Abstract
Interactions between different phytoplankton taxa and heterotrophic bacterial communities within aquatic environments can differentially support growth of various heterotrophic bacterial species. In this study, phytoplankton diversity was studied using traditional microscopic techniques and the bacterial communities associated with phytoplankton bloom were studied using High Throughput Sequencing (HTS) analysis of 16S rRNA gene amplicons from the V1-V3 and V3-V4 hypervariable regions. Samples were collected from Lake Akersvannet, a eutrophic lake in South Norway, during the growth season from June to August 2013. Microscopic examination revealed that the phytoplankton community was mostly represented by Cyanobacteria and the dinoflagellate Ceratium hirundinella. The HTS results revealed that Proteobacteria (Alpha, Beta, and Gamma), Bacteriodetes, Cyanobacteria, Actinobacteria and Verrucomicrobia dominated the bacterial community, with varying relative abundances throughout the sampling season. Species level identification of Cyanobacteria showed a mixed population of Aphanizomenon flos-aquae, Microcystis aeruginosa and Woronichinia naegeliana. A significant proportion of the microbial community was composed of unclassified taxa which might represent locally adapted freshwater bacterial groups. Comparison of cyanobacterial species composition from HTS and microscopy revealed quantitative discrepancies, indicating a need for cross validation of results. To our knowledge, this is the first study that uses HTS methods for studying the bacterial community associated with phytoplankton blooms in a Norwegian lake. The study demonstrates the value of considering results from multiple methods when studying bacterial communities.
Introduction
In recent years, reports of cyanobacterial blooms in freshwater ecosystems attributed to eutrophication and global warming have increased [1–6]. Cyanobacteria rapidly form massive water blooms under favorable conditions, causing economic, ecological and health problems [7–9]. Many bloom-forming species produce toxins and the types of toxin produced are dependent on the species composition within the bloom [10–12]. Cyanobacteria coexist and interact with heterotrophic bacterial groups commonly found in freshwater ecosystems [13, 14]. They may produce various dissolved organic compounds and fix atmospheric nitrogen, supplying heterotrophic bacteria with substrates for growth [15–17]. Such compounds can differentially support growth of heterotrophic bacteria species [18]. Therefore, it is likely that the composition of cyanobacterial blooms will structure the associated heterotrophic bacterial populations [14].
Studies on phytoplankton cultures demonstrated a larger difference in bacterial community structure between diatom and cyanobacteria as compared to different cyanobacterial cultures. This is dependent on phytoplankton growth phase and the habitat of the associated bacterial community (free-living within bloom vs attached to the phytoplankton cell surface) [18]. In freshwater ecosystems, environmental conditions, including water temperature, residence time and mixing, light intensity and quality, nutrient concentrations and grazing pressure affect the development of cyanobacterial population [19].
Blooms in temperate regions, occur mostly during late summer and early autumn [6, 20] and are dominated by Microcystis, Aphanizomenon, Anabaena together with the invasive species e.g. Cylindrospermopsis raciborskii [21, 22]. Freshwater lakes are numerous in the Nordic countries: Norway (65,000 lakes), Sweden (95,700 lakes) and Finland (187,888 lakes) [23]. Almost 40% of Norwegian water bodies are at risk from human influences such as acidification and eutrophication; climate change is likely to exacerbate these problems in the future (www.environment.no, Freshwater 2015). Reports have shown that increase in water temperature in major European lakes, coupled with decreased duration of ice cover, has caused changes in the life cycle of phytoplankton and promoted the invasion of toxic species from warmer regions [3, 24].
Previous, culture-based, studies in Finland and Sweden have identified freshwater bacterial groups Bacteriodetes, Proteobacteria, Firmicutes, Planctomycetes, Verrucomicrobia, Acidobacteria, Chloroflexi, and Thermomicrobia in association with cyanobacteria, along with potentially pathogenic bacterial groups such as Pseudomonas, Aeromonas and Vibrio [13, 14].
Culture-dependent methods have shortcomings, as up to 95% of the microorganisms may be uncultivable [25–27]. High throughput sequencing, in contrast to culture dependent studies, can detect a large majority of microbial taxa present. This helps generate a deeper understanding when comparing bacterial communities [18, 28]. In this study we have used HTS to assess the bacterial community associated with phytoplankton during the blooming season in the eutrophic Norwegian lake Akersvannet by targeting two hypervariable regions, V1-V3 and V3-V4, of the 16S rRNA gene. To the best of our knowledge, this is the first study of the bacterial community in a freshwater ecosystem in Norway using HTS 16S rRNA amplicon sequencing.
Materials and methods
Study site and sampling
Lake Akersvannet is situated in Stokke Municipality, Vestfold County (South Norway). The lake has a mean depth of 6 m, maximum depth of 13 m and a total area of 2.4 km2 (Fig 1). It is an important wetland habitat for birds during the spring and autumn migration. During summer and fall cyanobacterial blooms occur frequently [29]. The lake is ice-covered during the winter. The lake is surrounded by agricultural land and is used for recreational activities including water sports and fishing. The first documented bloom at this lake was identified as Aphanizomenon flos-aquae (NIVA 1959). Lowering of the water level due to canalization during the year 1968 led to a shift in species composition. By the year 1994, the phytoplankton community was dominated by species of Anabaena and Aphanizomenon and the dinoflagellate Ceratium hirundinella [30].
Fig 1. Map showing Lake Akersvannet.
The sampling point is indicated by a blue circle [source; Mapping Authority (Norwegian Geographical Survey), www.norgeskart.no]. The insert in the right-hand corner shows the position of the lake on the map of southern Norway.
This study was conducted in close cooperation with Vestfold County administration and the group of land owners surrounding the lake. In accordance with “Section IV” of the “Preservation regulations for Lake Akersvannet nature reserve” (www.lovdata.no) and “Section 3.3” of the “Management plan for Lake Akersvannet” (www.fylkesmannen.no), specific permission for taking water samples for research and educational purposes is not needed. Water samples were collected monthly from June to August 2013. Samples were taken at the sampling point indicated in Fig 1 (59.25° N 10.33° E). Separate one liter water samples were collected at depths of 0 m, 2 m and 4 m respectively. Net samples (25 μm pore size) for microscopic analysis were collected at the same time and location. Samples were immediately transferred to a cooled chamber and stored at 4°C.
Chemical analysis
The chemical analysis of water quality parameters was performed in accordance with Norwegian standards (NS): pH (NS 4720), oxygen (NS 5813), total nitrogen (TN) (NS 4743), total phosphorus (TP) (NS 4725, 1984) and chlorophyll a (NS 4766 (1/1983)). Ammonia (NH4 +) and nitrate (NO3-) were analyzed on Dionex ICS 1100 ion chromatograph (Thermo Fisher Scientific, California, USA) according to the manufacturer’s instructions.
Toxin analyses
Enzyme-Linked Immunosorbent Assay technique (ELISA) with Microcystin ADDA ELISA Kit (product No. 520011) from Abraxis (Abraxis LLC, Warminster PA) was used for the microcystin analyses. All samples were freeze-thawed twice to lyse the cells and release the microcystins into the water. An AccuReader (Metertech, Taiwan) model 965 was used to read the absorbance at 450 nm.
Qualitative and quantitative analyses of phytoplankton
Net samples were fixed with Lugol solution and examined under 100 and 400 x magnification for qualitative determination of the phytoplankton community (S1 Table). Phytoplankton enumeration was performed by Utermöhl inverted microscope technique using a 10ml sedimentation chamber. Species identification was performed according to Tikkanen and Willén [31] (S2 Table).
DNA extraction
For each sampling date, three 1L samples, collected at depths of 0 m, 2 m and 4 m were available. Each sample was filtered through four to five 1.2 μm pore size, 47 mm diameter GF/C filters (Whatman, Norway). Each set of filters was cut into fragments in mm-sized pieces with a disposable sterile scalpel blade and incubated in XS buffer for 120 min and subjected to DNA extraction using the Xanthogenate-SDS (XS) method [32]. DNA pellets were washed twice with 70% ethanol, air-dried and suspended in 50μl TE buffer (10 mM Tris-HCL, 1 mM EDTA pH 8). The DNA extract from each set of filters were pooled to prepare one DNA pool per sampling depth. DNA pools from each sampling depth (0, 2 and 4 m) were then combined, giving an integrated sample. This integrated sample was then split into two equal volumes (replicates), and each replicate was used as a template for amplification and sequencing of V1-V3 and V3-V4 target regions of the 16S rRNA gene.
PCR amplification and paired-end illumina sequencing
PCR amplification and sequencing was performed at Biomedicum Functional Genomics Unit, University of Helsinki, Finland. Each DNA replicate was used as template for amplifying two hypervariable regions of the 16S rRNA gene, using primers pairs V1-V3-27F (AGAGTTTGATCMTGGCTCAG), V1-V3-519R (GWATTACCGCGGCKGCTG) [33], V3-V4-341F (CCTACGGGNGGCWGCAG) and V3-V4-805R (GACTACHVGGGTATCTAATCC) [34]. Illumina adapter overhang sequences were attached to the 5’ end of each primer. PCR amplification and sequencing was performed according to Illumina’s 16S Metagenomic Sequencing Library Preparation guide (Part # 15044223 Rev. B). After PCR amplification the libraries were quantified using a Qubit Fluorometer (dsDNA HS) and the size of each library was estimated using the Agilent 2100 Bioanalyzer (DNA HS). The PCR libraries were normalized, pooled and then sequenced using the Illumina MiSeq System. Sequencing was performed using MiSeq 2 × 300 bp paired-end strategy and MiSeq Reagent Kit V3. For each replicate, as PCR amplicons from two different 16S hypervariable regions were pooled together and sequenced, no further barcoding was required [35].
Data analysis
The sequence reads were analyzed using UPARSE v8.0.1623 [36], which allows accurate identification of Operational Taxonomic Units (OTU`s) (http://drive5.com/usearch/manual/uparse_pipeline.html). The raw paired-end sequence reads were merged using the command usearch -fastq_mergepairs with maximum expected error threshold set at 1.0. The merged reads were filtered on the basis of sequence quality using fastq_filter program, setting maximum expected error threshold to 0.5 and minimum sequence length to 250 bp. After quality filtering, the reads were aligned against the 16S rRNA sequences of the RDP database using the Aligner tool [37] in the RDP pipeline available at Ribosomal Database Project (RDP Release 11, Update 4), and endpoints were identified in order to allow their sorting into V1-V3 and V3-V4 reads. For both classes of reads, dereplication was performed to collapse identical reads into one single sequence. Following dereplication, abundance sorting was done and singletons were discarded. OTU`s were clustered at 3% divergence threshold and chimeras were eliminated using UCHIME [38]. Representative sequences for each OTU were selected and RDP classifier (16S rRNA training set 14) (S1 and S2 Texts) [39] was used to assign taxonomy. OTU’s assigned to Archaea and Chloroplast were filtered out and OTU tables were generated. Random subsampling of the OTU tables for both target regions (S3 and S4 Texts) was performed using script single_rarefaction.py in QIIME (Quantitative Insights Into Microbial Ecology package 1.9.1)[40]. Shared OTU’s between replicates were computed using script shared_phylotypes.py (S5 and S6 Texts). Diversity indices (Chao1, ACE and Shannon diversity) were calculated for the subsampled OTU tables using script alpha_diversity.py. The OTU’s were further classified using BLAST analysis [41] using nucleotide database (nt/nr) with uncultured/environmental sample sequences excluded. OTU’s were only assigned to a species, if the sequence similarity was ≥ 98% and no other species showed the same level of similarity. Where this was not achieved, the RDP taxonomic assignment was retained.
Accession numbers
The raw 16S rRNA dataset from Lake Akersvannet sample are available at Sequence Read Archive (SRA) under study SRP095055.
Results
The environmental conditions and phytoplankton biomass in Lake Akersvannet during the sampling season 2013 are shown in Table 1. TN ranged from 1.5 to 1.8 mg/l and TP varied between 90 and 148 μg/l. The highest water temperature (20.7°C) was recorded in August. The highest concentration of microcystin (0.5 μg/l) was detected in August. In June and July, the concentration of microcystin was below the detection limit of 0.15 μg/l.
Table 1. Environmental parameters and phytoplankton biomass in Lake Akersvannet during the sampling season 2013.
| Environmental Parameters | June | July | August |
|---|---|---|---|
| Temp (°C) * | 17.3 (± 0.3) | 19.1 (± 0.6) | 20.7 (± 2) |
| pH* | 8.2 (± 0.1) | 8.7 (± 0.3) | 8.3 (± 0.5) |
| O2 (%)* | 90 (± 2) | 103 (± 9) | 90 (± 15) |
| Total Nitrogen (mg/l) * | 1.8 (± 164) | 1.7 (± 30) | 1.5 (± 300) |
| Total Phosphorus (μg/l) * | 111(± 12) | 90 (± 47) | 148 (± 57) |
| Ammonia (μg/l) * | 53 (± 92) | 0 | 223 (± 76) |
| Nitrate (μg/l) * | 1590 (± 310) | 1530 (± 62) | 710 (± 16) |
| Chlorophyll a (μg/l) ** | 9 (± 1) | 30 (± 3) | 52 (± 0) |
| Total phytoplankton biomass (mg/l) *** | 0.4 | 1.4 | 16 |
| Biomass of Cyanobacteria (mg/l) *** | 0.1 | 1.2 | 0.1 |
| Cyanobacteria (% of total phytoplankton) | 24 | 77 | 1 |
| Biomass of Ceratium hirundinella (mg/l) *** | 0.04 | 0.05 | 15.4 |
| Ceratium hirundinella (% of total phytoplankton) | 11 | 3.5 | 96 |
| Microcystin (μg/l) ** | <0.15 | <0.15 | 0.5 (± 0.8) |
* mean values for depth 0-4m,
** mean values from photic zone,
*** mixed sample from photic zone.
Numbers in parentheses show standard deviation
Phytoplankton community composition: Microscopic identification
Total phytoplankton biomass varied between 0.4 and 16 mg/l (Table 1) with mean biomass of 5.9 mg/l. Microscopic examination revealed variation in species composition of the phytoplankton community throughout the sampling season. Cyanobacteria represented 77% of total phytoplankton biomass in July, whereas the dinoflagellate Ceratium hirundinella dominated the community in August (96%). Species level morphological identification of Cyanobacteria showed the presence of Aphanizomenon flos-aquae (June-August), Microcystis aeruginosa (June-August), Chroococcous turgidus (June) and Woronichinia naegeliana (August). Aphanizomenon dominated the cyanobacterial community throughout the season. The proportion of Microcystis was increased in August (S2 Table).
Bacterial community composition based on 16S rRNA amplicon sequencing
In total, 16,548 V1-V3 and 35,028 V3-V4 reads were obtained using RDP classifier (38). These were assigned to 255 and 502 bacterial OTU’s respectively.
Further analysis using BLAST allowed identification of 14 V1-V3 OTU’s at species level and three at genus level; similarly, 66 and nine V3-V4 OTU’s could be identified at species and genus level respectively (S3 Table). Six V1-V3 and 30 V3-V4 OTU’s, were highly similar (≥ 99%) to unclassified bacterial strain reported in other studies (S4 Table). Taxonomic classification using RDP classifier and BLAST were largely in agreement. There were two discrepancies at phylum level.
Bacterial richness and diversity indices (Alpha diversity) are shown in Table 2. Diversity was highest in July. Diversity was higher for the V3-V4 region than for V1-V3. Replicates gave comparable diversity estimates for each month, except August; August A had fewer OTU’s and lower diversity estimates. Comparison of phylotypes revealed that 76% of V1-V3 and 82% of V3-V4 OTU’s detected in August A were shared with August B (S5 and S6 Texts).
Table 2. Richness and Biodiversity indices for L. Akersvannet during the sampling season 2013.
| Sampling month | Observed OTU’s (V1-V3) |
Observed OTU’s (V3-V4) |
Chao1 (V1-V3) |
Chao1 (V3-V4) |
ACE (V1-V3) |
ACE (V3-V4) |
Shannon diversity (V1-V3) |
Shannon diversity (V3-V4) |
|---|---|---|---|---|---|---|---|---|
| June A | 138 | 272 | 152 | 340 | 151 | 324 | 5.6 | 6.4 |
| June B | 141 | 272 | 157 | 333 | 161 | 329 | 5.5 | 6.5 |
| July A | 188 | 385 | 215 | 422 | 211 | 415 | 5.8 | 7.2 |
| July B | 179 | 389 | 201 | 403 | 202 | 407 | 5.8 | 7.3 |
| August A | 46 | 107 | 63 | 137 | 66 | 137 | 1.9 | 2.2 |
| August B | 86 | 187 | 161 | 246 | 158 | 242 | 2.7 | 3.5 |
After completion of taxonomic analysis, 255 V1-V3 OTU’s were assigned to 11 phyla and one group of “Unclassified Bacteria”, and 502 V3-V4 OTU’s were assigned to 17 phyla and one group of “Unclassified Bacteria”. A comparison of taxonomic distribution of reads between the two 16S rRNA target region libraries is shown in Tables 3 and 4. This comparison showed that 10 phyla were shared between both target regions, whereas respectively 1 and 7 phyla were exclusively identified in V1-V3 and V3-V4 sequence library. Phylum level distribution of reads for both the target regions is shown in Fig 2. The proportion of reads belonging to “Unclassified Bacteria” and Proteobacteria-alpha is higher in V1-V3 than V3-V4, whereas the proportion of Verrucomicrobia and Bacteriodetes is higher in V3-V4 sequence library. Overall proportions of unclassified OTUs were higher in the V1-V3 library (71%) than the V3-V4 (61%) (S5 Table).
Table 3. Comparison of taxonomic distribution of reads between V1-V3 and V3-V4 target regions.
| Phylum | V1-V3 region | V3-V4 region | ||||
|---|---|---|---|---|---|---|
| OTU count | Read count | Percent | OUT count | Read count | Percent | |
| Acidobacteria | 5 | 167 | 1.01% | 20 | 640 | 1.83% |
| Actinobacteria | 15 | 844 | 5.10% | 31 | 2303 | 6.57% |
| Armatimonadetes | 2 | 10 | 0.06% | 5 | 113 | 0.32% |
| Bacteroidetes | 23 | 259 | 1.57% | 124 | 5732 | 16.36% |
| Candidate division WPS-1 | 0 | 0 | 0% | 1 | 13 | 0.04% |
| Candidatus Saccharibacteria | 0 | 0 | 0% | 1 | 20 | 0.06% |
| Chlamydiae | 0 | 0 | 0% | 1 | 2 | 0.01% |
| Chloroflexi | 0 | 0 | 0% | 3 | 6 | 0.02% |
| Cyanobacteria | 6 | 6958 | 42.05% | 10 | 10834 | 30.93% |
| Firmicutes | 1 | 65 | 0.39% | 1 | 378 | 1.08% |
| Gemmatimonadetes | 0 | 0 | 0% | 3 | 96 | 0.27% |
| Hydrogenedentes | 0 | 0 | 0% | 1 | 37 | 0.11% |
| Nitrospirae | 0 | 0 | 0% | 1 | 12 | 0.03% |
| Parcubacteria | 2 | 10 | 0.06% | 6 | 28 | 0.08% |
| Planctomycetes | 8 | 144 | 0.87% | 24 | 1230 | 3.51% |
| Proteobacteria* | 121 | 6201 | 37.47% | 185 | 9534 | 27.22% |
| Spirochaetes | 1 | 9 | 0.05% | 0 | 0 | 0% |
| Unclassified Bacteria | 69 | 1852 | 11.19% | 47 | 1039 | 2.97% |
| Verrucomicrobia | 2 | 29 | 0.18% | 38 | 3011 | 8.60% |
| Total | 255 | 16548 | 100% | 502 | 35028 | 100% |
* Please refer to Table 4 for class level breakup of phylum Proteobacteria.
Table 4. Comparison of class level taxonomic distribution of proteobacterial reads between V1-V3 and V3-V4 target regions.
| Phylum Proteobacteria | V1-V3 region | V3-V4 region | ||||
|---|---|---|---|---|---|---|
| OTU count | Read count | Percent | OTU count | Read count | Percent | |
| Proteobacteria- Oligoflexia | 0 | 0 | 0% | 1 | 1 | 0.01% |
| Proteobacteria-α | 82 | 5502 | 88.73% | 66 | 2718 | 28.51% |
| Proteobacteria-β | 14 | 346 | 5.58% | 56 | 2644 | 27.73% |
| Proteobacteria-γ | 8 | 185 | 2.98% | 35 | 2665 | 27.95% |
| Proteobacteria-δ | 2 | 30 | 0.48% | 15 | 386 | 4.05% |
| Unclassified "Proteobacteria" | 15 | 138 | 2.23% | 12 | 1120 | 11.75% |
| Total | 121 | 6201 | 100% | 185 | 9534 | 100% |
Fig 2. Phylum level distribution of reads for V1-V3 and V3-V4 hypervariable region.
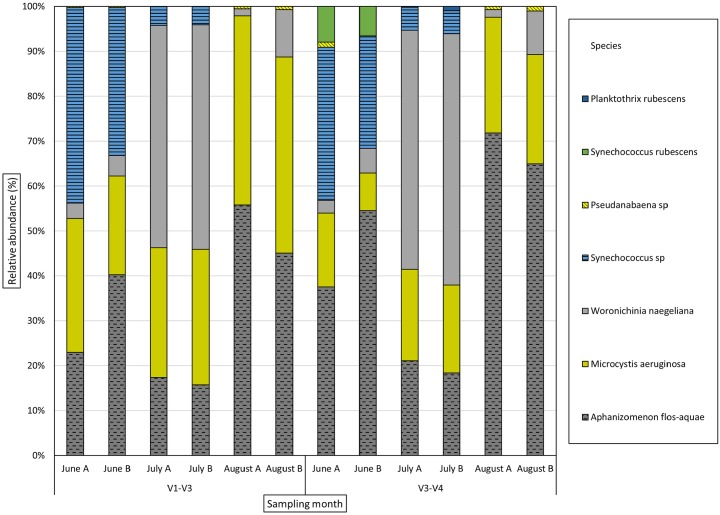
The phylum and genus level bacterial community distribution is shown in Figs 3 and 4 respectively. The bacterial community was dominated by 5 phyla; namely Actinobacteria, Proteobacteria, Cyanobacteria, Verrucomicrobia and Bacteriodetes along with “Unclassified Bacteria” in varying proportions. Cyanobacteria were detected throughout the sampling period, but dominated both sequence libraries in August. Genera identified were Aphanizomenon (GpI), Microcystis (GpXI), Synechococcus (GpIIa), Pseudanabaena (GpVI), Planktothrix and Woronichinia. Species level distribution of total cyanobacterial reads is shown in Fig 5. The same dominant cyanobacterial genera and species were identified from both the 16S rRNA target regions (data in S3 Table).
Fig 3. Phylum level distribution (%) of bacterial community in Lake Akersvannet from June to August 2013.
Fig 4. Genus level distribution (%) of bacterial community in Lake Akersvannet from June to August 2013.
Genera with abundance <0.5% were combined and represented as “Others”.
Fig 5. Species level distribution (%) of cyanobacterial community in Lake Akersvannet from June to August 2013.
Comparison of microscopic and sequence-based cyanobacterial species distribution
The cyanobacterial species distribution derived from microscopic and sequence-based analysis is compared in Table 5. The same major species were found with both methods, but relative species abundance estimates differed.
Table 5. Comparison of microscopic and sequence-based cyanobacterial species distribution (L. Akersvannet 2013).
| Identified | June | July | August | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mic | 16S | 16S | Mic | 16S | 16S | Mic | 16S | 16S | |
| Cyanobacterial Species | (V1-V3) | (V3-V4) | (V1-V3) | (V3-V4) | (V1-V3) | (V3-V4) | |||
| Aphanizomenon flos-aquae | 95.80% | 31.5% | 47.9% | 99.80% | 16.5% | 19.7% | 77.30% | 50.62% | 68.7% |
| Microcystis aeruginosa | 2.10% | 26% | 11.5% | 0.20% | 29.5% | 19.9% | 14% | 42.86% | 25.1% |
| Chroococcous turgidus | 2.10% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% |
| Woronichinia naegeliana | 0% | 4.0% | 4.4% | 0% | 49.8 | 54.6% | 8.70% | 5.90% | 5.4% |
| Synechococcus sp. | 0% | 38.3% | 28.6% | 0% | 4.2 | 5.1% | 0% | 0% | 0% |
| Pseudanabaena sp. | 0% | 0.3% | 0.4% | 0% | 0% | 0% | 0% | 0.62% | 0.9% |
| Unclassified Cyanobacteria | 0% | 0% | 0% | 0% | 0% | 0.1% | 0% | 0% | 0% |
| Synechococcus rubescens | 0% | 0% | 7.1% | 0% | 0% | 0.1% | 0% | 0% | 0% |
| Planktothrix rubescens | 0% | 0% | 0% | 0% | 0% | 0.5% | 0% | 0% | 0% |
| Total | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
(Mic) Abundance (percentage of cyanobacteria) based on microscopic biomass estimation.
(16S) Abundance (percentage of cyanobacteria) based on 16S gene abundance data. Only cyanobacterial reads are included in the analysis. The values shown are the average of two replicates.
Discussion
The mean phytoplankton biomass over the study period was 5.9 mg/l, making this lake hypereutrophic according to Brettum and Andersen [42]. Many factors regulate the composition of phytoplankton communities, but TN, TP and the ratio between TN and TP are considered particularly important [43, 44]. These parameters fluctuated moderately around means of TN 1.6 mg/l (SD ± 0.1 mg/l) and TP 116.3 μg/l (SD ± 23 μg/l) in this study. Although the low TN: TP ratios observed (10 to 18), were favorable for cyanobacterial blooms, the blooming organism was instead C. hirundinella, which also favors low TN: TP (i.e. < 7) as has been reported earlier in the literature [45–48]. It plausible that once a Ceratium bloom was established, it suppressed cyanobacterial bloom by competition for light and nutrients or by grazing [48, 49].
The increased proportions of Cyanobacteria observed in the 16S sequence library in August, could be due to the increase in water temperature (20.7°C) (Table 1). This temperature is optimal for growth of Aph. flos-aquae but suboptimal for M. aeruginosa. The lowest temperature at which Aph. flos-aquae can grow is reported to be 8°C [50]; optimum being 15 to 28°C [51], whereas M. aeruginosa can grow slowly at 15°C [52]; optimum being 27.5 to 32°C [51, 53]. Aph. flos-aquae has higher photosynthetic ability at lower temperatures as compared to M. aeruginosa [52]. In August, ammonium levels rose markedly and this was accompanied by an increase of M. aeruginosa (which favors ammonium as nitrogen source) [54, 55]. This would in turn explain the increase of microcystin in August.
Comparison between the two target regions
Although less discriminatory than full length 16S rRNA sequencing, high throughput sequencing of shorter hypervariable regions (V1 to V9) using universal primers has become common practice in microbial community studies [35, 56]. Different hypervariable regions have been targeted in different studies with no one region being preferred over others [57, 58]. In our study we have utilized two of the most commonly used primer pairs, targeting the V1-V3 and V3-V4 region of the 16S rRNA gene respectively [59]. Of the ten phyla detected by both primer sets, five (Cyanobacteria, Proteobacteria, Actinobacteria, Acidobacteria and Parcubacteria) showed similar abundance profiles in both target sequence libraries, while the other five (Bacteriodetes, Verrucomicrobia, Armatimonadetes, Firmicutes and Planctomycetes) were markedly more abundant in the V3-V4 library. Eight phyla were detected by only one of the primer sets. This suggests that the primers used in this study selectively detect certain phyla and neither is truly universal [34]. For cyanobacteria, however, the results were highly concordant, suggesting that both primers sets have similar coverage for this phylum. Although the overall proportion of phylum Proteobacteria in both libraries was comparable, Alphaproteobacteria were higher in proportion in the V1-V3 library. This variation might be explained by higher coverage of V1-V3 primers for class Alphaproteobacteria as seen in studies conducted in other environment, but will require further experimentation to confirm these findings for freshwater environment [60]. The proportion of unclassified Bacteria and overall unclassified genera was greater in the V1-V3 library. Thus, use of different sets of universal primers can lead to different apparent community structures. During PCR amplification, target sequences that do not precisely match the universal primers will be either less efficiently amplified or lost completely [61] and use of particular sets of universal primers can lead to under- or over-estimation of taxa [35].
Comparison of community richness and diversity estimates showed greater diversity for V3-V4 than for V1-V3 libraries. Such discrepancies have been reported in other studies [35, 61]. Replicates gave comparable diversity estimates, except for samples August A and B. Even after random subsampling was performed, August A had fewer OTU’s than August B. Such discrepancies have been reported in other studies and attributed to errors in PCR amplification, library preparation or sequencing steps [61]. In our case as both target regions were equally depleted of OTUs in August A, the discrepancy is most probably due to differences in sequencing efficiency as V1-V3 and V3-V4 were sequenced together but amplified separately.
The bacterial community was dominated by five phyla; Proteobacteria, Bacteriodetes, Cyanobacteria, Actinobacteria and Verrucomicrobia in varying proportions throughout the study period. Genus level taxonomic identifications showed the presence of a great diversity of low abundance taxa. Previous studies refer to these low abundance taxa as “rare biosphere”. It has been suggested that members of the rare biosphere may play important roles in the bacterial community, despite their scarcity [18, 62]. In addition to the rare biosphere, many unclassified OTU’s were detected, suggesting that as-yet undescribed bacterial groups with unknown metabolic capabilities are an important part of the lake microbiota, a matter which warrants further investigation. The large proportion of unclassified bacterial taxa also suggests that taxa peculiar to the Norwegian freshwater environment may be absent from the cultured and sequenced species represented by commonly used 16S rRNA databases.
Bacterial community distribution
Bacteriodetes, predominantly Flavobacteriia and Sphingobacteriia dominated the bacterial community throughout the sampling season. These are mostly chemoorganotrophs, that are well adapted to bloom conditions due to their ability to degrade complex biomolecules [18, 63, 64] such as algal-derived material [65] including cyanobacterial toxins [13]. Flavobacteriia and Sphingobacteriia have been found in high numbers during episodes of phytoplankton blooms [66], particularly cyanobacterial blooms in freshwater ecosystems [14, 67]. A study conducted in Sweden in 2007 reported multiple populations of Flavobacteriia in a bloom of cyanobacteria, without any seasonal succession and suggested that resource availability (organic matter) might be the driving force behind Flavobacteriia community structuring [64]. The genus Flavobacterium, is known to contain species that can lyse Microcystis cells [68] and degrade cyanobacterial hepatotoxins [13]. Member of this genus are also able to degrade complex organic molecules [64]. The dominant Flavobacterium taxon in June (OTU_11, V3-V4 library) showed high sequence similarity to a microcystin-degrading strain (Flavobacterium sp. AKB-2008 TE28) isolated in a previous study [13]. Fluviicola, another dominant genus of class Flavobacteriia detected in June and August is reported to contain species responsible for denitrification [69].
Actinobacteria have been found in various aquatic habitats and are considered to be ubiquitous members of the freshwater ecosystem, especially in the lake epilimnia [66, 70, 71]. In this study, the phylum was represented by Actinomycetales and Acidimicrobiales, two of the most abundant freshwater Actinobacterial orders [66, 72]. The ratio of allochthonous to autochthonous carbon sources, which may vary greatly during period of phytoplankton bloom, can be selective for different Actinobacterial species [73]. Actinobacteria have been previously associated with blooms of diatoms and cyanobacteria [14, 18, 71, 74, 75]. In this study, their abundance declined markedly from June to July while the cyanobacterial abundance increased. This might be due to water temperature increasing above the optimum for Actinobacteria [75] and/or competition from other, faster growing, bacterial phyla [76]. Ilumatobacter, one of the dominant Actinobacteria genera in June (V3-V4 library), are known to thrive in environments with elevated phosphorous levels [77], as found in L. Akersvannet.
In this study, Verrucomicrobia comprised 8.6% of the bacteria detected in the V3-V4 library. Although most studies report low abundance of Verrucomicrobia, much higher abundance was reported in a recent culture independent study [66, 78]. Verrucomicrobia abundance may have previously been underestimated due to poor cultivability and methodological biases [66, 78–80]. They are known degraders of algal polysaccharides and organic matter [81–83]. Hence their presence during a phytoplankton bloom is not unexpected [84]. A representative of Subdivision3 (OTU_8, V3-V4 library), which dominated in July, showed high sequence similarity to a strain known to degrade laminarin, one of the most abundant algal polysaccharides in nature [83].
Most of the OTU’s belonging to Actinobacteria and Verrucomicrobia, detected in this study were highly similar to isolates previously identified in North American lakes [83, 85], which suggests that these strains are geographically widespread. Comparable reports, suggesting that unique capabilities of these groups allow them to compete successfully in lakes with diverse trophic states have been published [70, 86].
Proteobacteria represented a significant portion of the bacterial community, which is in agreement with other studies [74, 87, 88]. Many of the most prevalent and well-studied freshwater bacterial groups belong to Betaproteobacteria [66, 89]. Paucibacter (Betaproteobacteria) is represented by a single dominant OTU (OTU_6) which showed high sequence similarity to Paucibacter toxinivorans, a novel species isolated from lake sediment. P. toxinivorans can degrade the cyclic cyanobacterial hepatotoxins and mineralize organic phosphorous compounds [90]. However, the abundance of P. toxinivorans was higher in July (with toxin level below detection threshold) than in August (high toxin concentration detected), suggesting that the toxin presence is not critical for the growth of this species. Limnohabitans, one the betaproteobacterial genus identified in July (V1-V3 library), are opportunistic freshwater bacteria that prefer non-acidic environments [91–93]. Their ability to use low molecular weight algal exudates as sole organic carbon source (DOC) and rapid growth rate gives them a competitive advantage during phytoplankton bloom [76, 91, 94].
Alphaproteobacteria were the dominant proteobacterial group in this study. In contrast to Betaproteobacteria, Alpha and Gammaproteobacteria rarely dominate freshwater bacterial communities, except under conditions of elevated organic and inorganic inputs during phytoplankton blooms [13, 66, 76].
One of the Gammaproteobacteria OTU's found in this study was a Rheinheimera species. Rheinheimera are abundant in marine, freshwater and estuarine environments and are able to grow on and degrade organic matter [95–97]. Other Gammaproteobacteria were Pseudomonas and Xanthomonadaceae. These groups include opportunistic pathogens of humans, fish and plants [98, 99]. Pseudomonas and Rheinheimera are thought to enhance the growth of Microcystis by modulating phosphate exchange in the cyanobacterial mucilage capsule [13, 100].
Sphingorhabdus and Novosphingobium (Alphaproteobacteria) are physiologically diverse generalists capable of aerobic anoxygenic photosynthesis and degrading polycyclic aromatic compounds [75, 101–105]. Candidatus Pelagibacter, one of the dominant genera in the V1-V3 library, belongs to the SAR11 lineage which is known to play important role in phosphate metabolism in aquatic ecosystems [106]. Roseomonas, a member of family Acetobacteraceae, has been previously been associated with cyanobacterial blooms and attaches to the cell surface of M. aeruginosa [14, 18]. Another Alphaproteobacteria species was Caulobacter profundus [107]. Caulobacter are known to be associated with cyanobacterial blooms and enhance the growth of Microcystis [13, 108]. The highest cyanotoxin concentration, in August, coincided with the greatest diversity of potentially cyanotoxin-degrading species like Caulobacter, Novosphingobium, Sphingorhabdus (Alphaproteobacteria), Paucibacter, Flavobacterium (Betaproteobacteria) and Pseudomonas (Gammaproteobacteria). It has been suggested that cyanotoxins provide a competitive advantage to cyanotoxin-degraders by serving as nutritional supplement [109–111].
Comparison of microscopic and sequence based results
The microscopic findings and 16S rRNA sequence analysis were in agreement in regard to the major species present. The quantitative results however were not in concordance. Similar findings have been reported previously [112, 113]. This discrepancy needs to be addressed, as it is commonly assumed that read counts are semi-quantitative and will provide meaningful relative abundance estimates [114]. A recent study has shown that Cyanobacteria were overrepresented in HTS data as compared to microscopic biovolume data. Thus, for the moment both methods are needed to obtain a picture of the cyanobacterial community structure [115].
The discrepancies observed indicate that there is significant bias in one or both of the methods used. Microscopy is a subjective method, dependent on the skill and experience of the microscopist and overestimation or underestimation of phytoplankton diversity may occur [112, 116, 117]. In addition, differential losses of cells may occur during fixation, depending on the species and cell size [113, 118, 119]. Smaller cells that do not sink fast enough within the counting chamber can be underrepresented or missed completely [115]. 16S amplicon read counts are subject to biases caused by cell lysis efficiency, DNA extraction efficiency, PCR amplification efficiency, varying 16S rRNA gene copy number, quality filtering criteria and the sequencing method used [14, 113, 114, 120–123]. Our results illustrate the importance of using multiple methods while characterizing freshwater microbiota. Additional experimentation is required to resolve the observed discrepancies.
Conclusion
To our knowledge, this study is the first use of 16S rRNA targeted amplicon sequencing to characterize the bacterial communities associated with phytoplankton blooms in a eutrophic lake in Norway. Our study revealed that choice of 16S rRNA region can affect the apparent community structure and this needs to be taken into account in the planning and interpretation of such studies. Our results showed a highly complex and fluctuating microbial flora, mostly comprised of taxa previously found in association with phytoplankton blooms. A significant proportion of the community was represented by low abundance bacterial groups and unclassified taxa that might be performing hitherto unidentified ecological functions. Our study also showed that the resolving power and the quantitative results of microscopic and 16S rRNA targeted sequence analysis may differ, which needs further investigation.
The 16S rRNA results, provided a snapshot of the phytoplankton-associated bacterial community, at three individual time points. As phytoplankton communities are known to be highly dynamic, more frequent sampling will be needed to identify trends.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
Acknowledgments
The authors would like to thank Karin Brekke Li for technical assistance and guidance and Brett Neilan (University of New South Wales, Australia) for providing the DNA extraction protocol. We acknowledge funding from SIU, Norway and UGC, India under the Indo-Norwegian Cooperation Program (INCP-2014). None of the funds received from the Indo-Norwegian Cooperation Program were used specifically for this study.
Data Availability
All relevant data are within the paper and its Supporting Information files. All raw 16S rRNA dataset are available from the SRA database under study SRP095055.
Funding Statement
We acknowledge funding from SIU, Norway and UGC, India under the Indo-Norwegian Cooperation Program (INCP-2014). None of the funds received from the Indo-Norwegian Cooperation Program were used specifically for this study.
References
- 1.Paerl HW, Fulton RS 3rd, Moisander PH, Dyble J. Harmful freshwater algal blooms, with an emphasis on cyanobacteria. ScientificWorldJournal. 2001;1:76–113. 10.1100/tsw.2001.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paerl HW, Huisman J. Climate. Blooms like it hot. Science. 2008;320(5872):57–8. 10.1126/science.1155398 [DOI] [PubMed] [Google Scholar]
- 3.Sinha R, Pearson LA, Davis TW, Burford MA, Orr PT, Neilan BA. Increased incidence of Cylindrospermopsis raciborskii in temperate zones- is climate change responsible? Water Res. 2012;46(5):1408–19. 10.1016/j.watres.2011.12.019 [DOI] [PubMed] [Google Scholar]
- 4.Dokulil MT, Teubner K. Cyanobacterial dominance in lakes. Hydrobiologia. 2000;438(1–3):1–12. [Google Scholar]
- 5.Shatwell T, Kohler J, Nicklisch A. Warming promotes cold-adapted phytoplankton in temperate lakes and opens a loophole for Oscillatoriales in spring. Glob Chang Biol. 2008;14(9):2194–200. [Google Scholar]
- 6.Chorus EI, Bartram J. Toxic cyanobacteria in water: a guide to their public health consequences, monitoring and management. New York: E & FN Spon, published on behalf of the World Health Organization; 1999. p. 1–13. [Google Scholar]
- 7.Jochimsen EM, Carmichael WW, An JS, Cardo DM, Cookson ST, Holmes CE, et al. Liver failure and death after exposure to microcystins at a hemodialysis center in Brazil. N Engl J Med. 1998;338(13):873–8. 10.1056/NEJM199803263381304 [DOI] [PubMed] [Google Scholar]
- 8.Falconer IR, Humpage AR. Health risk assessment of cyanobacterial (blue-green algal) toxins in drinking water. Int J Environ Res Public Health. 2005;2(1):43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miguens D, Valerio E. The impact of some microcystins on the growth of heterotrophic bacteria from Portuguese freshwater reservoirs. Limnetica. 2015;34(1):215–25. [Google Scholar]
- 10.Vareli K, Pilidis G, Mavrogiorgou MC, Briasoulis E, Sainis I. Molecular characterization of cyanobacterial diversity and yearly fluctuations of Microcystin loads in a suburban Mediterranean Lake (Lake Pamvotis, Greece). J Environ Monit. 2009;11(8):1506–12. 10.1039/b903093j [DOI] [PubMed] [Google Scholar]
- 11.Blaha L, Babica P, Marsalek B. Toxins produced in cyanobacterial water blooms—toxicity and risks. Interdiscip Toxicol. 2009;2(2):36–41. 10.2478/v10102-009-0006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolman AM, Rucker J, Pick FR, Fastner J, Rohrlack T, Mischke U, et al. Cyanobacteria and cyanotoxins: the influence of nitrogen versus phosphorus. PloS One. 2012;7(6):e38757 10.1371/journal.pone.0038757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg KA, Lyra C, Sivonen K, Paulin L, Suomalainen S, Tuomi P, et al. High diversity of cultivable heterotrophic bacteria in association with cyanobacterial water blooms. ISME J. 2009;3(3):314–25. 10.1038/ismej.2008.110 [DOI] [PubMed] [Google Scholar]
- 14.Eiler A, Bertilsson S. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ Microbiol. 2004;6(12):1228–43. 10.1111/j.1462-2920.2004.00657.x [DOI] [PubMed] [Google Scholar]
- 15.Cole JJ, Likens GE, Strayer DL. Photosynthetically produced dissolved organic carbon: An important carbon source for planktonic bacteria. Limnol Oceanogr. 1982;27(6):1080–90. [Google Scholar]
- 16.Bertilsson S, Jones J. Supply of dissolved organic matter to aquatic ecosystems: autochthonous sources In: Findlay S, Sinsabaugh R, editors. Aquatic ecosystems: interactivity of dissolved organic matter: Academic Press; 2003. p. 3–25. [Google Scholar]
- 17.Louati I, Pascault N, Debroas D, Bernard C, Humbert JF, Leloup J. Structural diversity of bacterial communities associated with bloom-forming freshwater cyanobacteria differs according to the cyanobacterial genus. PloS One. 2015;10(11):e0140614 10.1371/journal.pone.0140614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagatini IL, Eiler A, Bertilsson S, Klaveness D, Tessarolli LP, Vieira AA. Host-specificity and dynamics in bacterial communities associated with Bloom-forming freshwater phytoplankton. PloS One. 2014;9(1):e85950 10.1371/journal.pone.0085950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynolds CS. The ecology of phytoplankton. Cambridge: Cambridge University Press; 2006. [Google Scholar]
- 20.Oliver RL, Ganf GG. Freshwater Blooms In: Whitton BA, Potts M, editors. The ecology of cyanobacteria: their diversity in time and space. Dordrecht: Springer Netherlands; 2002. p. 149–94. [Google Scholar]
- 21.Miller TR, McMahon KD. Genetic diversity of cyanobacteria in four eutrophic lakes. FEMS Microbiol Ecol. 2011;78(2):336–48. 10.1111/j.1574-6941.2011.01162.x [DOI] [PubMed] [Google Scholar]
- 22.Miller TR, Beversdorf L, Chaston SD, McMahon KD. Spatiotemporal molecular analysis of cyanobacteria blooms reveals Microcystis-Aphanizomenon interactions. PloS One. 2013;8(9):e74933 10.1371/journal.pone.0074933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henriksen A, Skjelkvale BL, Mannio J, Wilander A, Harriman R, Curtis C, et al. Northern European Lake Survey, 1995—Finland, Norway, Sweden, Denmark, Russian Kola, Russian Karelia, Scotland and Wales. Ambio. 1998;27(2):80–91. [Google Scholar]
- 24.Füssel H-M, Jol A. Climate change, impacts and vulnerability in Europe 2012 an indicator-based report. EEA, Copenhagen: 2012;1725–9177. [Google Scholar]
- 25.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(16):10494–9. 10.1073/pnas.142680199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torsvik V, Ovreas L, Thingstad TF. Prokaryotic diversity-magnitude, dynamics, and controlling factors. Science. 2002;296(5570):1064–6. 10.1126/science.1071698 [DOI] [PubMed] [Google Scholar]
- 27.Vinas M, Sabate J, Guasp C, Lalucat J, Solanas AM. Culture-dependent and -independent approaches establish the complexity of a PAH-degrading microbial consortium. Can J Microbiol. 2005;51(11):897–909. 10.1139/w05-090 [DOI] [PubMed] [Google Scholar]
- 28.Brooks JP, Edwards DJ, Harwich MD Jr., Rivera MC, Fettweis JM, Serrano MG, et al. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015;15:66 10.1186/s12866-015-0351-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schartau AK, Skjelbred B, Edvardsen H, Fløystad L, Jensen TC, Mjelde M, et al. Utprøving av system for basisovervåking i henhold til vannforskriften. Resultater for utvalgte innsjøer 2012. Miljøovervåking i vann 2013–14: 2013. Norwegian.
- 30.Utkilen H, Skulberg OM, Underdal B, Gjølme N, Skulberg R, Kotai J. The rise and fall of a toxigenic population of Microcystis aeruginosa (Cyanophyceae/Cyanobacteria)- a decade of observations in Lake Akersvatnet, Norway. Phycologia. 1996;35(6):189–97. [Google Scholar]
- 31.Tikkanen T, Willén T. Växtplanktonflora: Statens naturvårdsverk; 1992. Swedish. [Google Scholar]
- 32.Tillett D, Neilan BA. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J Phycol. 2000;36(1):251–8. [Google Scholar]
- 33.Winsley T, van Dorst JM, Brown MV, Ferrari BC. Capturing greater 16S rRNA gene sequence diversity within the domain Bacteria. Appl Environ Microbiol. 2012;78(16):5938–41. 10.1128/AEM.01299-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res Suppl. 2013;41(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claesson MJ, Wang Q, O'Sullivan O, Greene-Diniz R, Cole JR, Ross RP, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res Suppl. 2010;38(22):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–8. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- 37.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25(10):1335–7. 10.1093/bioinformatics/btp157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–7. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 42.Brettum P, Andersen T. The use of phytoplankton as indicators of water quality. Norwegian Institute for Water Research SNO Report. 2004;(4818):33. [Google Scholar]
- 43.Xie L, Xie P, Li S, Tang H, Liu H. The low TN:TP ratio, a cause or a result of Microcystis blooms? Water Res. 2003;37(9):2073–80. 10.1016/S0043-1354(02)00532-8 [DOI] [PubMed] [Google Scholar]
- 44.Wang HJ, Liang XM, Jiang PH, Wang J, Wu SK, Wang HZ. TN: TP ratio and planktivorous fish do not affect nutrient-chlorophyll relationships in shallow lakes. Freshw Biol. 2008;53(5):935–44. [Google Scholar]
- 45.de Tezanos Pinto P, Litchman E. Interactive effects of N:P ratios and light on nitrogen-fixer abundance. Oikos. 2010;119(3):567–75. [Google Scholar]
- 46.Pick FR, Lean DRS. The role of macronutrients (C, N, P) in controlling cyanobacterial dominance in temperate lakes. New Zeal J Mar Fresh. 1987;21(3):425–34. [Google Scholar]
- 47.Smith VH. Low nitrogen to phosphorus ratios favor dominance by blue-green algae in lake phytoplankton. Science. 1983;221(4611):669–71. 10.1126/science.221.4611.669 [DOI] [PubMed] [Google Scholar]
- 48.Moore VL, Moss B. Dinophyta characterise nitrogen scarcity more strongly than cyanobacteria in moderately deep lakes. Acta Protozool. 2015;52(3):203–16. [Google Scholar]
- 49.Wehr JD, Sheath RG. Freshwater algae of North America: ecology and classification. USA: Academic Press; 2003. [Google Scholar]
- 50.Tsujimura S, Ishikawa K, Tsukada H. Effect of temperature on growth of the cyanobacterium Aphanizomenon flos-aquae in Lake Biwa and Lake Yogo. Phycol Res. 2001;49(4):275–80. [Google Scholar]
- 51.Robarts RD, Zohary T. Temperature effects on photosynthetic capacity, respiration, and growth rates of bloom-forming cyanobacteria. New Zeal J Mar Fresh. 1987;21(3):391–9. [Google Scholar]
- 52.Wu WJ, Li GB, Li DH, Liu YD. Temperature may be the dominating factor on the alternant succession of Aphanizomenon flos-aquae and Microcystis aeruginosa in Dianchi lake. Fresen Environ Bull. 2010;19(5):846–53. [Google Scholar]
- 53.Imai H, Chang K-H, Nakano S-i. Growth responses of harmful algal species Microcystis (cyanophyceae) under various environmental conditions. Interdisciplinary Studies on Environmental Chemistry- Environmental Research in Asia. 2009:269–75. [Google Scholar]
- 54.Dai GZ, Shang JL, Qiu BS. Ammonia may play an important role in the succession of cyanobacterial blooms and the distribution of common algal species in shallow freshwater lakes. Glob Chang Biol. 2012;18(5):1571–81. [Google Scholar]
- 55.Harris TD, Smith VH, Graham JL, Van de Waal DB, Tedesco LP, Clercin N. Combined effects of nitrogen to phosphorus ratios and nitrogen speciation on cyanobacterial metabolite concentrations in eutrophic Midwestern USA reservoirs. Inland Waters. 2016;6(2):199–210. [Google Scholar]
- 56.Tremblay J, Singh K, Fern A, Kirton ES, He S, Woyke T, et al. Primer and platform effects on 16S rRNA tag sequencing. Front Microbiol. 2015;6(771):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PloS One. 2011;6(12):e27310 10.1371/journal.pone.0027310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fredriksson NJ, Hermansson M, Wilen BM. The choice of PCR primers has great impact on assessments of bacterial community diversity and dynamics in a wastewater treatment plant. PloS One. 2013;8(10):e76431 10.1371/journal.pone.0076431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng W, Tsompana M, Ruscitto A, Sharma A, Genco R, Sun Y, et al. An accurate and efficient experimental approach for characterization of the complex oral microbiota. Microbiome. 2015;3(48):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Albertsen M, Karst SM, Ziegler AS, Kirkegaard RH, Nielsen PH. Back to basics- The influence of DNA extraction and primer choice on phylogenetic analysis of activated sludge communities. PloS One. 2015;10(7):e0132783 10.1371/journal.pone.0132783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mao DP, Zhou Q, Chen CY, Quan ZX. Coverage evaluation of universal bacterial primers using the metagenomic datasets. BMC Microbiol. 2012;12(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao Y, Williams DD, Williams NE. How important are rare species in aquatic community ecology and bioassessment? Limnol Oceanogr. 1998;43(7):1403–9. [Google Scholar]
- 63.Cottrell MT, Kirchman DL. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66(4):1692–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eiler A, Bertilsson S. Flavobacteria blooms in four eutrophic lakes: linking population dynamics of freshwater bacterioplankton to resource availability. Appl Environ Microbiol. 2007;73(11):3511–8. 10.1128/AEM.02534-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Riemann L, Steward GF, Azam F. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl Environ Microbiol. 2000;66(2):578–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev. 2011;75(1):14–49. 10.1128/MMBR.00028-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Riemann L, Winding A. Community dynamics of free-living and particle-associated bacterial assemblages during a freshwater phytoplankton bloom. Microb Ecol. 2001;42(3):274–85. 10.1007/s00248-001-0018-8 [DOI] [PubMed] [Google Scholar]
- 68.Yamamoto Y, Niizuma S, Kuroda N, Sakamoto M. Occurrence of heterotrophic bacteria causing lysis of cyanobacteria in a eutrophic lake. Japanese journal of phycology Sapporo. 1993;41(3):215–20. [Google Scholar]
- 69.Cheng C, Zaichao Z, Aizhong D, Jiayan W, Jingfa X, Yujiao S. Bar-coded pyrosequencing reveals the bacterial community during Microcystis water bloom in Guanting reservoir, Beijing. Procedia Eng. 2011;18:341–6. [Google Scholar]
- 70.Zwart G, van Hannen EJ, Kamst-van Agterveld MP, Van der Gucht K, Lindstrom ES, Van Wichelen J, et al. Rapid screening for freshwater bacterial groups by using reverse line blot hybridization. Appl Environ Microbiol. 2003;69(10):5875–83. 10.1128/AEM.69.10.5875-5883.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salcher MM, Pernthaler J, Posch T. Spatiotemporal distribution and activity patterns of bacteria from three phylogenetic groups in an oligomesotrophic lake. Limnol Oceanogr. 2010;55(2):846–56. [Google Scholar]
- 72.Newton RJ, Jones SE, Helmus MR, McMahon KD. Phylogenetic ecology of the freshwater Actinobacteria acI lineage. Appl Environ Microbiol. 2007;73(22):7169–76. 10.1128/AEM.00794-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones SE, Newton RJ, McMahon KD. Evidence for structuring of bacterial community composition by organic carbon source in temperate lakes. Environ Microbiol. 2009;11(9):2463–72. 10.1111/j.1462-2920.2009.01977.x [DOI] [PubMed] [Google Scholar]
- 74.Pope PB, Patel BK. Metagenomic analysis of a freshwater toxic cyanobacteria bloom. FEMS Microbiol Ecol. 2008;64(1):9–27. 10.1111/j.1574-6941.2008.00448.x [DOI] [PubMed] [Google Scholar]
- 75.Dziallas C, Grossart HP. Temperature and biotic factors influence bacterial communities associated with the cyanobacterium Microcystis sp. Environ Microbiol. 2011;13(6):1632–41. 10.1111/j.1462-2920.2011.02479.x [DOI] [PubMed] [Google Scholar]
- 76.Simek K, Hornak K, Jezbera J, Nedoma J, Vrba J, Straskrabova V, et al. Maximum growth rates and possible life strategies of different bacterioplankton groups in relation to phosphorus availability in a freshwater reservoir. Environ Microbiol. 2006;8(9):1613–24. 10.1111/j.1462-2920.2006.01053.x [DOI] [PubMed] [Google Scholar]
- 77.Eiler A, Heinrich F, Bertilsson S. Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J. 2012;6(2):330–42. 10.1038/ismej.2011.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, et al. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem. 2011;43(7):1450–5. 10.1016/j.soilbio.2011.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kielak A, Rodrigues JL, Kuramae EE, Chain PS, van Veen JA, Kowalchuk GA. Phylogenetic and metagenomic analysis of Verrucomicrobia in former agricultural grassland soil. FEMS Microbiol Ecol. 2010;71(1):23–33. 10.1111/j.1574-6941.2009.00785.x [DOI] [PubMed] [Google Scholar]
- 80.Sangwan P, Kovac S, Davis KE, Sait M, Janssen PH. Detection and cultivation of soil verrucomicrobia. Appl Environ Microbiol. 2005;71(12):8402–10. 10.1128/AEM.71.12.8402-8410.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cardman Z, Arnosti C, Durbin A, Ziervogel K, Cox C, Steen AD, et al. Verrucomicrobia are candidates for polysaccharide-degrading bacterioplankton in an Arctic fjord of Svalbard. Appl Environ Microbiol. 2014;80(12):3749–56. 10.1128/AEM.00899-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cai H, Jiang H, Krumholz LR, Yang Z. Bacterial community composition of size-fractioned aggregates within the phycosphere of cyanobacterial blooms in a eutrophic freshwater lake. PloS One. 2014;9(8):e102879 10.1371/journal.pone.0102879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martinez-Garcia M, Brazel DM, Swan BK, Arnosti C, Chain PS, Reitenga KG, et al. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PloS One. 2012;7(4):e35314 10.1371/journal.pone.0035314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kolmonen E, Sivonen K, Rapala J, Haukka K. Diversity of cyanobacteria and heterotrophic bacteria in cyanobacterial blooms in Lake Joutikas, Finland. Aquat Microb Ecol. 2004;36(3):201–11. [Google Scholar]
- 85.Martinez-Garcia M, Swan BK, Poulton NJ, Gomez ML, Masland D, Sieracki ME, et al. High-throughput single-cell sequencing identifies photoheterotrophs and chemoautotrophs in freshwater bacterioplankton. ISME J. 2012;6(1):113–23. 10.1038/ismej.2011.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zwart G, Hiorns WD, Methe BA, Van Agterveld MP, Huismans R, Nold SC, et al. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst Appl Microbiol. 1998;21(4):546–56. 10.1016/S0723-2020(98)80067-2. [DOI] [PubMed] [Google Scholar]
- 87.Shi L, Cai Y, Kong F, Yu Y. Specific association between bacteria and buoyant Microcystis colonies compared with other bulk bacterial communities in the eutrophic Lake Taihu, China. Environ Microbiol Rep. 2012;4(6):669–78. 10.1111/1758-2229.12001 [DOI] [PubMed] [Google Scholar]
- 88.Cai HY, Yan ZS, Wang AJ, Krumholz LR, Jiang HL. Analysis of the attached microbial community on mucilaginous cyanobacterial aggregates in the eutrophic Lake Taihu reveals the importance of Planctomycetes. Microb Ecol. 2013;66(1):73–83. 10.1007/s00248-013-0224-1 [DOI] [PubMed] [Google Scholar]
- 89.Zwisler W, Selje N, Simon M. Seasonal patterns of the bacterioplankton community composition in a large mesotrophic lake. Aquat Microb Ecol. 2003;31(3):211–25. [Google Scholar]
- 90.Rapala J, Berg KA, Lyra C, Niemi RM, Manz W, Suomalainen S, et al. Paucibacter toxinivorans gen. nov., sp. nov., a bacterium that degrades cyclic cyanobacterial hepatotoxins microcystins and nodularin. Int J Syst Evol Microbiol. 2005;55(Pt 4):1563–8. 10.1099/ijs.0.63599-0 [DOI] [PubMed] [Google Scholar]
- 91.Simek K, Hornak K, Jezbera J, Masin M, Nedoma J, Gasol JM, et al. Influence of top-down and bottom-up manipulations on the R-BT065 subcluster of beta-proteobacteria, an abundant group in bacterioplankton of a freshwater reservoir. Appl Environ Microbiol. 2005;71(5):2381–90. 10.1128/AEM.71.5.2381-2390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Simek K, Kasalicky V, Jezbera J, Jezberova J, Hejzlar J, Hahn MW. Broad habitat range of the phylogenetically narrow R-BT065 cluster, representing a core group of the Betaproteobacterial genus Limnohabitans. Appl Environ Microbiol. 2010;76(3):631–9. 10.1128/AEM.02203-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Glockner FO, Zaichikov E, Belkova N, Denissova L, Pernthaler J, Pernthaler A, et al. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of Actinobacteria. Appl Environ Microbiol. 2000;66(11):5053–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Simek K, Kasalicky V, Zapomelova E, Hornak K. Alga-derived substrates select for distinct Betaproteobacterial lineages and contribute to niche separation in Limnohabitans strains. Appl Environ Microbiol. 2011;77(20):7307–15. 10.1128/AEM.05107-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pinhassi J, Berman T. Differential growth response of colony-forming alpha- and gamma-proteobacteria in dilution culture and nutrient addition experiments from Lake Kinneret (Israel), the eastern Mediterranean Sea, and the Gulf of Eilat. Appl Environ Microbiol. 2003;69(1):199–211. 10.1128/AEM.69.1.199-211.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brettar I, Christen R, Hofle MG. Rheinheimera perlucida sp. nov., a marine bacterium of the Gammaproteobacteria isolated from surface water of the central Baltic Sea. Int J Syst Evol Microbiol. 2006;56(9):2177–83. [DOI] [PubMed] [Google Scholar]
- 97.Chen WM, Yang SH, Young CC, Sheu SY. Rheinheimera tilapiae sp. nov., isolated from a freshwater culture pond. Int J Syst Evol Microbiol. 2013;63(4):1457–63. [DOI] [PubMed] [Google Scholar]
- 98.Bleves S, Viarre V, Salacha R, Michel GP, Filloux A, Voulhoux R. Protein secretion systems in Pseudomonas aeruginosa: A wealth of pathogenic weapons. Int J Med Microbiol. 2010;300(8):534–43. 10.1016/j.ijmm.2010.08.005 [DOI] [PubMed] [Google Scholar]
- 99.Ferguson HW, Collins RO, Moore M, Coles M, MacPhee DD. Pseudomonas anguilliseptica infection in farmed cod, Gadus morhua L. J Fish Dis. 2004;27(4):249–53. 10.1111/j.1365-2761.2004.00537.x [DOI] [PubMed] [Google Scholar]
- 100.Jiang L, Yang L, Xiao L, Shi X, Gao G, Qin B. Quantitative studies on phosphorus transference occuring between Microcystis aeruginosa and its attached bacterium (Pseudomonas sp.) In: Qin B, Liu Z, Havens K, editors. Eutrophication of shallow lakes with special reference to Lake Taihu, China. Developments in Hydrobiology. 194: Springer; Netherlands; 2007. p. 161–5. [Google Scholar]
- 101.Yabuuchi E, Yano I, Oyaizu H, Hashimoto Y, Ezaki T, Yamamoto H. Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol Immunol. 1990;34(2):99–119. [DOI] [PubMed] [Google Scholar]
- 102.Kim MK, Schubert K, Im WT, Kim KH, Lee ST, Overmann J. Sphingomonas kaistensis sp. nov., a novel alphaproteobacterium containing pufLM genes. Int J Syst Evol Microbiol. 2007;57(7):1527–34. [DOI] [PubMed] [Google Scholar]
- 103.Feng X, Ou LT, Ogram A. Plasmid-mediated mineralization of carbofuran by Sphingomonas sp. strain CF06. Appl Environ Microbiol. 1997;63(4):1332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cai M, Xun L. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J Bacteriol. 2002;184(17):4672–80. 10.1128/JB.184.17.4672-4680.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fujii K, Satomi M, Morita N, Motomura T, Tanaka T, Kikuchi S. Novosphingobium tardaugens sp. nov., an oestradiol-degrading bacterium isolated from activated sludge of a sewage treatment plant in Tokyo. Int J Syst Evol Microbiol. 2003;53(1):47–52. [DOI] [PubMed] [Google Scholar]
- 106.Wilhelm LJ, Tripp HJ, Givan SA, Smith DP, Giovannoni SJ. Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data. Biol Direct. 2007;2(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jin L, La HJ, Lee HG, Lee JJ, Lee S, Ahn CY, et al. Caulobacter profunda sp. nov., isolated from deep freshwater sediment. Int J Syst Evol Microbiol. 2014;64(3):762–7. [DOI] [PubMed] [Google Scholar]
- 108.Jin L, Lee HG, Kim HS, Ahn CY, Oh HM. Caulobacter daechungensis sp. nov., a stalked bacterium isolated from a eutrophic reservoir. Int J Syst Evol Microbiol. 2013;63(7):2559–64. [DOI] [PubMed] [Google Scholar]
- 109.Mou X, Lu X, Jacob J, Sun S, Heath R. Metagenomic identification of bacterioplankton taxa and pathways involved in microcystin degradation in lake Erie. PloS One. 2013;8(4):e61890 10.1371/journal.pone.0061890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kormas KA, Lymperopoulou DS. Cyanobacterial toxin degrading bacteria: who are they? Biomed Res Int. 2013;2013:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhu L, Wu Y, Song L, Gan N. Ecological dynamics of toxic Microcystis spp. and microcystin-degrading bacteria in Dianchi Lake, China. Appl Environ Microbiol. 2014;80(6):1874–81. 10.1128/AEM.02972-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xiao X, Sogge H, Lagesen K, Tooming-Klunderud A, Jakobsen KS, Rohrlack T. Use of high throughput sequencing and light microscopy show contrasting results in a study of phytoplankton occurrence in a freshwater environment. PloS One. 2014;9(8):e106510 10.1371/journal.pone.0106510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Medinger R, Nolte V, Pandey RV, Jost S, Ottenwalder B, Schlotterer C, et al. Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol Ecol. 2010;19(1):32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Amend AS, Seifert KA, Bruns TD. Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol Ecol. 2010;19(24):5555–65. 10.1111/j.1365-294X.2010.04898.x [DOI] [PubMed] [Google Scholar]
- 115.Eiler A, Drakare S, Bertilsson S, Pernthaler J, Peura S, Rofner C, et al. Unveiling distribution patterns of freshwater phytoplankton by a next generation sequencing based approach. PloS One. 2013;8(1):e53516 10.1371/journal.pone.0053516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karl DM, Dobbs FC. Molecular approaches to microbial biomass estimation in the sea In: Cooksey KE, editor. Molecular Approaches to the Study of the Ocean. Dordrecht: Springer Netherlands; 1998. p. 29–89. [Google Scholar]
- 117.Olenina I, Hajdu S, Edler L, Andersson A, Wasmund N, Busch S, et al. Biovolumes and size-classes of phytoplankton in the Baltic Sea. HELCOM Baltic Sea Environ Proc. 2006;106:1–144. [Google Scholar]
- 118.Ngando TS, Groliere CA. Effets quantitatifs des Fixateurs sur la Conservation des Cillés Planctoniques d'Eau Douce. Archiv für Protistenkunde. 1991;140(2):109–20. French. [Google Scholar]
- 119.Zarauz L, Irigoien X. Effects of Lugol's fixation on the size structure of natural nano-microplankton samples, analyzed by means of an automatic counting method. J Plankton Res. 2008;30(11):1297–303. [Google Scholar]
- 120.Kembel SW, Wu M, Eisen JA, Green JL. Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS computational biology. 2012;8(10):e1002743 10.1371/journal.pcbi.1002743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vetrovsky T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PloS One. 2013;8(2):e57923 10.1371/journal.pone.0057923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66(4):1328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schirrmeister BE, Dalquen DA, Anisimova M, Bagheri HC. Gene copy number variation and its significance in cyanobacterial phylogeny. BMC Microbiol. 2012;12:177 10.1186/1471-2180-12-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All raw 16S rRNA dataset are available from the SRA database under study SRP095055.