shown in EMSA may therefore be due, at least in part, to increased E2F1 protein levels. Importantly, the E2F inhibitor HLM006474 blocked IMiDs-induced Bim upregulation in a dose-dependent fashion (supplemental Figure 18C), confirming that IMiD-induced upregulation of Bim is mediated via E2F1 activation. Taken together, these results suggest that IMiD-induced cytotoxicity in MM is mediated via apoptosis through inhibition of the TP53RK-p53-E2F1-Bim axis.
Pom modulates TP53RK activity independent of CRBN
We further examined whether IMiDs modulate TP53RK activity independent of CRBN. CRBN knockdown did not alter TP53RK, p53, or p-p53 expression in H929 cells (supplemental Figure 19A); conversely, TP53RK downregulation did not modulate CRBN expression (Figure 2I). Moreover, TP53RK, but not CRBN, knockdown triggered MM cell growth inhibition (supplemental Figure 19B). These results strongly suggest that Pom independently binds and regulates TP53RK and CRBN.
Discussion
Our GSEA identified that TP53RK, located on chromosome 20q13.12, increases with progression from SMM to active MM, as well as within MM, to identify patients with particularly poor prognosis. Here we validate TP53RK as a novel therapeutic target in this patient subset. Importantly, we show that IMiDs also bind to and inhibit TP53RK activity. Both TP53RK knockdown and IMiDs trigger MM cell apoptosis associated with downregulation of c-Myc and E2F-1–mediated upregulation of pro-apoptotic Bim. We also identified novel downstream targets of TP53RK, including ribonucleotide reductase-1, telomerase reverse transcriptase, and CDKN2C. These studies provide the framework to develop selective TP53RK inhibitors to improve outcome in this subset of MM patients with poor prognosis.
TP53RK positively regulates p53 function by phosphorylating its serine residue Ser155; conversely, siRNA knockdown of TP53RK enhanced sensitivity to taxanes in HeLa cells.6 Here we show that TP53RK knockdown triggers MM cell growth inhibition associated with downregulation of phosphorylation of p53 at Ser15. Knockdown of p53 similarly triggers MM cell apoptosis in MM.1S cells with wt-p53. p53 can promote cell survival by inducing reversible cell-cycle arrest and allow for DNA repair; conversely, p53 downregulation can induce apoptosis in this setting.34 We observed that oncogenic c-Myc and its target IKZF1/3 are downregulated by p53 knockdown. Previous studies have defined crosstalk between p53 and E2F135 and shown that E2F1 activity is essential for induction of apoptosis.36 In our study, we observed that p53 knockdown enhanced expression of both E2F1 and its target pro-apoptotic Bim.20 Taken together, these results indicate that the p53-E2F1-Bim axis mediates, at least in part, apoptotic MM cell death resulting from TP53RK inhibition.
Importantly, we observed cell growth inhibition by TP53RK knockdown even in p53-deficient KMS11 cells; therefore, we examined molecular mechanisms of TP53RK-mediated MM cell growth/survival independent of p53. Although p53 is the only known downstream target of TP53RK, we identified novel MM-relevant interacting proteins/pathways including RRM1, p18 (CDKN2C), ubiquitin/proteasome, and hTERT. The biologic function of RRM1 has not totally been elucidated; however, it is a prognostic marker in non–small cell lung cancer37 and other cancers,38 as well as a promoter of metastasis.39 Moreover, previous RNA interference lethality screening studies in MM identified RRM1,24 and ongoing studies are further evaluating its biologic significance in MM. p18 (CDKN2C) interacts with CDK4/6 and controls G1 cell-cycle and tumor progression.40 Interestingly, CDKN2C gene is localized on chromosome 1p32.3, which is deleted in patient MM cells and associated with poor outcome.41 Moreover, p18 (CDKN2C) is regulated by E2F1,42 and we show that p53 knockdown increases E2F1, indicating that TP53RK knockdown-induced upregulation of p18 (CDKN2C) is mediated, at least in part, via the TP53-p53-E2F1 pathway in MM cells. We also observed that TP53RK knockdown downregulates ubiquitin-proteasome pathway genes. Specifically, TP53RK knockdown induces inhibition of proteasome activity, evidenced by accumulation of polyubiquitinated proteins. The ubiquitin-proteasome pathway plays a crucial role in MM cell survival, and proteasome inhibitors bortezomib, carfilzomib, and ixazomib show remarkable clinical efficacy.43-45 These studies therefore further validate TP53RK as a therapeutic target and identify several novel downstream targets—RRM1, p18 (CDKN2C), and ubiquitin/proteasome—with biologic and clinical relevance in MM.
No small molecule inhibitor selectively targeting TP53RK is currently available. IMiDs show remarkable antitumor activity in MM46-48 and their target CRBN has recently been identified.11 Specifically, Len binds to CRBN and triggers proteasomal degradation of IKZF1/3, followed by inhibition of interferon regulatory factor 4 and MM cell growth.14,15 Conversely, downregulation or mutations of CRBN confer IMiD resistance.12,16,17 Because we have previously shown that IMiDs trigger MM cell growth inhibition associated with downregulated p21CIP1 in MM.1S MM cells with wt-p53,7 we examined whether IMiDs, either directly or indirectly, inhibit the TP53RK-p53 axis. Other studies have shown that Len downregulates p53 by proteasomal degradation of p53 in Namalwa cells with a 5q deletion and in myelodysplastic syndrome patient samples.29 Using Pom-beads with a distinct linker position from previous Thal-11 or Len-beads,12 we identified TP53RK as a novel binding protein of Pom that is greater than Len. As a positive control for CRBN binding, we generated Thal-beads as in a previous report11 and observed that Thal-beads bind to CRBN, but not to TP53RK. TP53RK binding to Pom and Len was further confirmed by NMR spectrometry, DSF, and ITC. Moreover, NMR and ITC showed that binding of Pom and Len to TP53RK is enhanced in the presence of TPRKB. Importantly, upon binding to TP53RK, Pom potently downregulates autophosphorylation of TP53RK, suggesting that it may bind to the kinase domain of TP53RK and inhibit its activity. To identify this site, our ongoing studies are resolving the TP53RK crystal structure.
We also show that IMiDs selectively induce downregulation of Ser15 on p53. Although p53 is phosphorylated by DNA damage-associated kinases including ATM, ATR, or DNA-PK at Ser15 and Ser37,49 our results show that IMiDs do not inhibit Ser37 or Ser20 phosphorylation, suggesting that their effect is independent of DNA damage-associated kinases and solely mediated via TP53RK. Importantly, Ser15 phosphorylation is required for p53 function,30 and our studies indicate that IMiDs inhibit p53 function via binding and inactivation of TP53RK.
Because IMiDs inhibit TP53RK-p53, we further examined the effect of IMiDs on novel downstream targets identified in our knockdown experiments. Consistent with TP53RK or p53 knockdown, IMiDs enhance the E2F1-Bim axis, downregulate RRM1 and hTERT, and upregulate p18. Interestingly, IMiDs also downregulate proteasome activity, suggesting that the synergistic activity noted in preclinical and clinical studies when combining IMiDs with proteasome inhibitors may be associated, at least in part, with enhanced proteasome inhibition. Finally, of great interest is the identification of TERT as a novel downstream target of TP53RK. Specifically, the impact of downregulation of both p53 and TERT function by IMiDs is under active investigation because of the increased risk of secondary cancers observed in MM patients who are treated with both IMiDs and alkylating agents.50
In conclusion, our results identify TP53RK as a marker of poor prognosis in MM, validate TP53RK as a novel therapeutic target in this patient subset, and delineate a novel mechanism whereby IMiDs bind TP53RK, inhibit its activity, and trigger apoptotic MM cell death. Ongoing efforts are directed to developing selective TP53RK inhibitors to improve patient outcome in this patient subgroup.
Acknowledgments
The authors thank S. DeAngelo (Department of Cancer Biology, Dana-Farber Cancer Institute) for assistance in the expression of the bicistronic construct; Satoru Ito (Tsukuba Research Center, TAIHO Pharmaceutical Co., Ltd.) for generating IMiDs-conjugated beads; and Geppo Sartori (Universita of Padua, Italy) for kindly providing us TP53RK wild-type and mutant D163A constructs.
This study was supported by grants from the National Institutes of Health, Specialized Programs of Research Excellence-P50100707 (K.C.A.) and the National Institutes of Health, National Cancer Institute (P01-CA078378 and R01-CA050947) (K.C.A.) (R01-CA178264) (T.H. and K.C.A.). K.C.A. is an American Cancer Society Clinical Research Professor.
Footnotes
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: T.H. coordinated activities from all authors and performed western blot, silver staining, Pom/Thal-beads pull-down experiments, siRNA (p53, TP53RK, Bim, and c-Myc) transfection, cell toxicity assay (3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide and cell count), and electrophoretic mobility shift assay, and wrote the manuscript; F.C. performed transfection of wild-type and mutant TP53RK, real-time quantitative polymerase chain reaction (c-Myc, p53, IKZF1, IKZF3, PSMB5, PSMB7, PSMA3), and gene expression profiling (GEP)/gene set enrichment analysis analyses; Y.N., Y.I., and T.U. generated Pom/Thal-beads and performed Pom-beads-TP53RK/TPRKB binding assay as well as in vitro TP53RK auto-phosphorylation assay; H.-S.S. and S.D.-P. performed differential scanning fluorimetry, isothermal titration calorimetry, and wrote the manuscript; H.O. performed TP53RK short hairpin RNA infection experiments; M.K.S. analyzed GEP; D. Cirstea performed p53 short hairpin RNA infection and flow cytometry; N.M. performed real-time quantitative polymerase chain reaction (c-Myc); P.G.R. and N.C.M. collected the human samples; D. Chauhan designed the experiments; W.M. performed nuclear magnetic resonance and wrote the manuscript; and K.C.A. managed the project and wrote the manuscript.
Conflict-of-interest disclosure: K.C.A. serves on advisory boards to Celgene and Millennium. P.G.R. serves on advisory boards to Celgene, Millennium, and Johnson & Johnson. N.C.M. serves on advisory boards to Celgene, Millennium, and Novartis. T.H. is a consultant for Acetylon Pharmaceuticals. T.U., Y.I., and Y.N. are employees of TAIHO Pharmaceutical Co., Ltd. The remaining authors declare no competing financial interests.
Correspondence: Kenneth C. Anderson, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: kenneth_anderson@dfci.harvard.edu.
References
- 1.Anderson KC. The rapid evolution of novel therapies in multiple myeloma. J Natl Compr Canc Netw. 2016;14(5):493-496. [DOI] [PubMed] [Google Scholar]
- 2.Lohr JG, Stojanov P, Carter SL, et al. ; Multiple Myeloma Research Consortium. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25(1):91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolli N, Avet-Loiseau H, Wedge DC, et al. . Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5(5):2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cottini F, Hideshima T, Suzuki R, et al. . Synthetic lethal approaches exploiting DNA damage in aggressive myeloma. Cancer Discov. 2015;5(9):972-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abe Y, Matsumoto S, Wei S, et al. . Cloning and characterization of a p53-related protein kinase expressed in interleukin-2-activated cytotoxic T-cells, epithelial tumor cell lines, and the testes. J Biol Chem. 2001;276(47):44003-44011. [DOI] [PubMed] [Google Scholar]
- 6.Peterson D, Lee J, Lei XC, et al. . A chemosensitization screen identifies TP53RK, a kinase that restrains apoptosis after mitotic stress. Cancer Res. 2010;70(15):6325-6335. [DOI] [PubMed] [Google Scholar]
- 7.Hideshima T, Chauhan D, Shima Y, et al. . Thalidomide and its analogs overcome drug resistance of human multiple myeloma cells to conventional therapy. Blood. 2000;96(9):2943-2950. [PubMed] [Google Scholar]
- 8.Mitsiades N, Mitsiades CS, Poulaki V, et al. . Apoptotic signaling induced by immunomodulatory thalidomide analogs in human multiple myeloma cells: therapeutic implications. Blood. 2002;99(12):4525-4530. [DOI] [PubMed] [Google Scholar]
- 9.LeBlanc R, Hideshima T, Catley LP, et al. . Immunomodulatory drug costimulates T cells via the B7-CD28 pathway. Blood. 2004;103(5):1787-1790. [DOI] [PubMed] [Google Scholar]
- 10.Görgün G, Samur MK, Cowens KB, et al. . Lenalidomide enhances immune checkpoint blockade-induced immune response in multiple myeloma. Clin Cancer Res. 2015;21(20):4607-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito T, Ando H, Suzuki T, et al. . Identification of a primary target of thalidomide teratogenicity. Science. 2010;327(5971):1345-1350. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Girona A, Mendy D, Ito T, et al. . Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26(11):2326-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chamberlain PP, Lopez-Girona A, Miller K, et al. . Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat Struct Mol Biol. 2014;21(9):803-809. [DOI] [PubMed] [Google Scholar]
- 14.Krönke J, Udeshi ND, Narla A, et al. . Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343(6168):301-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu G, Middleton RE, Sun H, et al. . The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343(6168):305-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu YX, Braggio E, Shi CX, et al. . Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood. 2011;118(18):4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kortüm KM, Mai EK, Hanafiah NH, et al. . Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood. 2016;128(9):1226-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Facchin S, Ruzzene M, Peggion C, et al. . Phosphorylation and activation of the atypical kinase p53-related protein kinase (PRPK) by Akt/PKB. Cell Mol Life Sci. 2007;64(19-20):2680-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faber AC, Ebi H, Costa C, Engelman JA. Apoptosis in targeted therapy responses: the role of BIM. Adv Pharmacol. 2012;65:519-542. [DOI] [PubMed] [Google Scholar]
- 20.Biswas SC, Liu DX, Greene LA. Bim is a direct target of a neuronal E2F-dependent apoptotic pathway. J Neurosci. 2005;25(37):8349-8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hideshima T, Bergsagel PL, Kuehl WM, Anderson KC. Advances in biology of multiple myeloma: clinical applications. Blood. 2004;104(3):607-618. [DOI] [PubMed] [Google Scholar]
- 22.Chesi M, Bergsagel PL. Molecular pathogenesis of multiple myeloma: basic and clinical updates. Int J Hematol. 2013;97(3):313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin CY, Lovén J, Rahl PB, et al. . Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiedemann RE, Zhu YX, Schmidt J, et al. . Identification of molecular vulnerabilities in human multiple myeloma cells by RNA interference lethality screening of the druggable genome. Cancer Res. 2012;72(3):757-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Besse B, Olaussen KA, Soria JC. ERCC1 and RRM1: ready for prime time? J Clin Oncol. 2013;31(8):1050-1060. [DOI] [PubMed] [Google Scholar]
- 26.Leone PE, Walker BA, Jenner MW, et al. . Deletions of CDKN2C in multiple myeloma: biological and clinical implications. Clin Cancer Res. 2008;14(19):6033-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyd KD, Ross FM, Walker BA, et al. ; NCRI Haematology Oncology Studies Group. Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clin Cancer Res. 2011;17(24):7776-7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Downey M, Houlsworth R, Maringele L, et al. . A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell. 2006;124(6):1155-1168. [DOI] [PubMed] [Google Scholar]
- 29.Wei S, Chen X, McGraw K, et al. . Lenalidomide promotes p53 degradation by inhibiting MDM2 auto-ubiquitination in myelodysplastic syndrome with chromosome 5q deletion. Oncogene. 2013;32(9):1110-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loughery J, Cox M, Smith LM, Meek DW. Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res. 2014;42(12):7666-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mimura N, Hideshima T, Shimomura T, et al. . Selective and potent Akt inhibition triggers anti-myeloma activities and enhances fatal endoplasmic reticulum stress induced by proteasome inhibition. Cancer Res. 2014;74(16):4458-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18(53):7644-7655. [DOI] [PubMed] [Google Scholar]
- 33.Cottini F, Hideshima T, Xu C, et al. . Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat Med. 2014;20(6):599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukin DJ, Carvajal LA, Liu WJ, Resnick-Silverman L, Manfredi JJ. p53 Promotes cell survival due to the reversibility of its cell-cycle checkpoints. Mol Cancer Res. 2015;13(1):16-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9(10):738-748. [DOI] [PubMed] [Google Scholar]
- 36.Lazzerini Denchi E, Helin K. E2F1 is crucial for E2F-dependent apoptosis. EMBO Rep. 2005;6(7):661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bepler G, Sharma S, Cantor A, et al. . RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22(10):1878-1885. [DOI] [PubMed] [Google Scholar]
- 38.Jordheim LP, Sève P, Trédan O, Dumontet C. The ribonucleotide reductase large subunit (RRM1) as a predictive factor in patients with cancer. Lancet Oncol. 2011;12(7):693-702. [DOI] [PubMed] [Google Scholar]
- 39.Gautam A, Li ZR, Bepler G. RRM1-induced metastasis suppression through PTEN-regulated pathways. Oncogene. 2003;22(14):2135-2142. [DOI] [PubMed] [Google Scholar]
- 40.Bai F, Pei XH, Godfrey VL, Xiong Y. Haploinsufficiency of p18(INK4c) sensitizes mice to carcinogen-induced tumorigenesis. Mol Cell Biol. 2003;23(4):1269-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyd KD, Ross FM, Tapper WJ, et al. ; NCRI Haematology Oncology Studies Group. The clinical impact and molecular biology of del(17p) in multiple myeloma treated with conventional or thalidomide-based therapy. Genes Chromosomes Cancer. 2011;50(10):765-774. [DOI] [PubMed] [Google Scholar]
- 42.Blais A, Monté D, Pouliot F, Labrie C. Regulation of the human cyclin-dependent kinase inhibitor p18INK4c by the transcription factors E2F1 and Sp1. J Biol Chem. 2002;277(35):31679-31693. [DOI] [PubMed] [Google Scholar]
- 43.Richardson PG, Sonneveld P, Schuster MW, et al. ; Assessment of Proteasome Inhibition for Extending Remissions (APEX) Investigators. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med. 2005;352(24):2487-2498. [DOI] [PubMed] [Google Scholar]
- 44.Stewart AK, Rajkumar SV, Dimopoulos MA, et al. ; ASPIRE Investigators. Carfilzomib, lenalidomide, and dexamethasone for relapsed multiple myeloma. N Engl J Med. 2015;372(2):142-152. [DOI] [PubMed] [Google Scholar]
- 45.Moreau P, Masszi T, Grzasko N, et al. ; TOURMALINE-MM1 Study Group. Oral ixazomib, lenalidomide, and dexamethasone for multiple myeloma. N Engl J Med. 2016;374(17):1621-1634. [DOI] [PubMed] [Google Scholar]
- 46.Dimopoulos M, Spencer A, Attal M, et al. ; Multiple Myeloma (010) Study Investigators. Lenalidomide plus dexamethasone for relapsed or refractory multiple myeloma. N Engl J Med. 2007;357(21):2123-2132. [DOI] [PubMed] [Google Scholar]
- 47.Weber DM, Chen C, Niesvizky R, et al. ; Multiple Myeloma (009) Study Investigators. Lenalidomide plus dexamethasone for relapsed multiple myeloma in North America. N Engl J Med. 2007;357(21):2133-2142. [DOI] [PubMed] [Google Scholar]
- 48.Richardson PG, Siegel DS, Vij R, et al. . Pomalidomide alone or in combination with low-dose dexamethasone in relapsed and refractory multiple myeloma: a randomized phase 2 study. Blood. 2014;123(12):1826-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91(3):325-334. [DOI] [PubMed] [Google Scholar]
- 50.Palumbo A, Bringhen S, Kumar SK, et al. . Second primary malignancies with lenalidomide therapy for newly diagnosed myeloma: a meta-analysis of individual patient data. Lancet Oncol. 2014;15(3):333-342. [DOI] [PubMed] [Google Scholar]