ABSTRACT
Isolated islets used for transplantation are known to be stressed, which can result from the circumstances of death, in particular brain death, the preservation of the pancreas with its warm and cold ischemia, from the trauma of the isolation process, and the complex events that occur during tissue culture. The current study focused upon the events that occur before the islet isolation procedure. Pancreases were obtained from brain dead donors (n = 7) with mean age 50 (11) and normal pancreatic tissue obtained at surgery done for pancreatic neoplasms (n = 7), mean age 69 (9). Frozen sections were subjected to laser capture microdissection (LCM) to obtain β-cell rich islet tissue, from which extracted RNA was analyzed with microarrays. Gene expression of the 2 groups was evaluated with differential expression analysis for genes and pathways. Marked changes were found in pathways concerned with endoplasmic reticulum stress with its unfolded protein response (UPR), apoptotic pathways and components of inflammation. In addition, there were changes in genes important for islet cell identity. These findings advance our understanding of why islets are stressed before transplantation, which may lead to strategies to reduce this stress and lead to better clinical outcomes.
KEYWORDS: Beta cell stress, islets, islet transplantation, laser capture microdissection, pancreatic beta cells
Introduction
The efficacy of islet transplantation continues to improve,1 but concerns remain about changes found in islets isolated from deceased donors that make them vulnerable to injury and loss of β cells with the transplantation. It has long been known that kidneys and livers obtained from deceased donors undergo deleterious changes including inflammation that lead to less successful transplant outcomes.2,3 Similar findings have been noted in isolated islets. For example, isolated human islets produce tissue factor (TF), which can trigger a thrombin reaction, called the instant blood-mediated inflammatory reaction (IBMIR), which is associated with cell loss when islets are transplanted into the portal vein.4,5 There has been particular concern about the health of islets obtained from pancreases of brain-dead donors. Toyama et al.6 found that after brain injury in rats the isolated islets were infiltrated with macrophages and had increased gene expression of interleukin 1- β (IL1-β), interleukin 6 (IL6), Tumor necrosis factor-α (TNF-α), and monocyte chemoattractant protein-1 (MCP-1). Contreras et al. (7) performed similar experiments in rats and found that plasma measurements of TNF-α, IL1-β and IL6 were elevated and that gene expression of these same factors was elevated in pancreatic tissue. Moreover, there were increased numbers of apoptotic events in β cells as determined by Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) staining. In another study, Saito et al.8 found that isolated islets from brain dead rats had increased gene expression of tissue factor (TF) and mono chemoattractant protein-1 (MCP-1) in isolated islets compared with islets obtained from control rats not subjected to brain injury. Isolated human islets have also been found to produce MCP-1, which could contribute to inflammation.9
To understand the threat to isolated islets, 4 points of variability must be considered: 1) brain injury and the stress of end of life care, 2) the combined detrimental effects of warm and cold ischemia of the harvested pancreas, 3) the trauma of the islet isolation process, and 4) the changes that occur when the isolated islets are cultured before transplantation.
To better understand the contribution of these stages, we sought to understand the changes in islets that occur before the isolation procedure, that is to determine the combined effects of end-of-life events including brain death and organ preservation. This question was examined by comparing gene expression in islets contained in deceased human donor pancreases with those in fresh pancreatic tissue obtained at surgery. A particular strength of this study is that LCM was used to obtain β cell rich tissue samples from frozen sections, which could then be analyzed for gene expression using Affymetrix gene arrays.
Methods and materials
Characteristics and preparation of pancreas specimens
Frozen blocks of 7 surgical pancreas specimens were provided by the Department of Pathology of Massachusetts General Hospital, and 7 cadaver pancreas were obtained from the New England Organ Bank. All of the samples were processed and evaluated at the Joslin Diabetes Center. The patients underwent pancreatic surgery for concerns about pancreatic cancer. We excluded specimens with evidence of pancreatitis, history of diabetes or glucose intolerance. The patient characteristics are shown in Table 1. All subjects were Caucasian; their mean age was 69.4 (8.7) years and body mass index (BMI) was 25.7 (4.3) kg/m2. To select the 7 pancreas specimens from deceased donors, donors who were hyperglycemic before isolation of pancreas were excluded. One subject had a glucose of 248 mg/dl upon taken at admission, but the peak after this was recorded as 159 mg/dl. All donors were Caucasian, and their mean age was 50.4 (11.4) years and BMI was 25.6 (4.8) kg/m2. Frozen pancreatic tissue was sectioned at 8 μm in a cryostat, mounted on uncoated glass slides at −20°C, and immediately stored at −80°C.
Table 1.
Surgical samples and cadaveric samples.
| Donor | Age | Sex | BMI | BG | Diagnosis / Cause of Death |
|---|---|---|---|---|---|
| 1 | 54 | M | 34 | 121 | Intracerebral Hemorrhage |
| 2 | 38 | F | 22 | 117 | Motor Vehicle Accident & CA |
| 3 | 57 | M | 33 | 133 | Head trauma & CA |
| 4 | 44 | M | 27 | 248 | Asphyxia |
| 5 | 63 | M | 29 | 135 | Intracerebral Hemorrhage |
| 6 | 62 | F | 22 | 136 | Subarachnoid Hemorrhage |
| 7 | 35 | F | 23 | 137 | Intracerebral Hemorrhage |
| 9* | 81 | M | 23 | 92 | Adenocarcinoma |
| 8* | 60 | M | 24 | 85 | Adenocarcinoma |
| 10* | 67 | F | 26 | NA | Cystadenoma |
| 11* | 72 | M | 35 | 92 | Adenocarcinoma |
| 12* | 72 | M | 22 | 81 | Tubular Adenoma |
| 13* | 57 | F | 24 | 90 | Neoplasm |
| 14* | 77 | F | 26 | 91 | Neoplasm |
Starred samples are the surgical specimens. Blood glucose (BG mg/dL) Diagnosis/Cause of Death: Cardiac Arrest (CA), BG in Cadavers are the initial recording.
Laser capture microdissection (LCM)
Prior to LCM, frozen pancreatic sections were dehydrated in 70% ethanol for 30 seconds, diethylpyrocarbonate (DEPC)-treated water for 30 seconds, 70% ethanol for 30 seconds, 100% ethanol twice for 1 min, and xylene for 4 minutes. Slides were immediately air-dried. LCM was performed using PixCell II Laser Capture Microdissection System (Arcturus Engineering, Mountain View, CA). The dissections were performed by melting thermoplastic films mounted on transparent LCM caps (Arcturus) targeting selected populations of cells with intrinsic fluorescence.10 To obtain optimal size of the laser pulse, the system parameters were set as follows: laser power 35 mW; pulse duration 3.0 msec; and spot size 7.5 μm. Each LCM session was completed within 30 minutes to avoid RNA degradation. The cells attached to the thermoplastic transfer film were incubated with 10 μl of a guanidine thiocyanate and polyethylene glycol octylphenol ether based buffer for 30 minutes at 42°C. Each section typically had 3–10 islets, and each islet typically contained 2–4 clumps of intrinsic fluorescence representing β-cells. On average, 6–7 sections were used to obtain 800 pulses, which were needed to obtain sufficient RNA for arrays.
RNA extraction, amplification and labeling
Total RNA was extracted from the laser-captured cells using PicoPure RNA Isolation Kit (Arcturus). DNase treatment was performed for 15 minutes using the RNase-free DNase Set (Qiagen, Valencia, CA) to reduce risk of DNA interference. The extracted RNA was amplified by T7-based linear amplification using T7-oligo-dT-primers. Two rounds of amplification were performed using RiboAmp HS RNA Amplification Kits (Arcturus). The quantity and quality of amplified RNA were determined by measuring absorbance at 260 nm and 280 nm using NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE). Biotinylated complementary RNA (cRNA) was generated from cDNA by in vitro transcription reaction using the BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics, Farmingdale, NY). RNA products were then purified using the MiraCol Purification Columns (Arcturus).
Microarray profiling
For comparison of gene expression profiles, single RNA preparations were run on the GeneChip® Human X3P Array (Affymetrix, Santa Clara, CA), containing 61,000 probe sets representing 47,000 transcripts. Ten μg of biotinylated RNA were fragmented to nucleotide stretches of 30–200 nucleotides and hybridized to the chips by the Joslin Diabetes Center DRC Genomics Core.
Determination of β-cell enrichment
To clarify the degree of β–cell enrichment in β–cell samples isolated by LCM, insulin, glucagon, and somatostatin transcripts were quantified by real-time PCR. We measured the expression of these genes in samples obtained by dissected clumps of auto fluorescent cells (β–cells) from within islets compared with samples obtained from randomly dissected islet cells. The LCM avoided acinar tissue. Three pancreases were used for this comparison. Total RNA was extracted as described previously,1 and RNA quantity and purity were evaluated by NanoDrop ND-1000 Spectrophotometer. cDNA templates were prepared from 35.5 ng of total RNA with random hexamers using TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA). The housekeeping gene ribosomal protein S18 (RPS18) transcript was selected as reference. ΔCT values of glucagon or somatostatin in the β–cell samples were used to calculate the differences between glucagon or somatostatin and insulin gene expression in β–cell or islet samples using the ΔΔCT method. The fold changes obtained were used to calculate ratios of expression of glucagon or somatostatin to insulin in β–cell enriched and randomly selected islet cells.
Data analysis
All array data were normalized by Robust Multi-Array Average (RMA).11 Control probes (whose label begins with “AFFX”) were removed. To visualize whether the data significantly clustered into cadaver and donor groups, we plotted the samples according to their first 2 principal components from principal component analysis (PCA), but visual inspection was inconclusive (Fig. 1). To test the clustering formally, we calculated a clustering score (using the Calinski-Harabasz Index12), and compared this score to those scores calculated from 10,000 permutations of the group labels. This analysis showed that the arrangement of the 2 groups is unlikely to be random (Fig. 2). We moved forward with the analysis by identifying differentially expressed genes between these 2 groups using the limma package13 accounting for empirical array quality weights.14 Then, we looked for genes in particular that were related to stress and inflammation. In addition, we ran a pathway analysis against the Broad Institute's Molecular Signature Database's (MSigDB)15 Hallmark pathways, GO terms gene sets, C2 curated gene sets, and C2: Biological Processes independently using the sigPathway package.16 Analyses were conducted in the R/Bioconductor software (www.r-project.org) by the Joslin DRC Bioinformatics Core.
Figure 1.

Principal component analysis did not show differential clustering.
Figure 2.
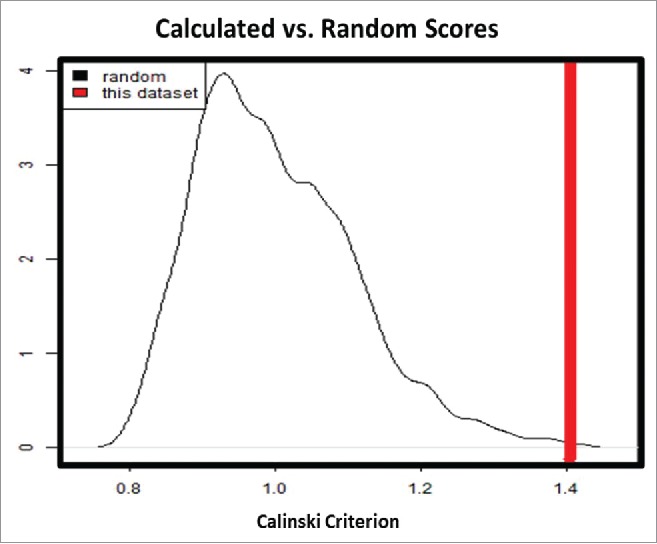
Calinski-Harabasz index describes the ratio of between-cluster variance to within-cluster variance. calinski-Harabasz clustering index between our data and 10,000 permutations of the data showed that although these groups are close, the arrangement of the groups is far from random. Calinski-Harabasz Index describes the ratio of between-cluster variance to within cluster variance.
Data presentation
Data are presented as mean and standard error of the mean [mean(SEM)]. In table 3–5, upregulation is designated with a plus symbol and means upregulated in cadaver pancreases as compared with surgical pancreases.
Table 3.
Genes associated with er stress, UPR, inflammation, and apoptosis.
| Gene Title | Gene Name | p-value | Up/ Down | Function | INF | ER/ UPR | APO | ||
|---|---|---|---|---|---|---|---|---|---|
| CD9 | tetraspanin-29 | 3.92E-04 | 4.27 | regulates and prevents inflammation | + | + | |||
| HSP90B1 | heat shock protein 90 Da | 1.13E-04 | 4.25 | molecular chaperone, PI3K-AKT-mTOR survival signaling | + | ||||
| HSPA5 | heat shock protein family A (Hsp70) member 5 | 1.56E-04 | 3 | folding proteins, monitors protein transport, ER stress | + | ||||
| CEBPD | CCAAT/enhancer binding protein (C/EBP), delta | 1.00E-04 | 2.47 | inflammation, growth arrest, apoptosis | + | + | |||
| RHOB | Ras homolog | 1.06E-03 | 2.38 | promotes intraluminal vesicle delivery | + | + | |||
| ARNTL | aryl hydrocarbon receptor nuclear translocator-like | 1.90E-04 | 2.36 | heterodimer with CLOCK | + | ? | |||
| ZFP36 | ZFP36 ring finger protein | 7.29E-03 | 2.2 | regulated by NF κ B, regulates TNF | + | ||||
| CARD16 | caspase recruitment domain family, member 16 | 8.97E-03 | 2.11 | inflammation response and NF κ B activator, cell death, caspase 1 | + | + | |||
| NAIP | NLR family, apoptosis inhibitory protein | 0.013 | 2.1 | apoptosis inhibition | + | ||||
| KDELR3 | endoplasmic reticulum protein retention receptor 3 | 3.92E-03 | 2.09 | retains proteins in ER | + | ||||
| CHAC1 | ChaC glutathione specific gamma-glutamylcyclotransferase 1 | 2.78E-04 | 2.06 | downstream of ATF4, ATF3 and CHOP, proapoptotic | + | + | |||
| DnaJC3 | heat shock protein family (Hsp40) member C3 | 0.015 | 2.02 | stress inducible protein | + | ||||
| IL17RB | interleukin 17 receptor B | 2.81E-04 | 1.99 | ER induced apoptosis | + | + | |||
| SDF2L1 | stromal cell derived factor 2 like 1 | 1.41E-03 | 1.96 | ER stress-induced gene | + | ||||
| SLC33A1 | ER membrane transporter for acetyl-CoA | 3.99E-03 | 1.93 | regulates autophagy | + | ||||
| DDIT4 | DNA damage inducible transcript 4 | 1.06E-03 | 1.92 | modulates apoptosis, mTOR related ∼ p53 | + | ||||
| ISG20 | interferon stimulated exonuclease gene 20kDa | 3.62E-03 | 1.88 | degrades single stranded DNA and RNA, apoptotic agent | + | ||||
| ETS2 | vets avian erythroblastosis | 3.48E-03 | 1.88 | regulates apoptosis | + | ||||
| IFNGR1 | interferon gamma receptor 1 | 0.020 | 1.86 | inflammation | + | + | |||
| F3 | coagulation factor III (thromboplastin, tissue factor) | 0.032 | 1.79 | inflammation and cellular stress | + | + | |||
| ATF3 | activating transcription factor 3 –cellular stress response | 0.040 | 1.74 | cellular stress response | + | ||||
| DnaJB9 | DnaJ heat shock protein family (Hsp40) member B9 | 6.00E-03 | 1.73 | induced by ER stress, ER chaperone, related to NF κ B | + | ||||
| HYOU 1 | hypoxia upregulated 1 -HSP 70 family | 1.29E-03 | 1.72 | UPR pathway | + | + | |||
| NFKBIZ | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta | 0.020 | 1.7 | inflammation | + | ||||
| PDIA6 | protein disulfide isomerase family A member 6 | 3.86E-04 | 1.7 | formation, reduction and isomerization of disulfide bonds | + | ||||
| GADD45G | growth arrest and DNA-damage-inducible, gamma | 8.95E-03 | 1.68 | can activate JNK pathways | + | ||||
| EIF2AK3 | eukaryotic translation initiation factor 2-α kinase 3 | 7.96E-03 | 1.68 | induced by ER stress | + | ||||
| DERL 2 | derlin 2 | 2.70E-04 | 1.66 | detects misfolded proteins, ERAD complex | + | ||||
| KDELR2 | endoplasmic reticulum protein retention receptor 2 | 3.54E-04 | 1.65 | retains proteins in ER | + | ||||
| KDELR2 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 | 3.54E-04 | 1.65 | ER associated gene | + | ||||
| DNAJB11 | DnaJ (Hsp40) homolog, subfamily B, member 11 | 0.015 | 1.62 | ER chaperone complex subunit | + | ||||
| HILPDA | hypoxia inducible lipid droplet-associated | 3.22E-03 | 1.59 | stress response gene | + | + | |||
| IL13RA1 | interleukin 13 receptor | 4.63E-04 | 1.59 | linked to IL4 ∼ related to JAK/STAT signaling | + | + | |||
| NFKBIA | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, α | 0.011 | 1.58 | Inflammation | + | ||||
| LMAN1 | lectin-mannose binding | 2.02E-04 | 1.57 | cycles between ER and Golgi | + | ||||
| HIF3A | hypoxia inducible factor 3, α subunit | 4.94E-03 | 1.56 | mitochondrion response to hypoxia and stress | + | + | |||
| SERP1 | stress-associated endoplasmic reticulum protein 1 | 0.017 | 1.55 | UPR and ER and Golgi transport | + | ||||
| IL6ST | interleukin 6 signal transducer | 2.48E-03 | 1.55 | transducer shared by many cytokines regulates apoptosis | + | ||||
| SLC9A6 | solute carrier family 9 member A6 | 0.018 | 1.54 | mediator of ER stress-induced apoptosis | + | + | |||
| TNFRSF1A | tumor necrosis factor receptor superfamily member 1A | 7.93E-03 | 1.51 | regulates inflammation, mediate apoptosis | + | + | |||
| MCL1 | myeloid cell leukemia 1 | 2.16E-03 | 1.51 | anti-apoptotic gene – member of Bcl−2 family | + | ||||
| PDCD4 | programmed cell death 4 (neoplastic transformation inhibitor) | 0.021 | 1.49 | apoptosis and translation inhibitor | + | ||||
| CASP6 | caspase 6, apoptosis-related cysteine peptidase | 0.019 | 1.45 | regulates ER stress and apoptosis | + | ||||
| VIMP | VCP interacting membrane protein | 2.36E-03 | 1.45 | central to ER stress and ERAD | + | + | |||
| SRPRB | signal recognition particle receptor | 2.76E-04 | 1.44 | ER response to apoptosis | + | + | |||
| BBC3 | BCL2 binding component 3 | 5.92E-03 | 1.43 | essential mediator of apoptosis with BCL−2 | + | + | |||
| CALR | calreticulin, major calcium binding protein in the ER | 5.51E-03 | 1.43 | major Calcium binding protein in the ER | + | ||||
| CALR | calreticulin | 5.51E-03 | 1.43 | ER associated gene | + | ||||
| LDHA | lactate dehydrogenase A | 0.048 | 1.42 | LDHA is not expressed in normal β cells | + | ||||
| DERL1 | derlin 1 | 9.73E-03 | 1.4 | detects misfolded proteins, ERAD complex | + | ||||
| TNIP2 | TNFAIP3 interacting protein 2 | 4.13E-03 | 1.38 | inhibitor NF κ B and involved in apoptosis | + | + | |||
| COPB2 | coatomer protein complex subunit β 2 | 2.32E-03 | 1.38 | Important for Golgi budding | + | ||||
| ERGIC1 | endoplasmic reticulum-Golgi intermediate compartment (ERGIC) 1 | 1.03E-03 | 1.35 | transportation between ER and Golgi | + | ||||
| DAD1 | defender against cell death | 3.19E-03 | 1.27 | anti-apoptotic | + | ||||
| MAPK8IP3 | mitogen-activated protein kinase 8 interacting protein 3 | 9.63E-03 | −1.23 | JNK related protein | + | ||||
| WFS1 | Wolfram Syndrome 1 (Wolframin) | 0.015 | −1.29 | link to apoptosis | + | ||||
| GGA2 | golgi associated, gamma adaptin ear containing, ARF binding protein 2 | 2.64E-03 | −1.49 | regulates Golgi's traffic and lysosome | + | ||||
| MX1 | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | 2.54E-04 | −1.62 | anti-cell-death and ER stress | + | + | |||
| IGF1R | insulin-like growth factor 1 receptor | 1.74E-06 | −1.88 | anti-apoptotic enhancing the cell survival | + |
ER stress and unfolded protein response (UPR) gene expression increased in β cells of cadaver pancreases compared those from surgical pancreases. These are designated as plus and minus symbols for fold change. These upregulated genes indicate major changes of genes involved with ER mechanisms, including ER Stress and the unfolded protein response (UPR). Also, a variety of gene concerned with apoptosis (APO) and cell viability are markedly upregulated in the β cells of cadaver pancreases. Genes associated with Inflammation upregulated in β cells of cadaver pancreases. Furthermore, genes that are associated with inflammation (INF) and stress response were observed to be affected in the cadaver group. A group of these genes is related to BCL2 and NFκB inflammatory molecules.
Table 4.
Islet cell identity genes.
| Gene Title | Gene.Name | p-value | FC | Function |
|---|---|---|---|---|
| Differentially Expressed Genes | ||||
| SLCO5A1 | solute carrier organic anion transporter family, member 5A1 | 1.69E-05 | 2.98 | differentiation |
| CHGA | chromogranin A (parathyroid secretory protein 1) | 2.86E-03 | −1.23 | secretory cell marker |
| SYT13 | synaptotagmin XIII | 1.04E-04 | −1.29 | exocytosis |
| ABCC8 | ATP-binding cassette, sub-family C (CFTR/MRP), member 8 | 0.011 | −1.31 | sulfonylurea – SUR1 receptor |
| STXBP1 | syntaxin binding protein 1 | 1.62E-03 | −1.33 | exocytosis |
| PPDPF | pancreatic progenitor cell differentiation and proliferation factor | 0.024 | −1.33 | cell differentiation and exocrine regeneration |
| SCAMP | secretory carrier membrane protein 1 | 4.96E-03 | −1.38 | secretory carrier membrane proteins |
| ISL1 | ISL LIM homeobox 1 | 0.045 | −1.39 | differentiation |
| PAX6 | paired box 6 | 0.013 | −1.46 | differentiation |
| NKX6–1 | NK6 homeobox 1 | 1.77E-03 | −1.51 | β cell maturation |
| MAFB | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B | 4.18E-04 | −1.52 | β and α cell identity |
| KCNG3 | potassium voltage-gated channel modifier subfamily G member 3 | 3.11E-03 | −1.82 | cell viability, nuclear migration speed, ER |
| ARX | aristaless related homeobox | 1.17E-03 | −2.02 | α-cell marker |
| GRIN3A | glutamate receptor, ionotropic, N-methyl-D-aspartate 3A | 3.90E-06 | −3.76 | related to ion channels |
| SLC6A4 | solute carrier family 6 (neurotransmitter transporter), member 4, serotonin | 3.38E-03 | −3.92 | insulin secretion |
| Non-differentially Expressed Genes | ||||
| NKX2–2 | NK2 homeobox 2 | 0.149 | 1.33 | β cell development |
| SLC2A2 | solute carrier family 2 (facilitated glucose transporter), member 2 | 0.625 | 1.11 | glucose transporter 2 (GLUT2) |
| AKT3 | v-akt murine thymoma viral oncogene homolog 3 | 0.805 | 1.1 | key signaling pathway |
| NEUROD1 | neuronal differentiation 1 | 0.816 | −1.02 | β cell identity |
| FOXA2 | forkhead box A2 | 0.729 | −1.03 | important β cell transcription factor |
| SST | somatostatin | 0.84 | −1.03 | Inhibits insulin and glucagon secretion |
| INS | insulin | 0.515 | −1.05 | β cell product |
| GCG | glucagon | 0.324 | −1.06 | stimulates gluconeogenesis and glycogenolysis |
| PCSK2 | proprotein convertase subtilisin/kexin type 2 | 0.504 | −1.07 | proinsulin cleavage |
| NEUROG3 | neurogenin 3 | 0.219 | −1.08 | Islet cell development |
| IAPP | islet amyloid polypeptide | 0.408 | −1.11 | β cell secretory product |
| G6PC2 | glucose-6-phosphatase, catalytic, 2 | 0.551 | −1.13 | important for neogenesis |
| GCK | glucokinase (hexokinase 4) | 0.053 | −1.34 | glucose metabolism control |
The expression of some of the key transcription factors for β cells are downregulated, but no changes were seen for insulin, glucagon or somatostatin expression.
Table 5.
Noteworthy genes.
| Gene Title | Gene Name | p-value | Up/Down | Function |
|---|---|---|---|---|
| REG1B | regenerating islet-derived 1 β | 2.78E-03 | 8.19 | regeneration and β cell proliferation |
| REG1A | regenerating islet-derived 1 α | 0.01 | 4.63 | regeneration and β cell proliferation |
| SYT16 | synaptotagmin XVI | 4.86E-06 | 3.44 | exocytosis |
| HYOU1 | hypoxia upregulated 1 | 1.29E-03 | 1.72 | related to heat shock 70 family, hypoxia-related stress and apoptosis |
| TM9SF3 | transmembrane 9 superfamily member 3 | 5.68E-06 | 1.66 | related to interferon |
| GRHPR | glyoxylate reductase/hydroxypyruvate reductase | 1.43E-03 | −1.41 | metabolism |
| MRPL10 | mitochondrial ribosomal protein L10 | 9.63E-05 | −1.57 | protein synthesis |
| NDRG1 | N-myc downstream regulated 1 | 6.47E-05 | −1.92 | stress responses |
| STAT4 | signal transducer and activator of transcription 4 | 1.15E-06 | −2.21 | important element for β cell dysfunction Induced by Inflammatory cytokines |
| ACSM1 | acyl-CoA synthetase medium-chain family member 1 | 1.14E-05 | −2.61 | fatty acid synthesis |
| CCNB1 | cyclin B1 | 6.41E-06 | −2.90 | cell cycle, cell division, growth arrest if downregulated |
Results
Quantity and quality of RNA
RNA extraction and amplification from samples obtained by LCM yielded 16.9 (3.9) μg of aRNA with A260:A280 ratios ranging from 2.44 to 2.58. The A260:A280 ratios of the aRNA were within the acceptable limits and not different between surgical and deceased donor pancreas specimens. After amplification and labeling, the integrity of the RNA was evaluated by 3′/M’ ratio of the housekeeping gene (glyceraldehyde-3-phosphate dehydrogenase, GAPDH). The mean 3′/M’ ratio of GAPDH measured from surgical and cadaver pancreas specimens was 8.4 (1.1) and 7.8 (1.5), which were within the acceptable ranges.
β cell enrichment by LCM
LCM samples were obtained from selected β cell rich autofluorescent areas of islets or from generalized non-selected areas LCM of islet tissue. Real time PCR was performed on unamplified RNA to avoid artifacts from amplification. The ratio of GLUCAGON/INSULIN expression in β cell selected tissue to GLUCAGON/INSULIN expression in the generalized non-selected islet tissue was 0.238 (0.132). The ratio of SOMATOSTATIN/INSULIN expression in β cell selected tissue to SOMATOSTATIN/INSULIN expression in the generalized non-selected islet tissue was 0.584 (0.277). This means that the selected LCM samples had 74% less GLUCAGON expression than randomly isolated tissue and 42% less SOMATOSTATIN expression. This result is mostly in agreement with our earlier report finding about an 80% reduction in GLUCAGON expression and a 60% reduction in SOMATOSTATIN expression.10
Pathways for differential expression
In Table 2, the 9 most differentially expressed pathways are listed. The most notable changes are those associated with the endoplasmic reticulum, apoptosis, inflammation, and β cell identity. The most differentially expressed pathway was Hallmark: Glycolysis, which identifies a variety of changes that include genes related to chaperones and apoptosis.
Table 2.
GSEA pathway analysis.
| Group |
Noteworthy Pathways |
Z-score |
FDR |
|---|---|---|---|
| Cell Function | Hallmark: Glycolysis | 4.05 | 0.003 |
| UPR | Hallmark: Unfolded Protein Response | 3.88 | 0.003 |
| Stress | Hallmark: mTORC1 Signaling | 3.62 | 0.004 |
| Cell Identity | Hallmark: Pancreas β Cells | −3.58 | 0.004 |
| Stress | Hallmark: TNFA Signaling via NF κ B | 3.09 | 0.007 |
| ER | GO: Endoplasmic Reticulum | 3.84 | 0.009 |
| Apoptosis | KEGG: Apoptosis | 3.1 | 0.011 |
| Hypoxia | Hallmark: Hypoxia | 2.75 | 0.013 |
| Apoptosis | Reactome: Intrinsic Pathway for Apoptosis | 2.88 | 0.016 |
Z-score represents “Nek* Stat” from sigPathway package, which is standardized enrichment score. Positive Z score means upregulated in cadaver pancreases and conversely negative means downregulated in cadaver pancreases. For instance, β Cell identity is downregulated in cadavers whereas unfolded protein response (UPR) is upregulated. False Discovery Rate (FDR).
Expression changes related to ER stress, UPR, inflammation, and apoptosis
As shown in Table 3, there are impressive increases in the number of upregulated genes in the β cells of deceased donor pancreases that are related to ER function and ER stress with its related Unfolded Protein Response (UPR).17 There were notable increases in chaperone genes including HSP90B1, HSPA5, DNAJC3, DNAJB9, and DNAJB11. There were also many upregulated genes related to apoptosis and inflammation although it is important to point out the complexity of these changes in that the genes related to apoptosis included a mixture of changes that were both pro- and anti-apoptotic. Likewise activation of NFκB can lead to changes that are pro-apoptotic or protective.18 For example, some proapoptotic genes that were notably upregulated include CHAC1,19 CASP6,20 BBC321 and CEBPD.22 With regard to NFκB signaling, IL17RB23 is thought to be an activator and TNIP224 is inhibitory. In spite of so many changes, islets isolated from cadaver pancreases usually function very well when used for clinical transplants. It is of interest that so many of the genes in these categories are upregulated while a much smaller number were downregulated.
Islet cell identity genes
As shown in Table 4, there were several surprising changes. There was notable downregulation in the deceased donor pancreases of MAF B25 and ARX,26 which are important for α cell identity. SYT1327 and STXBP1,28 which are important components of the β cell exocytotic machinery, were both downregulated. Also downregulated were the important β cell genes NKX6.1, a key transcription factor29; ABCC8, the sulfonylurea receptor30; and PAX6.31 On the other hand, most of the key genes for β cell identity including INS, IAPP, GCK, PCSK2, NKX2.2, and NEUROD1 were not differentially expressed.
Other noteworthy changes
As shown in Table 5, there were a variety of other notable changes. REG1A and REG1B have marked increases in expression in the β cells of the deceased donor pancreases. The roles of Reg family of proteins continue to be puzzling. They have been associated with regeneration in several tissues including β cells32 and a knockout of Reg1 led to reduced β cell mass.33 The expression of the Reg genes can undergo marked changes, as was found in the present study and in an earlier study when β cells gene expression from subjects with T2D were studied after LCM.34
Discussion
The use of LCM is a unique strength of this study, which makes it possible to obtain RNA from β cell enriched tissue of human pancreatic frozen sections. Most other studies measure gene expression from isolated islets, which can be expected have different patterns of gene expression. First, the trauma of islet isolation and time spent in tissue culture must lead to changes in β cell gene expression. Second, other cell types, notably duct and acinar cells, inevitably contaminate islet cell preparations. In particular, using electron microscopy for cell identification of islet preparations used for clinical islets transplants from 37 pancreases, we found that on average only 36% of cells were β cells.35
In the present study there were important changes in gene expression in β cells dissected from deceased donor pancreases compared with those dissected from fresh surgically obtained pancreatic tissue. These changes resulted from a combination of events occurring during the end-of-life events that occur before death is pronounced, including brain death, fluctuations of blood pressure, administration of drugs, etc. Then another set of variables occurs with warm ischemia as the pancreas is procured followed by cold ischemia during the period of organ preservation, which last for periods of hours. It was not possible to determine the effects of brain death alone because of the many other variables that were involved. Moreover, we could not determine potential effects of hyperglycemia. Even though we chose deceased donors without a recorded history of hyperglycemia, there could easily have been periods of hyperglycemia as so often happens in intensive care situations. This issue is important because hyperglycemia, even modest elevations of glucose concentrations, can induce changes in gene expression through a process often called glucotoxicity.36
Several pathways were very significantly differentially expressed, as shown in Table 2, including those related to glycolysis, the unfolded protein response (UPR), hypoxia, H-MTORC1, H-TNFA-NFkB, and the ER membrane. The most striking finding was how many gene related to the endoplasmic reticulum were differentially expressed. Many of these were genes were associated with UPR, including the heat shock proteins that serve a chaperone function, DERL1 and 2, which detect misfolded proteins, PDIA6, which is important for formation, reduction and isomerization of disulfide bonds, and ATF3, which is an important mediator of UPR.17 The UPR is mostly a protective response that is activated when the ER's capacity for handing the production of new proteins is exceeded, which is characterized by increased production of chaperones, which should enhance folding capacity and slowing down peptide synthesis by inhibiting both transcription and translation.37 As ER stress becomes more intense the protective effects of the UPR can be overwhelmed leading to apoptosis, with several proapoptotic gees including the so-called “executioner” gene CHOP and JNK being turned on,38 but in this case neither was differentially expressed. It is not clear why the UPR was activated. There was no reason to think insulin synthesis and secretion would be turned on by severe illness and organ preservation.
Pathways concerned with apoptosis were also differentially expressed, but it is not clear what these changes mean. Perhaps they indicate that the β cells are vulnerable to apoptosis. We know there is a low incidence of apoptosis as determined by TUNEL staining of β cells in paraffin sections of pancreases obtained at autopsy, and that the incidence is a little higher39 in type 2 diabetes. There are also data indicating that the incidence of apoptosis is low in pancreases obtained from organ banks,40 although it is possible that the cold preservation temperature inhibited the active process of apoptosis.
In conclusion, the present findings show that β cells obtained from deceased donor pancreases are stressed even before the trauma of islet isolation and culture. Finding ways to improve organ retrieval and preservation could lead to healthier β cells following the isolation process. Improvements at many points during the process including better pre-death care, organ procurement, organ preservation, islet isolation, and islet culture should lead to better clinical outcomes.
Data access
All the microarray data has been submitted to GEO under GSE89120 accession number.
Abbreviations
- ER
endoplasmic reticulum
- FDR
false discovery rate
- IBMIR
instant blood-mediated inflammatory reaction
- LCM
laser capture microdissection
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- UPR
unfolded protein response
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Grace Daher of the Advanced Genomics/Genetics Core, Chris Cahill of the Advanced Microscopy Core, and the Bioinformatics Core of the Joslin Diabetes Research Center [DRC].
Funding
This study was supported by grants from the NIH (R01 DK093909 [SBW], NCRR ICR U4Z 16606 (GCW), NCRR ICR U42 RR0023244–01(GCW), P30 DK036836 Joslin Diabetes Research Center (DRC), the Diabetes Research and Wellness Foundation and an important group of private donors. No conflicts of interest relevant to this article are reported.
References
- [1].Hering BJ, Clarke WR, Bridges ND, Eggerman TL, Alejandro R, Bellin MD, Chaloner K, Czarniecki CW, Goldstein JS, Hunsicker LG, et al.. Phase 3 Trial of Transplantation of Human Islets in Type 1 Diabetes Complicated by Severe Hypoglycemia. Diabetes Care 2016; 39(7):1230-40; PMID:27208344; http://dx.doi.org/ 10.2337/dc15-1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Terasaki PI, Cecka JM, Gjertson DW, Takemoto S. High survival rates of kidney transplants from spousal and living unrelated donors. N Engl J Med 1995; 333(6):333-6; PMID:7609748; http://dx.doi.org/ 10.1056/NEJM199508103330601 [DOI] [PubMed] [Google Scholar]
- [3].Jassem W, Koo DD, Cerundolo L, Rela M, Heaton ND, Fuggle SV. Leukocyte infiltration and inflammatory antigen expression in cadaveric and living-donor livers before transplant. Transplantation 2003; 75(12):2001-7; PMID:12829901; http://dx.doi.org/ 10.1097/01.TP.0000061605.30685.03 [DOI] [PubMed] [Google Scholar]
- [4].Moberg L, Johansson H, Lukinius A, Berne C, Foss A, Kallen R, Ø Østraat, Salmela K, Tibell A, Tufveson G, et al.. Production of tissue factor by pancreatic islet cells as a trigger of detrimental thrombotic reactions in clinical islet transplantation. Lancet 2002; 360(9350):2039-45. [DOI] [PubMed] [Google Scholar]
- [5].Johansson H, Lukinius A, Moberg L, Lundgren T, Berne C, Foss A, Felldin M, Källen R, Salmela K, Tibell A, et al.. Tissue factor produced by the endocrine cells of the islets of Langerhans is associated with a negative outcome of clinical islet transplantation. Diabetes 2005; 54(6):1755-62; PMID:15919797; http://dx.doi.org/ 10.2337/diabetes.54.6.1755 [DOI] [PubMed] [Google Scholar]
- [6].Toyama H, Takada M, Suzuki Y, Kuroda Y. Activation of macrophage-associated molecules after brain death in islets. Cell Transplant 2003; 12(1):27-32; PMID:12693661; http://dx.doi.org/ 10.3727/000000003783985205 [DOI] [PubMed] [Google Scholar]
- [7].Contreras JL, Eckstein C, Smyth CA, Sellers MT, Vilatoba M, Bilbao G, Rahemtulla FG, Young CJ, Thompson JA, Chaudry IH, et al.. Brain death significantly reduces isolated pancreatic islet yields and functionality in vitro and in vivo after transplantation in rats. Diabetes 2003; 52(12):2935-42; PMID:14633854; http://dx.doi.org/ 10.2337/diabetes.52.12.2935 [DOI] [PubMed] [Google Scholar]
- [8].Saito Y, Goto M, Maya K, Ogawa N, Fujimori K, Kurokawa Y, Satomi S. Brain death in combination with warm ischemic stress during isolation procedures induces the expression of crucial inflammatory mediators in the isolated islets. Cell Transplant 2010; 19(6):775-82; PMID:20573302; http://dx.doi.org/ 10.3727/096368910X508889 [DOI] [PubMed] [Google Scholar]
- [9].Piemonti L, Leone BE, Nano R, Saccani A, Monti P, Maffi P, Bianchi G, Sica A, Peri G, Melzi R, et al.. Human pancreatic islets produce and secrete MCP-1/CCL2: relevance in human islet transplantation. Diabetes 2002; 51(1):55-65; PMID:11756323; http://dx.doi.org/ 10.2337/diabetes.51.1.55 [DOI] [PubMed] [Google Scholar]
- [10].Marselli L, Thorne J, Ahn YB, Omer A, Sgroi DC, Libermann T, Otu HH, Sharma A, Bonner-Weir S, Weir GC. Gene expression of purified beta-cell tissue obtained from human pancreas with laser capture microdissection. J Clin Endocrinol Metab 2008; 93(3):1046-53; PMID:18073315; http://dx.doi.org/ 10.1210/jc.2007-0931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Irizarry R, Hobbs B, Collin F, Beazer-Barclay Y, Antonellis K, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics (Oxford, England) 2003; 4(2):249-64; PMID:12925520; http://dx.doi.org/ 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- [12].Caliń ski T, Harabasz J. A dendrite method for cluster analysis. Communications in Statistics 1974; 3(1):1-27. [Google Scholar]
- [13].Ritchie M, Phipson B, Wu D, Hu Y, Law C, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 2015; 43(7):gkv007-e47; http://dx.doi.org/ 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ritchie M, Diyagama D, Neilson J, van Laar R, Dobrovic A, Holloway A, Smyth GK. Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics 2006; 7(1):261; PMID:16712727; http://dx.doi.org/ 10.1186/1471-2105-7-261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Subramanian A, Tamayo P, Mootha V, Mukherjee S, Ebert B, Gillette M, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al.. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005; 102(43):15545-50; PMID:16199517; http://dx.doi.org/ 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tian L, Greenberg S, Kong S, Altschuler J, Kohane I, Park P. Discovering statistically significant pathways in expression profiling studies. Proceedings of the National Academy of Sciences of the United States of America 2005; 102(38):13544-9; PMID:16174746; http://dx.doi.org/ 10.1073/pnas.0506577102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem 2005; 74:739-89; PMID:15952902; http://dx.doi.org/ 10.1146/annurev.biochem.73.011303.074134 [DOI] [PubMed] [Google Scholar]
- [18].Darville MI, Eizirik DL. Regulation by cytokines of the inducible nitric oxide synthase promoter in insulin-producing cells. Diabetologia 1998; 41:1101-8; PMID:9754830; http://dx.doi.org/ 10.1007/s001250051036 [DOI] [PubMed] [Google Scholar]
- [19].Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol 2009; 182(1):466-76; PMID:19109178; http://dx.doi.org/ 10.4049/jimmunol.182.1.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang XJ, Cao Q, Zhang Y, Su XD. Activation and regulation of caspase-6 and its role in neurodegenerative diseases. Annual review of pharmacology and toxicology 2015; 55:553-72; PMID:25340928; http://dx.doi.org/ 10.1146/annurev-pharmtox-010814-124414 [DOI] [PubMed] [Google Scholar]
- [21].Ballar P, Zhong Y, Nagahama M, Tagaya M, Shen Y, Fang S. Identification of SVIP as an endogenous inhibitor of endoplasmic reticulum-associated degradation. The Journal of biological chemistry 2007; 282(47):33908-14; PMID:17872946; http://dx.doi.org/ 10.1074/jbc.M704446200 [DOI] [PubMed] [Google Scholar]
- [22].Balamurugan K, Sterneck E. The many faces of C/EBPdelta and their relevance for inflammation and cancer. International journal of biological sciences 2013; 9(9):917-33; PMID:24155666; http://dx.doi.org/ 10.7150/ijbs.7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schwandner R, Yamaguchi K, Cao Z. Requirement of tumor necrosis factor receptor-associated factor (TRAF)6 in interleukin 17 signal transduction. J Exp Med 2000; 191(7):1233-40; PMID:10748240; http://dx.doi.org/ 10.1084/jem.191.7.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Leotoing L, Chereau F, Baron S, Hube F, Valencia HJ, Bordereaux D, Demmers JA, Strouboulis J, Baud V. A20-binding inhibitor of nuclear factor-kappaB (NF-kappaB)-2 (ABIN-2) is an activator of inhibitor of NF-kappaB (IkappaB) kinase alpha (IKKalpha)-mediated NF-kappaB transcriptional activity. The Journal of biological chemistry 2011; 286(37):32277-88; PMID:21784860; http://dx.doi.org/ 10.1074/jbc.M111.236448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Guo S, Dai C, Guo M, Taylor B, Harmon JS, Sander M, Robertson RP, Powers AC, Stein R. Inactivation of specific beta cell transcription factors in type 2 diabetes. J Clin Invest 2013; 123(8):3305-16; PMID:23863625; http://dx.doi.org/ 10.1172/JCI65390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Collombat P, Hecksher-Sorensen J, Krull J, Berger J, Riedel D, Herrera PL, Serup P, Mansouri A. Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. J Clin Invest 2007; 117(4):961-70; PMID:17404619; http://dx.doi.org/ 10.1172/JCI29115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Andersson SA, Olsson AH, Esguerra JL, Heimann E, Ladenvall C, Edlund A, Salehi A, Taneera J, Degerman E, Groop L, et al.. Reduced insulin secretion correlates with decreased expression of exocytotic genes in pancreatic islets from patients with type 2 diabetes. Molecular and cellular endocrinology 2012; 364(1–2):36-45; PMID:22939844; http://dx.doi.org/ 10.1016/j.mce.2012.08.009 [DOI] [PubMed] [Google Scholar]
- [28].Lang H, Ai Z, You Z, Wan Y, Guo W, Xiao J, Jin X. Characterization of miR-218/322-Stxbp1 pathway in the process of insulin secretion. Journal of molecular endocrinology 2015; 54(1):65-73; PMID:25489007; http://dx.doi.org/ 10.1530/JME-14-0305 [DOI] [PubMed] [Google Scholar]
- [29].Schisler JC, Jensen PB, Taylor DG, Becker TC, Knop FK, Takekawa S, German M, Weir GC, Lu D, Mirmira RG, et al.. The Nkx6.1 homeodomain transcription factor suppresses glucagon expression and regulates glucose-stimulated insulin secretion in islet beta cells. Proc Natl Acad Sci U S A 2005; 102(20):7297-302; PMID:15883383; http://dx.doi.org/ 10.1073/pnas.0502168102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gloyn AL, Siddiqui J, Ellard S. Mutations in the genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) in diabetes mellitus and hyperinsulinism. Human mutation 2006; 27(3):220-31; PMID:16416420; http://dx.doi.org/ 10.1002/humu.20292 [DOI] [PubMed] [Google Scholar]
- [31].Gosmain Y, Katz LS, Masson MH, Cheyssac C, Poisson C, Philippe J. Pax6 is crucial for beta-cell function, insulin biosynthesis, and glucose-induced insulin secretion. Mol Endocrinol 2012; 26(4):696-709; PMID:22403172; http://dx.doi.org/ 10.1210/me.2011-1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lieu HT, Batteux F, Simon MT, Cortes A, Nicco C, Zavala F, Pauloin A, Tralhao JG, Soubrane O, Weill B, et al.. HIP/PAP accelerates liver regeneration and protects against acetaminophen injury in mice. Hepatology 2005; 42; 42(3. 3):618-26, -26; PMID:16116631; http://dx.doi.org/ 10.1002/hep.20845 [DOI] [PubMed] [Google Scholar]
- [33].Unno M, Nata K, Noguchi N, Narushima Y, Akiyama T, Ikeda T, Nakagawa K, Takasawa S, Okamoto H. Production and characterization of Reg knockout mice: reduced proliferation of pancreatic beta-cells in Reg knockout mice. Diabetes 2002; 51(Suppl 3):S478-83; PMID:12475793; http://dx.doi.org/ 10.2337/diabetes.51.2007.S478 [DOI] [PubMed] [Google Scholar]
- [34].Marselli L, Thorne J, Dahiya S, Sgroi DC, Sharma A, Bonner-Weir S, Marchetti P, Weir GC. Gene expression profiles of Beta-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PLoS ONE 2010; 5(7):e11499; PMID:20644627; http://dx.doi.org/ 10.1371/journal.pone.0011499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pisania A, Weir GC, O'Neil JJ, Omer A, Tchipashvili V, Lei J, Colton CK, Bonner-Weir S. Quantitative analysis of cell composition and purity of human pancreatic islet preparations. Lab Invest 2010; 90(11):1661-75; PMID:20697378; http://dx.doi.org/ 10.1038/labinvest.2010.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Weir GC, Bonner-Weir S. Islet beta cell mass in diabetes and how it relates to function, birth, and death. Annals of the New York Academy of Sciences 2013; 1281:92-105; http://dx.doi.org/ 10.1111/nyas.12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kennedy J, Katsuta H, Jung MH, Marselli L, Goldfine AB, Balis UJ, Sgroi D, Bonner-Weir S, Weir GC. Protective unfolded protein response in human pancreatic beta cells transplanted into mice. PLoS ONE 2010; 5(6):e11211; PMID:20585452; http://dx.doi.org/ 10.1371/journal.pone.0011211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kaufman RJ. Beta-cell failure, stress, and type 2 diabetes. N Engl J Med 2011; 365(20):1931-3; PMID:22087686; http://dx.doi.org/ 10.1056/NEJMcibr1109442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes 2003; 52(1):102-10; PMID:12502499; http://dx.doi.org/ 10.2337/diabetes.52.1.102 [DOI] [PubMed] [Google Scholar]
- [40].Marchetti P, Bugliani M, Lupi R, Marselli L, Masini M, Boggi U, Filipponi F, Weir GC, Eizirik DL, Cnop M. The endoplasmic reticulum in pancreatic beta cells of type 2 diabetes patients. Diabetologia 2007; 50(12):2486-94; PMID:17906960; http://dx.doi.org/ 10.1007/s00125-007-0816-8 [DOI] [PubMed] [Google Scholar]


