Version Changes
Revised. Amendments from Version 1
The changes to the text are as follows: 1) We addressed referee #1's question regarding the dbVar update cycle 2) We address referee #1's questions about the percent total reported 3) We added additional references for the manuscript 4) We added clearer images for the figures We would like to thank the reviewers for taking the time to review this paper!
Abstract
dbVar houses over 3 million submitted structural variants (SSV) from 120 human studies including copy number variations (CNV), insertions, deletions, inversions, translocations, and complex chromosomal rearrangements. Users can submit multiple SSVs to dbVAR that are presumably identical, but were ascertained by different platforms and samples, to calculate whether the variant is rare or common in the population and allow for cross validation. However, because SSV genomic location reporting can vary – including fuzzy locations where the start and/or end points are not precisely known – analysis, comparison, annotation, and reporting of SSVs across studies can be difficult. This project was initiated by the Structural Variant Comparison Group for the purpose of generating a non-redundant set of genomic regions defined by counts of concordance for all human SSVs placed on RefSeq assembly GRCh38 (RefSeq accession GCF_000001405.26). We intend that the availability of these regions, called structural variant clusters (SVCs), will facilitate the analysis, annotation, and exchange of SV data and allow for simplified display in genomic sequence viewers for improved variant interpretation. Sets of SVCs were generated by variant type for each of the 120 studies as well as for a combined set across all studies. Starting from 3.64 million SSVs, 2.5 million and 3.4 million non-redundant SVCs with count >=1 were generated by variant type for each study and across all studies, respectively. In addition, we have developed utilities for annotating, searching, and filtering SVC data in GVF format for computing summary statistics, exporting data for genomic viewers, and annotating the SVC using external data sources.
Keywords: NCBI, dbVar, Structural Variation Cluster, GVF, Genomics, Open-Source, Genome Annotation, Education, Software
Introduction
There is a growing body of evidence suggesting that genomic structural variants play an important role in the etiology of human disease and in determining individuals’ characteristics and phenotypes 1, 2. Structural variants are also important for understanding the evolution of species 3. dbVar is a database of large structural genomic variants that catalogs millions of records from both small and large studies and makes them freely available to the public 4, 5. The data are organized by submitted study, which makes for convenient comparisons between cases and controls. dbVar online search and browser tools make it easy to search and retrieve the data.
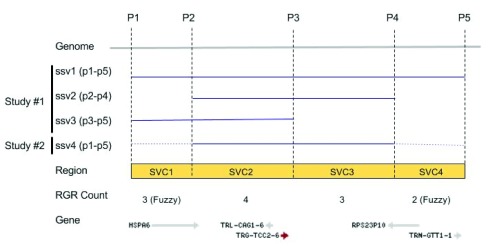
It is difficult to annotate novel SVs or to compute summary data without a reference record or exemplar when multiple SSV choices are available in the same genomic region, and there has been no publicly available resource to date that combines variants from all studies for integration into a bioinformatic pipeline for search, analysis, and comparison. We created structural variant clusters (SVC) to overcome these problems. Structural variant clusters ( Figure 1) are smaller discrete genomic features that include counts of the features shared between SSVs. In regions with fuzziness between overlapping SSVs, SCVs allow the calculation of annotation and frequency by either consensus overlapping regions or by user-defined limits.
Figure 1. An alignment of variants ssv1-ssv3 (blue lines) with the genome (grey line) between positions P1 and P5.
Reference genomic regions SVC1-SVC4 (yellow box) are demarcated by overlap and non-overlapping positions (P1-P2, P2-P3, etc.) between SSVs. The observed SVC counts and the genes are shown on the bottom.
Additional benefits of having a defined set of SVCs include:
improved data exchange, data mining, computation, and reporting;
better searching and matching of genomic coordinates across studies;
easier aggregation of annotations such as disease and phenotype, frequency, and genomic features that co-locate with a SVC;
a simplified display in the Sequence Viewer as an aggregated histogram or density track from all studies (currently dbVar display each study as a track, which can be slow to render and difficult to display on small screens); and
the ability to measure SSV concordance regions and validate across studies.
The Structural Variation Cluster project aimed to accomplish a number of goals. First, we generated a Genome Variant Format (GVF) file of SVC regions as defined above, based on RefSeq GRCh38 1. Each region is assigned a unique ID (SVC1, SVC2, etc.). The SVC VCF file is used as the basis for generating aggregated data, filtering, generating sequence viewer tracks, and for comparison with user data. We also generated a histogram track to show the frequency of the regions across studies in genomic context for the Sequence Viewer. In addition, we annotated SVC regions with Gene, colocated dbSNP 6 reference SNPs, ClinVar 7, and other colocated features. We aimed to create a tool for filtering SVC GVFs by variant type, region size, region count, chromosome, and additional user-defined splitting and filtering parameters. This tool would allow users to compare their data with SVC GVFs and report matching regions of overlap.
Methods
SVCs are defined as the union set of overlapping and non-overlapping regions for all SSVs aligned to the genome using HTSeq version 0.6.0 8, based on the genomic coordinates in RefSeq human genome assembly GRCh38 (RefSeq accession GCF_000001405.26) 1 ( Figure 1).
Structural variant cluster (SVC) from SSV
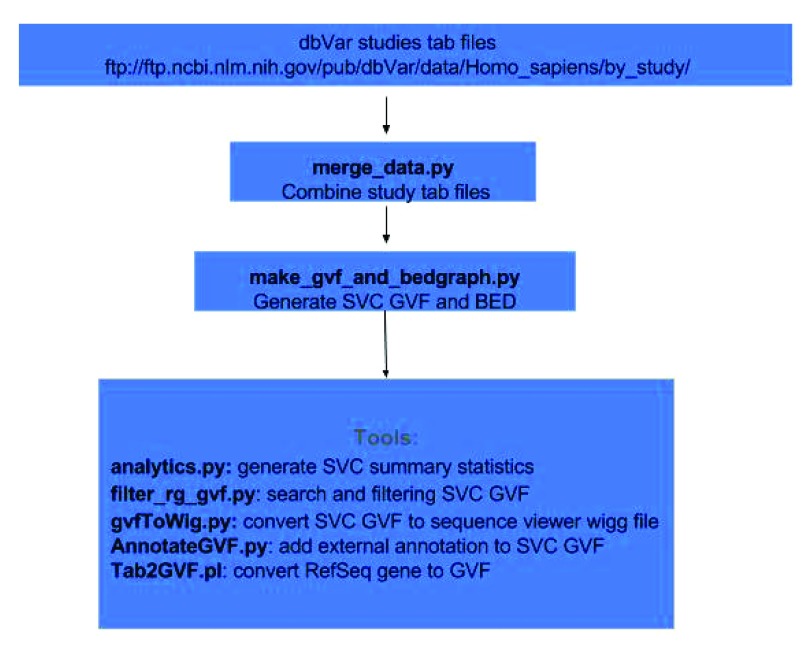
Figure 2 demonstrates the workflow for this analysis. dbVar SSV data by studies were obtained in tab delimited format from the FTPsite (ftp:// ftp.ncbi.nlm.nih.gov/pub/dbVar/data/Homo_sapiens/by_study/) and used as input. The study files were combined and sorted by chromosome positions into a single file using the script merge_data.py. SVC regions, including counts as shown in Figure 1, were generated from the merged file using the script make_gvf_and_bedgraph.py, which output SVC GVF and BED files. Since the approach in Figure 1 is similar to finding consensus regions or overlapping features between aligned reads make_gvf_and_bedgraph.py use HTSeq.GenomicInterval class to store SSV chr. start, and stop coordinates as genomic features and the HTSeq.GenomicArrayOfSets class to identify overlapping positions to generate SVC and counts.
Figure 2. Dataflow for generating SVC and tools for analysis.
Additional tools are available as scripts using SVC GVF as input to compute summary statistics, to search and filter, to generate WIG files for viewing in sequence viewer, and to annotate using external data sources. All scripts and examples are available on GitHub ( https://github.com/NCBI-Hackathons/Structural_Variant_Comparison/). For this study all coordinates reported are based on GRCh38.
Results
Computing structural variant cluster (SVC)
As shown in Figure 1, SVCs were created from overlapping and non-overlapping regions of two or more SSVs using the HTSeq.GenomicArrayOfSets class and output as GVF file format. Each SVC is counted for the number of times it is present as a subregion of a SSV, providing a total SVC count across studies. A single SSV by itself without any overlap between itself and another SSV in the region constitutes a single SVC with a feature count of 1. 3.6 million dbVar SSVs generated 3.4 million SVCs for all dbVar data (combined-set) by variant type ( Table 1).
Table 1. SVC count and percentage of total by variant types from combined-set across all studies.
The most common variant type was deletion followed by CNV. All CNV types combined (rows 1, 7, and 8 in Table 1) total 972,335. We also generated SVC for each variant type (ie. CNV, in/del, etc.) and by individual studies (study-set) for QA/QC and analysis between types and studies of interest. The study-set generated a total of 2.5 million SVCs versus 3.4 million SVCs from the combined-set.
| Variant Type | Percent
Total (%) |
SVC Count |
|---|---|---|
| deletion | 35% | 1194270 |
| copy number loss | 18% | 633268 |
| mobile element insertion | 15% | 517506 |
| duplication | 9% | 303790 |
| insertion | 7% | 232552 |
| indel | 5% | 186658 |
| copy number gain | 5% | 169876 |
| copy number variation | 5% | 169191 |
| Others | 1% | 17810 |
| Total | 100% | 3424921 |
Comparison of combined-set and study-set SVC derived from variant type CNV with ClinVar and genomic annotations
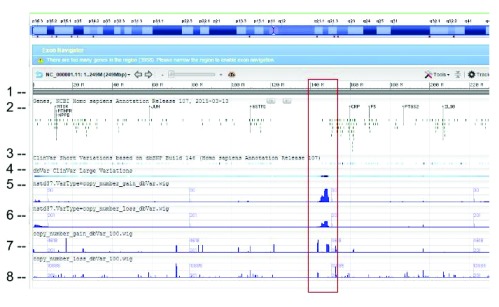
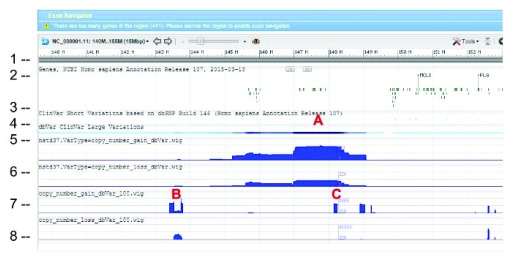
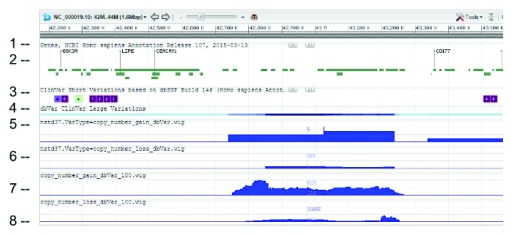
WIG files were generated from SVC GVF files to allow loading into sequence viewer for quick visual inspection as shown in Figure 3. The SVC sets used for inspections are the combined-set which includes 1000Genomes 9, as well as other large studies to provide frequently occurring or “common” SVC to compare with presumed curated variants that have clinical significance from study-set (dbVar:nstd37) submitted by ClinGen 10. The Variation Viewer 11 allows for quick navigation by genes, chromosome positions, and variations for visual comparison ( Figure 3, Figure 4, and Figure 5). Figure 3 and Figure 4 show a hotspot peak A in ClinVar (track 4) that corresponds with a peak in SVC from nstd37, suggesting that this region is critical for function and that variations in this region are rare. These conclusions are supported by the lack of corresponding SVC peaks in the combined-set “common” tracks 7 and 8. However, tracks 7 and 8 also contain peaks B and C that flank the ClinVar peak, which may demarcate the boundaries for the critical region peak A. In contrast, Figure 5 shows that there are corresponding SVC peaks in the nstd37 (rare) and in the combined-set (common), suggesting that variants in this region may have minimal or no clinical impact by themselves.
Figure 3. SVC (type=CNV) distribution on chromosome 1 as seen on NCBI Variation Viewer (Variation Viewer - NCBI).
Starting from the top: (1) chr 1 sequence, (2) Gene track, (3) ClinVar short variation for dbSNP SNV, (4) ClinVar large variation, (5) ClinGen SVC study-set (dbVar:nstd37) copy number gain, (6) ClinGen SVC study-set (dbVar:nstd37) copy number loss, (7) SVC combined-set for copy number gain with count >= 100, and (8) SVC combined-set for copy number loss with count >= 100. The red box highlights an SVC hotspot region found in ClinGen (dbVar:nstd37) tracks 5 and 6 that correspond with the variants in ClinVar. The scale for SVC count histogram are 1–90 (track 5), 1–20 (track 6), 1–4618 (track 7), and 1–10885 (track 8).
Figure 4. Zoomed in view of the red box region in Figure 1 showing dbVar:nstd37 SVC peak A corresponding to ClinVar variants and flanking peaks B and C from the common combined-set.
The track and histogram scales are as described in Figure 5.
Figure 5. Magnified region of Chr19 showing a region of high density of variants where SVCs presumed to be common are also found.
The tracks and histogram scales are as described in Figure 3.
Conclusions
The software tools we developed and provide here compute SVCs and provide counts of concordance regions across SSVs. We also developed tools to search, filter, annotate, and graphically view the results in sequence viewers or to incorporate them into custom analysis pipelines. Using these tools, we provide examples ( Figure 3) for comparing across different SVC data sets with other annotation (such as genes and ClinVar). Such comparisons will allow users to investigate across the genome - or near a gene of interest - and to look for concordance and conflicts between data, which may help users form hypotheses regarding the biological impact of observed variation in SVC regions. In future, we will conduct the work and analysis required for SVC data quality assurance. We believe that SVC data promise to improve the analysis and the elucidation of the biological impact of structural variants, and in future, will probably have uses beyond those described here. Potential uses for SVC data could include:
the evaluation of other SVC hot spot regions to determine if they occur biologically or are due to genome problem regions;
the use of study metadata to validate SVCs that are in concordance with regions across studies and different assay platforms;
the validation of rare SVCs (count =< 2) and common SVCs (count > 2);
identification of evidence of variations in all public SRA data;
combined analysis and annotation of SVCs to ClinVar, dbSNP, and other variation resources;
the creation of a reference dbVar “SV” number based on SVCs, which would be the equivalent to dbSNP’s RS number;
identification of population-specific SVCs to gain insight into the functional significance of structural variants and their evolution; and
determination of high-priority SVCs with significant functional impact and effects.
In addition, a “dbVar Beacon Service” could be developed to allow users to query dbVar if variants exists for a genomic location of interest using combined SVC data. The results would report the number of SVCs and associated SSV IDs and study IDs. Users could then download the study or SSV of interest from dbVar.
Software availability
Latest source code: https://github.com/NCBI-Hackathons/Structural_Variant_Comparison/
Archived source code as at time of publication: http://dx.doi.org/10.5281/zenodo.48201 12
Accompanying wiki: https://github.com/NCBI-Hackathons/Structural_Variant_Comparison/wiki
Manual: https://docs.google.com/document/d/1WBnEnShnw28ZFg17A3xUpWOyvxXjb2q-h1kF-XYVWEw/edit?usp=sharing
License: CC0 1.0 Universal
Acknowledgements
The authors thank Lisa Federer, NIH Library Writing Center, for manuscript editing assistance.
Funding Statement
Lon Phan, John Garner, John Lopez, and Ben Busby’s work on this project was supported by the Intramural Research Program of the National Institutes of Health (NIH)/National Library of Medicine (NLM)/NCBI.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 2 approved]
References
- 1. Saeed S, Bonnefond A, Manzoor J, et al. : Genetic variants in LEP, LEPR, and MC4R explain 30% of severe obesity in children from a consanguineous population. Obesity (Silver Spring). 2015;23(8):1687–95. 10.1002/oby.21142 [DOI] [PubMed] [Google Scholar]
- 2. Ross JS, Badve S, Wang K, et al. : Genomic profiling of advanced-stage, metaplastic breast carcinoma by next-generation sequencing reveals frequent, targetable genomic abnormalities and potential new treatment options. Arch Pathol Lab Med. 2015;139(5):642–9. 10.5858/arpa.2014-0200-OA [DOI] [PubMed] [Google Scholar]
- 3. Radke DW, Lee C: Adaptive potential of genomic structural variation in human and mammalian evolution. Brief Funct Genomics. 2015;14(5):358–68. 10.1093/bfgp/elv019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Home - dbVar - NCBI [Internet]: Home - dbVar - NCBI.[cited 2016 Feb 24]. Reference Source [Google Scholar]
- 5. Lappalainen I, Lopez J, Skipper L, et al. : DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res. 2013;41(Database issue):D936–41. 10.1093/nar/gks1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sherry ST, Ward MH, Kholodov M, et al. : dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–11. 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Landrum MJ, Lee JM, Benson M, et al. : ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–8. 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anders S, Pyl PT, Huber W: HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. estd214 - 1000 Genomes Consortium Phase 3 - dbVar Study - NCBI [Internet]: estd214 - 1000 Genomes Consortium Phase 3 - dbVar Study - NCBI.[cited 2016 Feb 24]. Reference Source [Google Scholar]
- 10. ClinGen - ClinGen Clinical Genome Resource [Internet]: ClinGen - ClinGen Clinical Genome Resource.[cited 2016 Feb 24]. Reference Source [Google Scholar]
- 11. Variation Viewer - NCBI [Internet]: Variation Viewer - NCBI.[cited 2016 Feb 24]. Reference Source [Google Scholar]
- 12. John G, TriLe965, Hsu J, et al. : Structural_Variant_Comparison: Initial Post-Hackathon Release. Zenodo. 2016. Data Source [Google Scholar]