To the editor:
Quantitative response evaluation in acute myeloid leukemia (AML) and myelodysplastic syndromes (MDSs) relies on the morphologic quantification of bone marrow (BM) blasts. This process is subject to the operator-dependent quality of BM collection and the interobserver variability among pathologists.1-9 Determining responses can be further complicated by hemodiluted sampling, declined BM biopsies, dry taps, and confounding drug toxicities that prevent count recovery or give the impression of persistent dysplasia.
We previously observed that the clonal architecture of diagnostic BM samples in AML patients with leukocytosis is recapitulated in their simultaneously obtained peripheral blood (PB).10 Other groups have observed concordant PB NPM1 mutation clearance and PB copy number abnormalities11-14 and have occasionally used PB for mutation discovery.15-17 Therefore, we sought to compare the mutation burden in paired serial PB and BM samples in patients with AML or MDS to determine whether sequencing of PB samples is a viable approach for determining clonal architecture and whether it might provide an adjunct, and less invasive, measure of response to therapy.
We quantified mutation burden in PB vs BM samples in a subset of patients treated at Washington University with 10-day courses of decitabine (NCT01687400).18 Twenty-seven patients were selected: 22 with AML and 5 with MDS. Cases were selected based on the presence of at least 2 somatic mutations in the BM sample from each patient using a panel of 264 recurrently mutated AML genes (see supplemental Table 1, available on the Blood Web site) and adequate DNA from matched PB samples at multiple times. PB DNA was analyzed using this recurrently mutated AML gene panel (supplemental Methods; Welch et al18 and Cancer Genome Atlas Research Network19). In total, 138 somatic mutations were detected (median of 4 mutations per patient) across 93 time points (median of 3 time points per patient), providing a total of 446 pairwise comparisons of mutation detection in the blood vs marrow. The median white blood cell count across all time points was 1500/µL (range, 100-75 000/µL). The median age of the patients was 73 years (range, 47-88). Clinical responses included 5 complete remissions, 9 complete remissions with incomplete count recovery, 2 marrow complete remissions, 3 partial remissions, 6 stable disease, and 2 progressive disease. The median read depth in PB samples was ×193; in BM samples, the median read depth was ×295. All patients consented to genome sequencing analysis and were treated in accordance with the Declaration of Helsinki.
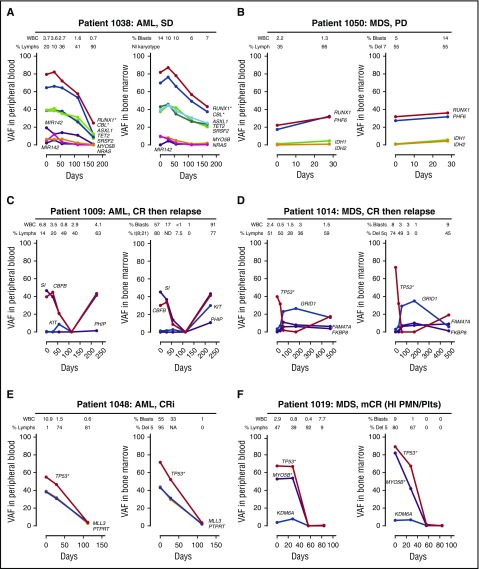
Mutation patterns observed in the PB strongly paralleled the BM results, including subclonal architecture (eg, 1012 and 1018), copy number variation (eg, 1038 and 1019), dynamic responses during decitabine therapy (eg, 1009 and 1048), early expansion of relapse subclones (eg, 1009 and 1021), and clonal hematopoiesis during remission unrelated to the malignant clone20 (eg, 1014) (Figure 1; supplemental Figures 1-4).
Figure 1.
Comparison of clonal architectures in PB vs BM samples. Each panel contains pairwise analysis of the VAF (the ratio of the variant reads vs total reads) in the PB and BM. (A) The 2 purple lines represent variants in miR142. *Indicates variants with evidence of loss of heterozygosity or copy number changes. NI, normal; SD, stable disease. (B) Del, deletion; PD, progressive disease. (C) CR, complete remission. (D) CR. (E) CRi, complete remission with incomplete count recovery; WBC, white blood cell. (F) mCR (HI PMN/Plts), marrow complete remission with hematologic improvement in neutrophils and platelets. Days, number of days since starting decitabine.
The sensitivity and specificity of PB sequencing to detect BM mutations from the same time point were calculated across the entire dataset. For BM mutations with variant allele frequencies (VAFs) >5% and read counts >100, PB sensitivity was 88% and specificity was 84%. A receiver operator curve was generated using a range of VAF and read-count thresholds (supplemental Figure 5A-B); the area under the curve increased only modestly with higher read-count thresholds (0.934 vs 0.943 for read-count cutoffs of 50 vs 200; VAFs >5%).
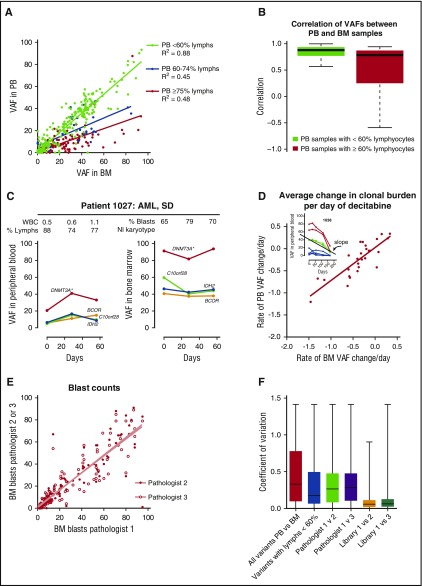
Across all mutations and time points, the PB VAFs were highly correlated with BM VAFs (linear regression, P < .001, R2 = 0.69); the correlation increased modestly when the analysis was restricted to mutations with higher read counts, consistent with binomial sampling probability (supplemental Figure 5C). We examined the concordance of PB and BM results across clinical subgroups defined by 3 clinical variables: peripheral white blood cells, percent PB lymphocytes, and BM blast counts. Of these 3, concordance between PB VAF and BM VAF was only associated with the percentage of PB lymphocytes, with decreased concordance noted in samples with >60% lymphocytes, in which small proportions of myeloid cells would be expected to underestimate the myeloid clonal burden (Figure 2A-B). However, patients with a high proportion of PB lymphocytes tended to maintain this relationship across collections (Figure 2C; supplemental Figure 1); thus, the rate of founding clone mutation clearance still strongly correlated between PB and BM (linear regression, P < .001, R2 = 0.69; Figure 2D).
Figure 2.
Measurements of disease burden using mutations vs morphologic evaluation. (A) Comparison of VAF in the PB vs BM samples. Samples with <60% lymphocytes (lymphs) are indicated in green, 60% to 74% in blue, and ≥75% in red. (B) Correlation between PB and BM VAFs based on the percent PB lymphocytes (see supplemental Methods). (C) Comparison of VAFs in patient 1027, who had a high percentage of PB lymphocytes at multiple time points. (D) Comparison of the rate of change in the tumor burden in the PB vs BM. Each data point represents the slope of VAF change during decitabine treatment of an individual patient. Because the total number of variants in the founding clone was small, the slope of the founding clone was determined using the mutation with the highest copy number–adjusted VAF on day 0. (Inset) Red, variants with copy number changes; green, founding clone variants; blue, subclonal variants; (main plot) red, average rate of change in the VAF. (E) Interobserver variability in blast count estimates between hematopathologists. (F) Coefficients of variation among all PB and BM VAFs, variants in cases with <60% blood lymphocytes, 200 cell morphologic blasts counts among 3 pathologists, and bone marrow VAFs among 3 replicate libraries created from the same sample (supplemental Figure 5D). The coefficient of variation differed between the replicate library VAFs and all other samples (P < .001) and between the blood and BM VAFs vs variants in cases with <60% blood lymphocytes (P < .05, 1-way analysis of variance Kruskal-Wallis test with Dunn’s posttest comparison of all pairs).
Having defined the VAF concordance and the variability between VAFs in the PB and BM samples, we compared these results with the variability in morphologic blast count estimates and the reproducibility of VAFs sequenced from replicate libraries. We evaluated interobserver variability in 200 manual cell differential counts by subjecting 128 BM biopsies to blinded morphologic evaluation by 3 board-certified hematopathologists. A high degree of concordance was observed among the pathologists (Figure 2E; linear regression, P < .001, R2 = 0.8). Next, we compared VAFs generated from 3 independent whole genome sequencing libraries made from the same AML marrow sample (AML31: 1276 mutations were detected with a median depth of 186, 231, and 503 reads in each respective library).21 Independent libraries were highly correlated (R2 = 0.92 and 0.91; supplemental Figure 5D), suggesting that VAFs were highly reproducible independent of sample preparations. The coefficient of variation was compared across blood and marrow VAFs, morphologic blast estimates, and VAFs from independent libraries generated from the same marrow (Figure 2F). Interlibrary VAFs varied the least; PB and BM VAFs varied more, but the variability was similar between blood and marrow VAFs and blast estimates from different pathologists examining the same slide.
What issues remain before serial PB mutation analysis could be integrated into clinical trials? First, the optimal read depth and sequence sensitivity required to measure “mutation clearance” has not been established; a study with deep sequencing, downsampling analysis, and correlation with response and survival will be required. Second, mutation distribution is not always uniform within different hematopoietic lineages (eg, DNMT3A variants may preferentially distribute into the lymphoid compartment, whereas NPM1 and FLT3 variants do not).22 This may cause certain discrepancies between PB and BM VAFs. Finally, it has been observed that subclonal mutations may clear even while founding clone mutations persist23; therefore, molecular monitoring methods will need to consider subclonal architecture.
Taken together, our data show that targeted PB sequencing recapitulates the major genomic events observed in the BM cells of AML and MDS patients, including subclonal architecture and rate of mutation clearance in response to therapy. PB analysis may be valuable in identifying a clonal hematopoietic process, especially when the physician or patient may be reluctant to perform a BM biopsy.24 PB mutation analysis may provide a less invasive alternative for monitoring response to treatment, which is not subject to the same issues of hemodilute sampling and declined sample collection. This approach quantifies the clearance of mutations as a distinct end point that is separate from the recovery of trilineage hematopoiesis, which may be influenced by drug toxicity or limited stem cell reserve. Whether PB mutation analysis may provide an adjunctive approach to assess response to treatment will ultimately require a larger set of samples that can correlate mutation clearance with overall survival.
Footnotes
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank Greg Malnassy, Nichole Helton, and the Washington University Tissue Procurement Core for sample storage and sample preparation, and Theresa Fletcher, Megan Haney, Shannon Kramer, Sharon Heath, Kierstin Luber, Megan Janke, and Paige Schnoebelen for assistance in patient enrollment, sample collection, and clinical data processing.
This work was supported by a National Institutes of Health National Cancer Institute Acute Myeloid Leukemia (AML)–Specialized Programs of Research Excellence (P50 CA171963), the Genomics of AML Program Project grant (P01 CA101937), and the Alliance for Clinical Trials in Oncology Foundation (G.L.U.).
Contribution: E.J.D., B.T., and Y.-S.L. analyzed morphologic bone marrow samples; A.A.P., C.A.M., C.C.F., M.O., R.S.F., and R.K.W. performed the sequencing; G.L.U., M.J.W., P.W., D.C.L, and J.F.D. collected clinical data; F.G. provided statistical analysis. T.J.L. and J.S.W. designed and oversaw the clinical trial and study design; the manuscript was written by E.J.D., G.L.U., and J.S.W; and all authors contributed to manuscript review.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John S. Welch, Department of Internal Medicine, Campus Box 8007, Washington University Medical School, 660 South Euclid Ave, St. Louis, MO 63110; e-mail: jwelch@wustl.edu.
References
- 1.Naqvi K, Jabbour E, Bueso-Ramos C, et al. . Implications of discrepancy in morphologic diagnosis of myelodysplastic syndrome between referral and tertiary care centers. Blood. 2011;118(17):4690-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Font P, Loscertales J, Benavente C, et al. . Inter-observer variance with the diagnosis of myelodysplastic syndromes (MDS) following the 2008 WHO classification. Ann Hematol. 2013;92(1):19-24. [DOI] [PubMed] [Google Scholar]
- 3.Senent L, Arenillas L, Luño E, Ruiz JC, Sanz G, Florensa L. Reproducibility of the World Health Organization 2008 criteria for myelodysplastic syndromes. Haematologica. 2013;98(4):568-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheson BD, Bennett JM, Kopecky KJ, et al. ; International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia [published correction appears in J Clin Oncol. 2004;22(3):576]. J Clin Oncol. 2003;21(24):4642-4649. [DOI] [PubMed] [Google Scholar]
- 5.Douglas DD, Risdall RJ. Bone marrow biopsy technic. Artifact induced by aspiration. Am J Clin Pathol. 1984;82(1):92-94. [DOI] [PubMed] [Google Scholar]
- 6.Loken MR, Chu SC, Fritschle W, Kalnoski M, Wells DA. Normalization of bone marrow aspirates for hemodilution in flow cytometric analyses. Cytometry B Clin Cytom. 2009;76(1):27-36. [DOI] [PubMed] [Google Scholar]
- 7.Font P, Loscertales J, Soto C, et al. . Interobserver variance in myelodysplastic syndromes with less than 5 % bone marrow blasts: unilineage vs. multilineage dysplasia and reproducibility of the threshold of 2 % blasts. Ann Hematol. 2015;94(4):565-573. [DOI] [PubMed] [Google Scholar]
- 8.Parmentier S, Schetelig J, Lorenz K, et al. . Assessment of dysplastic hematopoiesis: lessons from healthy bone marrow donors. Haematologica. 2012;97(5):723-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Souto Filho JT, Loureiro MM, Pulcheri W, Morais JC, Nucci M, Portugal RD. Evaluation of bone marrow aspirates in patients with acute myeloid leukemia at day 14 of induction therapy. Diagn Pathol. 2015;10:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klco JM, Spencer DH, Miller CA, et al. . Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014;25(3):379-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivey A, Hills RK, Simpson MA, et al. ; UK National Cancer Research Institute AML Working Group. Assessment of minimal residual disease in standard-risk AML. N Engl J Med. 2016;374(5):422-433. [DOI] [PubMed] [Google Scholar]
- 12.Mohamedali AM, Gäken J, Ahmed M, et al. . High concordance of genomic and cytogenetic aberrations between peripheral blood and bone marrow in myelodysplastic syndrome (MDS). Leukemia. 2015;29(9):1928-1938. [DOI] [PubMed] [Google Scholar]
- 13.Tong WG, Sandhu VK, Wood BL, et al. . Correlation between peripheral blood and bone marrow regarding FLT3-ITD and NPM1 mutational status in patients with acute myeloid leukemia. Haematologica. 2015;100(3):e97-e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohamedali AM, Alkhatabi H, Kulasekararaj A, et al. . Utility of peripheral blood for cytogenetic and mutation analysis in myelodysplastic syndrome. Blood. 2013;122(4):567-570. [DOI] [PubMed] [Google Scholar]
- 15.Papaemmanuil E, Gerstung M, Malcovati L, et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616-3627, quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thol F, Bollin R, Gehlhaar M, et al. . Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood. 2014;123(6):914-920. [DOI] [PubMed] [Google Scholar]
- 17.Merlevede J, Droin N, Qin T, et al. . Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat Commun. 2016;7:10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welch JS, Petti AA, Miller CA, et al. . TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med. 2016;375(21):2023-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong TN, Miller CA, Klco JM, et al. . Rapid expansion of preexisting nonleukemic hematopoietic clones frequently follows induction therapy for de novo AML. Blood. 2016;127(7):893-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffith M, Miller CA, Griffith OL, et al. . Optimizing cancer genome sequencing and analysis. Cell Syst. 2015;1(3):210-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shlush LI, Zandi S, Mitchell A, et al. ; HALT Pan-Leukemia Gene Panel Consortium. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia [published correction appears in Nature. 2014;508(7496):420]. Nature. 2014;506(7488):328-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klco JM, Miller CA, Griffith M, et al. . Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA. 2015;314(8):811-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwok B, Hall JM, Witte JS, et al. . MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood. 2015;126(21):2355-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]