Abstract
Background
A statistical methodology is available to estimate the proportion of cell types (cellular heterogeneity) in adult whole blood specimens used in epigenome-wide association studies (EWAS). However, there is no methodology to estimate the proportion of cell types in umbilical cord blood (also a heterogeneous tissue) used in EWAS.
Objectives
The objectives of this study were to determine whether differences in DNA methylation (DNAm) patterns in umbilical cord blood are the result of blood cell type proportion changes that typically occur across gestational age and to demonstrate the effect of cell type proportion confounding by comparing preterm infants exposed and not exposed to antenatal steroids.
Methods
We obtained DNAm profiles of cord blood using the Illumina HumanMethylation27k BeadChip array for 385 neonates from the Boston Birth Cohort. We estimated cell type proportions for six cell types using the deconvolution method developed by Houseman et al. (2012).
Results
The cell type proportion estimates segregated into two groups that were significantly different by gestational age, indicating that gestational age was associated with cell type proportion. Among infants exposed to antenatal steroids, the number of differentially methylated CpGs dropped from 127 to 1 after controlling for cell type proportion.
Discussion
EWAS utilizing cord blood are confounded by cell type proportion. Careful study design including correction for cell type proportion and interpretation of results of EWAS using cord blood are critical.
Key Words: Boston Birth Cohort, cell type proportion, DNA methylation, epigenomics, gestational age, neonates, nursing research, umbilical cord blood
Epigenome-wide association studies (EWAS) are increasingly being used to investigate birth weight, prematurity, and exposures in neonates with the hope of providing insight into molecular differences so that interventions can be developed. These studies utilize umbilical cord blood to measure DNA methylation (DNAm; Chen et al., 2014; Kile et al., 2014; Lee et al., 2012; Liu et al., 2014; Remy et al., 2014), but most of these studies do not take into account the fact that umbilical cord blood, like adult blood, is a heterogeneous tissue (Blackburn, 2013). For example, the proportion of leukocytes can vary depending on age, disease status, and other variables that impact the immune system. This means that any DNAm differences may be confounded by differences in the cellular proportions of the umbilical cord blood samples because EWAS may yield associations that are false positives; that is, findings may be the result of cell type proportion differences rather than an exposure of interest. Researchers have recognized that cellular heterogeneity is a significant issue for EWAS utilizing adult whole blood specimens, developed specialized statistical methods to account for that heterogeneity, and adapted those methods as tools for epigenetic research continue to evolve (Jaffe & Irizarry, 2014).
In a seminal paper, Houseman et al. (2012) developed an algorithm to estimate the proportion of immune cells in adult whole blood by identifying differentially methylated DNA regions (DMRs) for each of the principal immune components of whole blood—granulocytes, monocytes, and some subpopulations of lymphocytes (CD4 T cells, CD8 T cells, natural killer [NK] cells, and B cells). Using the Illumina HumanMethylation27k BeadChip Microarray (Illumina, Inc., San Diego, CA), the authors identified 500 CpG sites that are specific to these cell types and provided new analytical tools for estimating cell type proportions in EWAS. Despite the availability of this algorithm, few researchers have attempted to address potential confounding from cell type heterogeneity in their EWAS analyses in umbilical cord blood.
Three research teams have used the methodology created by Houseman et al. (2012) to examine the impact of exposures and DNAm while controlling for blood cell type proportions in cord blood. Most recently, the methodology was used to demonstrate that in utero arsenic exposure in term and preterm infants was associated with altered DNAm patterns after controlling for leukocyte proportions; in utero arsenic exposure had a dose-dependent effect on certain T-cell subpopulations in umbilical cord blood (Kile et al., 2014). A similar approach was used when studying DNAm patterns and birth weight in both term and preterm infants (Engel et al., 2014). Deconvolution analysis was also used to infer changes in the distribution of leukocytes in a study of arsenic exposure in utero and cord blood DNAm in term infants (Koestler, Avissar-Whiting, Houseman, Karagas, & Marsit, 2013). These research teams all used the methodology created by Houseman et al. (2012), including the reference data set comprising only adult peripheral blood samples; several noted that it is unclear whether this methodology is sufficient for correcting the cell-specific patterns of DNAm in umbilical cord blood.
The methodology developed by Houseman et al. (2012) may not be sufficient for leukocyte proportion adjustment when analyzing cord blood because blood cell type proportion changes naturally across gestational age. DNAm levels are measured on nucleated cells, and in cord blood, most of the cells with nuclei are the cells of the immune system (i.e., monocytes neutrophils, eosinophils, basophils, B cells, NK cells, and T cells). Because immunological changes occur in gestation, cord blood cell type proportions fluctuate greatly throughout fetal development and may vary significantly depending on the gestational age of the infant (Koenig & Yoder, 2004; Maheshwari & Christensen, 2004; Segel & Palis, 1995; Ygberg & Nilsson, 2012). Granulocytes (Remington, Klein, Wilson, Nizet, & Maldonado, 2001), CD4 T cells (Chabra et al., 1998; Duijts et al., 2009), and B cells (Correa-Rocha et al. 2012; Thilaganathan, Nicolaides, Mansur, Levinsky, & Morgan, 1993) increase during gestation, whereas NK cells vary throughout gestation (Correa-Rocha et al., 2012; Pérez, Gurbindo, Resino, Aguarón, & Muñoz-Fernández, 2007; Peoples et al., 2009; Ygberg & Nilsson, 2012). If cord blood cell type proportion differences between samples account for a certain amount of the overall DNAm variability, then cell type proportion fluctuations across gestational age should significantly affect DNAm findings.
As researchers continue to analyze the impact of environmental exposures on the fetal epigenome by studying cord blood, it is critical that we clarify whether cell type proportion is confounding findings. We hypothesize that researchers analyzing the DNAm of cord blood without correction for cell type proportions will identify differentially methylated CpGs, which they may attribute to an environmental exposure, when in fact, those differences between DNAm patterns may exist only because of cell type proportion changes across gestational age. Therefore, the objective of this study was to determine whether differences in DNAm patterns in umbilical cord blood are the result of blood cell type proportion changes that occur naturally across gestational age and to highlight the potential for confounded results by comparing a sample of preterm infants who were exposed and not exposed to antenatal steroids.
In this study, we used Houseman et al.’s (2012) deconvolution methodology to analyze the DNAm profiles of cord blood for 385 term and preterm infants from the Boston Birth Cohort, ranging from 30 to 44 weeks in gestational age. The aims were to:
measure the amount of overall DNAm variability that may be accounted for by differences in cell type proportions in the umbilical cord blood of term and preterm infants, relative to differences in gestational age and gender;
estimate cell type proportions for six cell types (granulocytes, monocytes, B cells, NK cells, CD4 T cells, and CD8 T cells) and identify differences in cell type proportion patterns across gestational age; and
determine if an analysis of the cord blood of preterm infants (gestational ages 30–36 weeks) exposed and not exposed to antenatal steroids reveals differences in DNAm patterns and whether some or all of those patterns can be explained by differences in cell type proportions as an illustrative example of potential confounding.
Glucocorticosteroids are a common exposure in preterm infants, and yet, research indicates that steroid application is associated with changes in DNAm (Carreno et al., 2011; Romero, Dey, & Fisher, 2014). The question is whether controlling for cell type proportions would impact this finding. As far as we are aware, this is the first study to use the DMRs identified by Houseman et al. (2012) to investigate how umbilical cord blood cell type proportion changes over gestation.
METHODS
Study Sample
The sample for this study was taken from an ongoing study called the Boston Birth Cohort (BBC) that enrolls mother–infant pairs from Boston Medical Center at birth and follows them prospectively. We studied a subsample of 385 African American, Haitian, or Cape Verdean mother–infant pairs; gestational ages ranged from 25 and 44 weeks. Cord blood was collected from all births at the time of delivery by trained nursing staff. Maternal interviews were conducted using a structured questionnaire. Gestational age was assessed with an algorithm based on the last menstrual period and early ultra sound before 20 weeks gestation. Further details concerning BBC design and methodology are available in D. Wang et al. (2012) and X. Wang et al. (2002).
Maternal consent was obtained for all cohort participants. The BBC was approved by the Boston Medical Center and Johns Hopkins University Institutional Review Boards. This study was approved by the University of Maryland Institutional Review Board.
DNAm Data Preprocessing
We obtained DNAm profiles of cord blood using the Illumina HumanMethylation27k BeadChip (Illumina, Inc.) array for 385 neonates from the BBC. Only probes in autosomes were used to avoid bias from large differences in sex chromosomes. Arrays with low total signal were filtered as part of quality assessment. We detected a strong batch effect for one of the 27k arrays and removed samples from that array. We restricted samples to those over 30 weeks of gestational age because only six samples were less than 30 weeks. After filtering the cord blood samples for total signal and batch correction, we dropped 38 samples, including some from infants born at later gestational ages because of quality assurance issues; we retained 341 samples, ranging from 30 to 43 weeks gestational age. Methylated and unmethylated signals were each quantile normalized across arrays using a modification of the minfi package (Aryee et al., 2014). We used the Illumina-provided annotation for the 27k BeadArray (HumanMethylation27 270596 v.1.2.bpm). All analyses were performed on methylation percentages computed after quantile normalization—except where noted.
Blood Cell Type-Specific CpGs
We obtained the top 500 CpGs for the 27k array that distinguish cell types in adult blood as described in Houseman et al. (2012) and implemented in code provided by that group using version 112 (http://people.oregonstate.edu/~housemae/software/).
Hierarchical Clustering
We performed hierarchical clustering analysis using Euclidean distance on methylation log ratio (M values) and estimated cell type estimates. Using both a two-sided t-test with unequal variance and a Wilcoxon rank-sum test, we completed tests for difference in gestational age across clusters, segregating samples according to the first partition obtained by hierarchical clustering. Tests for difference in variance on these two groups of samples were performed using an F-test.
Cell Type Proportion Estimation
We estimated cell type proportions for six cell types (granulocytes, monocytes, B cells, NK cells, CD4 T cells, and CD8 T cells) using Houseman et al.’s (2012) deconvolution method. We used the top 500 cell-specific CpGs as determined above and the methylation profiles provided by Houseman et al. for cell-sorted samples of adult blood. This method estimates a proportion of each cell type within each sample (i.e., interpreted as the proportion of cells of that type in each cord blood sample).
Statistical Analyses
Methylation Differences
We used statistical methods developed by Houseman et al. (2012) to study methylation in this cohort. We defined a mixed effects model of DNAm percentage for each CpG and cord blood sample, using gestational age and gender as fixed effects and proportion of each cell type (estimated as above) as random effects. We used variance components analysis to determine the percentage of total variation that is systematic (explained by both fixed and random effects) and the percentage of systematic variation explained by cell type proportions (explained by random effects). As part of this analysis, we estimate model parameters that summarize the association between cell type proportion on gestational age as described in Houseman et al. (2012).
Antenatal Steroid Application
We obtained data on antenatal steroid use for 60 preterm infants in our cohort; 19 of those infants were administered antenatal steroids. Their gestational ages ranged from 30 to 36 weeks. We used logistic regression to determine association between antenatal steroid administration and (a) gestational age, (b) infant gender, (c) granulocyte proportion, and (d) CD4 T-cell proportion. The latter two were estimated from DNAm measurements using the Houseman et al. (2012) deconvolution method and then transformed using an inverse logit transformation. All numerical covariates were centered and scaled for this analysis. We then fit a linear model regressing DNAm log ratio on antenatal steroid application and an interaction term between gestational age and CD4 T-cell proportion as above. We used empirical Bayes methods to shrink variance estimates across CpGs, and a moderated t-test was performed to determine differentially methylated CpGs, using the minfi package (Aryee et al., 2014). CpGs were reported as differentially methylated at 1% false discovery rate.
RESULTS
Detailed statistical results are available (see Figures, Supplemental Digital Content 1, http://links.lww.com/NRES/A236).
Changing Cell Type Proportions Across Gestational Age
After using the deconvolution method and performing cluster analysis on the resulting cell type proportion estimates, we observed that samples segregated into two groups based on cell type proportions (Figure 1A). One cluster is generally composed of granulocyte-rich samples; the other is generally composed of granulocyte-poor samples. There is also a significant difference in gestational age between samples in the two clusters (Figure 1B), with a mean of 39.0 (SD = 1.85) and 37.6 weeks (SD = 2.59) on the granulocyte-rich and -poor samples, respectively (t-test: p = .000001; Wilcoxon rank sum test: p = .000002), suggesting that differences in gestational age are associated with blood cell type proportions. The cell type proportions of the cluster corresponding to later gestational age is dominated by granulocytes, whereas the cluster corresponding to earlier gestational age is characterized by diminished presence of granulocytes and increased CD4 T cells, B cells, and NK cells (Figure 1A). Gestational age variability differed within each of the two clusters; the cluster corresponding to later gestational age had significantly lower variance (SD = 1.85 vs. 2.59; F-test: p = .000006).
FIGURE 1.
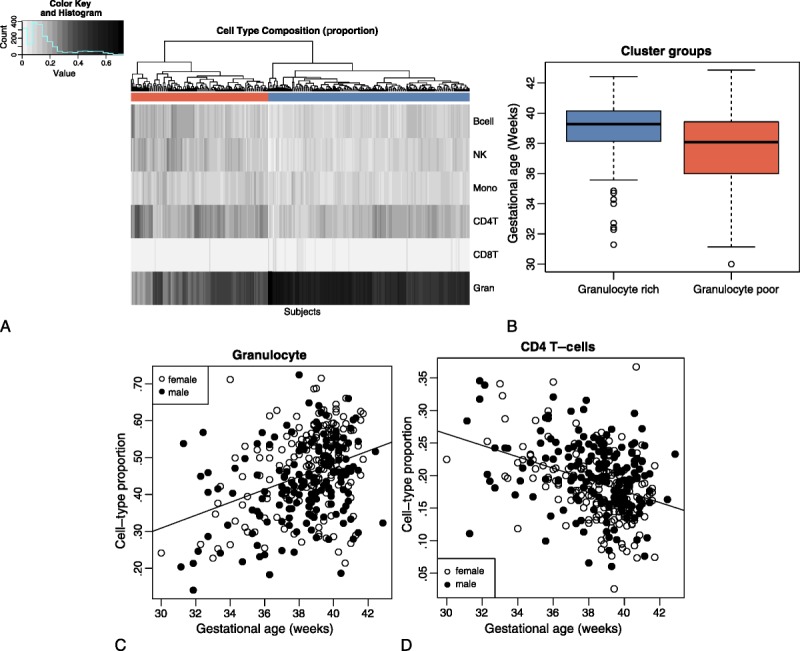
Differences in cell type proportions in cord blood are associated with gestational age. (A) Heatmap of blood cell type proportions estimated from DNAm for 341 neonate cord blood samples. Hierarchical clustering is based on blood cell type proportions; colors under the tree are used to indicate clustering of subjects into two groups (corresponding to granulocyte rich vs. granulocyte poor samples). (B) Gestational age distribution of subject clusters determined by blood cell type proportions (granulocyte rich or granulocyte poor). Granulocyte rich samples (blue) are predominantly full term; granulocyte poor samples (red) are predominantly preterm. (C) Proportion of granulocytes by gestational age. Marker types indicate subject gender, indicating no significant difference in proportion due to gender. (D) Proportion of CD4 T cell by gestational age. Marker types indicate subject gender, indicating no significant difference in proportion due to gender.
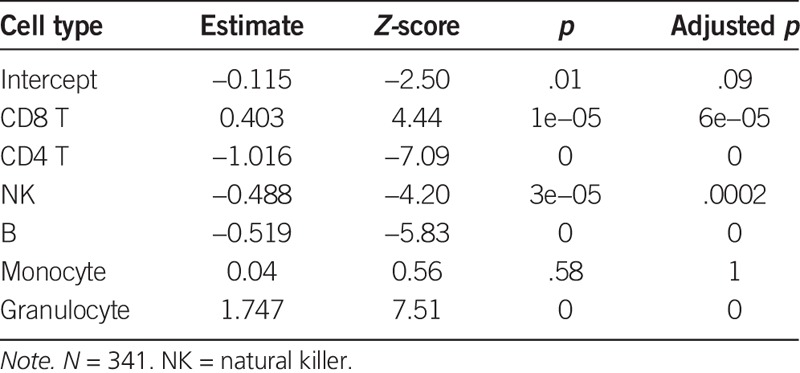
The regression analysis revealed the dynamics of cell type proportions in cord blood (as estimated by DNAm in cell type specific CpGs) when examined across a range of gestational age groups (Table 1; Figure 1C and D; also see Figures, Supplemental Digital Content 2, http://links.lww.com/NRES/A237). Granulocyte proportion varied most across gestational age; the proportion nearly doubled from 32 to 40 weeks (Z-score = 7.5; p = 6.08e−14; see Figure 1C). The second largest effect was for CD4 T cells, where the proportion dropped by half from 32 to 40 weeks (Z-score = −7.08; p = 1.37e−12; see Figure 1D). We observed smaller effects on B cells and NK cells, where the proportion decreased across gestation. No significant changes were observed for monocytes and CD8 T cells in this cohort.
TABLE 1.
Association Between Gestational Age and Blood Cell Type Proportions in Cord Blood
Explaining Methylation Variability After Accounting for Gestational Age
We sought to measure the amount of overall DNAm variability that may be accounted for by differences in cell type proportions relative to differences in gestational age and gender. Following the methods described by Houseman et al. (2012), we built a model including gestational age and gender as fixed effects, with estimated cell type proportion as a random effect, as described in the Statistical Analysis. We obtained estimates for total variation in methylation, along with an estimate of systematic variation, which was partitioned into variation explained by gestational age and gender and that explained by cell type proportions. The percentage of total variation explained by systematic variation was small (8.21%), but the percentage of systematic variation explained by cell type proportions was high (84.93%). This finding indicates that, although systematic variation contributes modestly to the total variation in methylation in this cohort, differences in cell type proportions make up a significant percentage of that variation. The percentages of DNAm variability explained by systematic variability and the percentages of systematic variability explained by cell type proportions are similar to those reported by Houseman et al. (2012) in their studies of adult cohorts.
Potential Confounding by Cell Type Proportions and Gestational Age
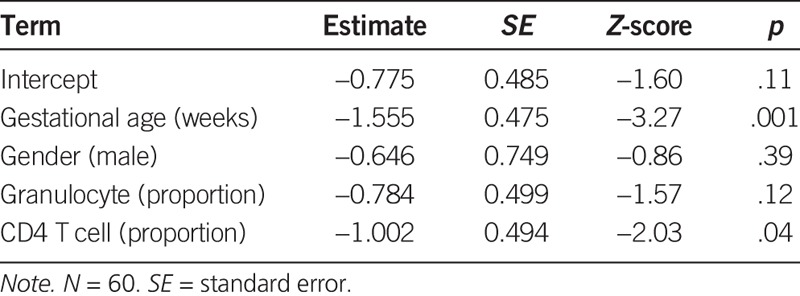
When we investigated a subsample of 19 infants exposed to antenatal steroids, along with 41 infants for which we had record of no exposure to antenatal steroids, we found a strong association between antenatal steroid exposure and both early gestational age (Table 2; Z-score = −3.27, p = .001) and lower CD4 T-cell concentration (Z-score = −2.03, p = .04). On the basis of this observation, we regressed the methylation log ratio for each CpG on antenatal steroid exposure, gestational age, and CD4 T-cell concentration. We compared two models to determine the association between antenatal steroid exposure and differences in DNAm: one model controlling for gestational age and CD4 T-cell concentration and one not controlling for these.
TABLE 2.
Association Between Antenatal Steroid Application, Gestational Age, Gender, and Cell Type Proportions in Cord Blood
When only considering antenatal steroid exposure in this cohort, we found 127 CpGs that showed significant differences. On the other hand, only one CpG—in the promoter region of FRA10AC1—showed a significant difference in DNAm when controlling for gestational age and CD4 T-cell concentration. We observed that subjects who received antenatal steroids had consistently higher methylation at similar gestational age (Figure 2).
FIGURE 2.
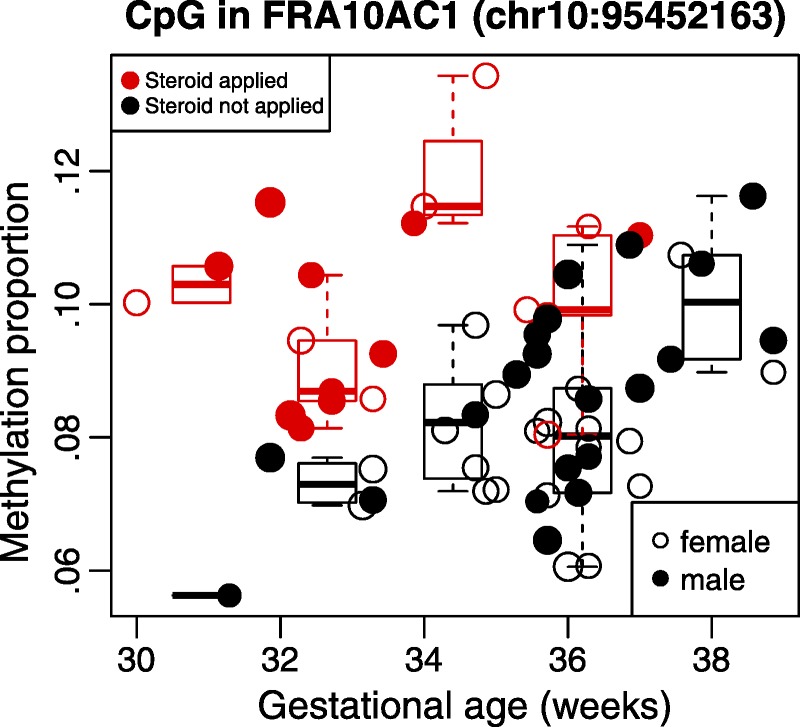
DNAm percentage for 60 neonate cord blood samples for a CpG in the promoter region of FRA10AC1 by gestational age. DNAm changes in promoter region of FRA10AC1 are associated with gestational age and antenatal steroid application. Points in red correspond to samples where antenatal steroids were applied. Points in black are samples where no antenatal steroids were applied. Point size is proportional to estimated proportion of CD4 T cells.
DISCUSSION
In this study, we observed that cell type proportions in the umbilical cord blood of term and preterm infants explain a significant portion of systematic variation in this cohort after accounting for gestational age in analyses of DNAm patterns; and although systematic variation only explains a modest portion of total methylation variability, it is sufficient to affect findings of DNAm differences associated to phenotypes of interest. Blood cell type methylation markers segregated the infant cohort by gestational age into two clusters: a cluster corresponding to later gestational age and a cluster corresponding to earlier gestational age. The clusters were characterized by distinct cord blood cell type proportion profiles based on proportions of granulocytes, lymphocytes, and B cells. Some of our findings are consistent with what is known about fetal cord blood cell type proportions and its changes during gestation, whereas others were unexpected.
The later gestational age cluster had significantly higher numbers of granulocytes than did the earlier gestational age cluster. This finding corresponds with the fact that normally granulocytes increase rapidly in a fetus during the last trimester (Maheshwari & Christensen, 2004). The number of circulating granulocytes is around 10% of the leukocyte population at 14–16 weeks gestation and increases to 50%–60% by term. Previous research has also shown that preterm infants tend to have a low proportion of granulocytes (Remington et al., 2001).
Lymphocyte proportion differed significantly between the two clusters. CD4 T cells were more abundant in the cluster corresponding to earlier gestational age, which was unexpected because these cells should typically increase as gestational age increases. Studies investigating CD4 T cells in preterm infants showed lower counts compared to term infants (Chabra et al., 1998; Duijts et al., 2009). NK cells were also more abundant in preterm infants than in term infants. The literature has mixed findings for NK cell numbers during gestation. NK cells have been found to increase over gestation (Correa-Rocha et al., 2012; Pérez et al., 2007) and correlate with gestational age (Ygberg & Nilsson, 2012). However, Peoples et al. (2009) found increased NK cells in preterm infant cord blood compared to term infant cord blood.
B cells were more abundant in the early gestational age cluster than in the late gestational age cluster, another unexpected finding. Correa-Rocha et al. (2012) found lower absolute counts of B cells in preterm infants, and there was a positive correlation with gestational age and birth weight. The total number of B cells exponentially increases over gestation and remains stable after 36 weeks (Thilaganathan et al., 1993) and are abundant at birth (Ygberg & Nilsson, 2012).
There are a number of potential explanations for why CD4 T, NK, and B cells were more abundant in the earlier gestational age cluster than in the later gestational age cluster in this sample. First, the proportion of granulocytes is significantly lower in the cord blood of preterm infants, making selection of other cell types during sampling more likely. The proportion of granulocytes compared to CD4 T, NK, and B cells may be higher in later gestational age but may not affect the absolute value of CD4 T, NK, and B cells. Second, the higher numbers of NK cells in the cord blood of the cluster associated with earlier gestational age may be the result of inflammation. Research indicates that congenital infection in the second trimester is associated with an increased count of NK cells (Remington et al., 2001). Infection and inflammation are regarded as one of the most important and potentially preventable causes of preterm birth (Burdet et al., 2014; Kallapur, Presicce, Rueda, Jobe, & Chougnet, 2014; Romero et al., 2014), but this biological mechanism would not explain the higher numbers of CD4 T and B cells. Third, a number of exposures experienced by preterm infants can also independently influence cord blood cell type proportions and may explain some of our unexpected study findings, as well the higher variability in blood cell type proportion in the cluster corresponding to earlier gestational age (Carreno et al., 2011; Romero et al., 2014). For example, fetal exposure to maternal smoking is associated with normal to higher counts of T cells (Remington et al., 2001).
Our analysis of antenatal steroid exposure demonstrates how the relationship between cell type proportions and gestational age can confound analyses of an epigenetic response to an exposure. The number of CpGs showing significant DNAm associated with antenatal steroid application in this cohort was reduced from 137 to a single CpG when controlling for gestational age and blood cell type proportions. This suggests that when the outcome under study, in this case, antenatal steroid exposure, correlates with gestational age and cell type proportions—an analysis without proper controls results in many false positives. Antenatal steroids are glucocorticoid steroids that have been found to be associated with changes in DNAm on specific genomic sites in rat models (Ewald et al., 2014). To the best of our knowledge, FRA10AC1 has not been reported in the literature as a specific site affected by glucocorticoid steroids. Hypermethylation in the region FRA10A C1 serves to counteract folate-sensitive fragility (Sarafidou et al., 2004). Our observation of increased methylation in this region associated with antenatal steroid application suggests that further investigation of the association between hypermethylation and genomic instabilities associated with depletion of folic acid and the role of antenatal steroids in this mechanism should be further explored.
Strengths and Limitations
The main strength of this study is that the BBC has a large sample size of preterm births, and a substantial number of umbilical cord blood samples were collected and analyzed using the Illumina HumanMethylation27k BeadChip. The main limitation of this study is that the DMRs we used to identify individual blood cell types were developed using a reference population of adult peripheral blood samples (see Houseman et al., 2012). We used this methodology because there are currently no specific DMRs identified for umbilical cord blood. The lymphocyte subtypes in cord blood may have different DNAm patterns because T-cell lineage epigenetically changes at midgestation during fetal development (Basha, Surendran, & Pichichero, 2014). It is possible that by utilizing the Houseman et al. (2012) correction, we did not identify all of the differences in cell type proportions that may occur across gestational age. It will be important to replicate this study once an umbilical cord blood correction is published.
Contributions to Nursing Science
Our study adds to nursing science by further elucidating the complexities involved in performing epigenetic research. Epigenetic researchers are still developing and refining methodologies, such as the cell type proportion correction we are suggesting that may be needed for the epigenetic analysis of umbilical cord blood. Nurses are actively involved in epigenetic research on a variety of health issues including pain, cardiovascular disease, childbirth, obesity, and metabolic syndrome and will continue to participate in the advancement of science. Understanding the latest advancements in this fast-developing field will be critical in conducting and advancing epigenetic research. Nurses play a key role in epigenetic research because of their unique insights into patients of all ages. This insight and rigorous research driven by multidisciplinary teams including nurses will help move the evolving science of epigenetics forward.
Conclusion
Our analysis suggests that systematic variance accounts for 8.21% of total variation; of this, cell type proportions explains 84.93% of the systematic variation in DNAm of the umbilical cord blood in this cohort. Researchers utilizing umbilical cord blood to perform EWAS must consider cord blood cell type proportion variability; however, the statistical methods currently used for studies based on adult whole blood are insufficient for this purpose. New statistical methods are needed to account for the complexities of the cellular proportions of cord blood—particularly preterm infant cord blood—which would need to include the presence of certain cell types, such as stem cells and nucleated red blood cells. EWAS have great potential to improve our understanding of the impact of exposures on the fetal epigenome, but we must develop appropriate statistical methods and conduct carefully designed studies in order to continue making significant progress.
Figure.

No caption available.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s web site (www.nursingresearchonline.com).
Editorial Note: Dr. Jacquelyn Taylor was Action Editor for this paper.
The authors acknowledge that this work was supported by the National Institutes of Health, Office of Research on Women’s Health, Building Interdisciplinary Research Careers in Women's Health (BIRCWH) Grant Number 2K12 HD043489-11.
The authors would like to thank Professor Xiaobin Wang, MD, MPH, ScD, and Assistant Scientist Xiumei Hong, MD, for providing access to the Boston Birth Cohort and reviewing the manuscript before publication, and Ms. Jen Bernstein, MPH, CCRP, for her thoughtful editing.
SB conceptualized and designed the study and drafted the initial manuscript. KO contributed substantially to the conception of the work and interpretation of data, drafted sections of the manuscript, and reviewed and revised the manuscript. AS contributed substantially to the conception of the work and interpretation of data. HCB contributed substantially to the conception of the work, carried out the initial analyses and interpretation of data, drafted sections of the manuscript, and reviewed and revised the manuscript. All authors approved the final manuscript as submitted and agreed to be accountable for all aspects of the work, ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
The authors have no conflicts of interest to report.
REFERENCES
- Aryee M. J., Jaffe A. E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A. P., Hansen K. D., & Irizarry R. A. (2014). Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics, 30, 1363–1369. doi:10.1093/bioinformatics/btu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha S., Surendran N., & Pichichero M. (2014). Immune responses in neonates. Expert Review of Clinical Immunology, 10, 1171–1184. doi:10.1586/1744666X.2014.942288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn S. (2013). Maternal, fetal, and neonatal physiology: A clinical perspective (4th ed). St. Louis, MO: Elsevier Saunders. [Google Scholar]
- Burdet J., Rubio A. P., Salazar A. I., Ribeiro M. L., Ibarra C., & Franchi A. M. (2014). Inflammation, infection and preterm birth. Current Pharmaceutical Design, 20, 4741–4748. doi:10.2174/1381612820666140130202224 [DOI] [PubMed] [Google Scholar]
- Carreno C. A., Costantine M. M., Holland M. G., Ramin S. M., Saade G. R., & Blackwell S. C. (2011). Approximately one-third of medically indicated late preterm births are complicated by fetal growth restriction. American Journal of Obstetrics and Gynecology, 204, 263.e1–263.e4. doi:10.1016/j.ajog.2010.12.004 [DOI] [PubMed] [Google Scholar]
- Chabra S., Cottrill C., Rayens M. K., Cross R., Lipke D., & Bruce M. (1998). Lymphocyte subsets in cord blood of preterm infants: Effect of antenatal steroids. Biology of the Neonate, 74, 200–207. doi:10.1159/000014025 [DOI] [PubMed] [Google Scholar]
- Chen D., Zhang A., Fang M., Fang R., Ge J., Jiang Y., … Dong M. (2014). Increased methylation at differentially methylated region of GNAS in infants born to gestational diabetes. BMC Medical Genetics, 15, 108 doi:10.1186/s12881-014-0108-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Rocha R., Pérez A., Lorente R., Ferrando-Martínez S., Leal M., Gurbindo D., & Muñoz-Fernández M. Á. (2012). Preterm neonates show marked leukopenia and lymphopenia that are associated with increased regulatory T-cell values and diminished IL-7. Pediatric Research, 71, 590–597. doi:10.1038/pr.2012.6 [DOI] [PubMed] [Google Scholar]
- Duijts L., Bakker-Jonges L. E., Labout J. A., Jaddoe V. W., Hofman A., Steegers E. A., … Moll H. A. (2009). Fetal growth influences lymphocyte subset counts at birth: The Generation R Study. Neonatology, 95, 149–156. doi:10.1159/000153099 [DOI] [PubMed] [Google Scholar]
- Engel S. M., Joubert B. R., Wu M. C., Olshan A. F., Håberg S. E., Ueland P. M., … London S. J. (2014). Neonatal genome-wide methylation patterns in relation to birth weight in the Norwegian mother and child cohort. American Journal of Epidemiology, 179, 834–842. doi:10.1093/aje/kwt433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald E. R., Wand G. S., Seifuddin F., Yang X., Tamashiro K. L., Potash J. B., … Lee R. S. (2014). Alterations in DNA methylation of Fkbp5 as a determinant of blood–brain correlation of glucocorticoid exposure. Psychoneuroendocrinology, 44, 112–122. doi:10.1016/j.psyneuen.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E. A., Accomando W. P., Koestler D. C., Christensen B. C., Marsit C. J., Nelson H. H., … Kelsey K. T. (2012). DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics, 13, 86 doi:10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A. E., & Irizarry R. A. (2014). Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biology, 15, R31 doi:10.1186/gb-2014-15-2-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallapur S. G., Presicce P., Rueda C. M., Jobe A. H., & Chougnet C. A. (2014). Fetal immune response to chorioamnionitis. Seminars in Reproductive Medicine, 32, 56–67. doi:10.1055/s-0033-1361823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile M. L., Houseman E. A., Baccarelli A. A., Quamruzzaman Q., Rahman M., Mostofa G., … Christiani D. C. (2014). Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics, 9, 774–782. doi:10.4161/epi.28153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig J. M., & Yoder M. C. (2004). Neonatal neutrophils: The good, the bad, and the ugly. Clinics in Perinatology, 31, 39–51. doi:10.1016/j.clp.2004.03.013 [DOI] [PubMed] [Google Scholar]
- Koestler D. C., Avissar-Whiting M., Houseman E. A., Karagas M. R., & Marsit C. J. (2013). Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environmental Health Perspectives, 121, 971–977. doi:10.1289/ehp.1205925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Jaffe A. E., Feinberg J. I., Tryggvadottir R., Brown S., Montano C., … Fallin M. D. (2012). DNA methylation shows genome-wide association of NFIX, RAPGEF2 and MSRB3 with gestational age at birth. International Journal of Epidemiology, 41, 188–199. doi:10.1093/ije/dyr237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Chen Q., Tsai H. J., Wang G., Hong X., Zhou Y., … Wang S. (2014). Maternal preconception body mass index and offspring cord blood DNA methylation: Exploration of early life origins of disease. Environmental and Molecular Mutagenesis, 55, 223–230. doi:10.1002/em.21827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari A., & Christensen R. (2004). Developmental granulocytopoiesis. Fetal and Neonatal Physiology, 2, 1388–1395. [Google Scholar]
- Peoples J. D., Cheung S., Nesin M., Lin H., Tatad A. M. F., Hoang D., … Cunningham-Rundles S. (2009). Neonatal cord blood subsets and cytokine response to bacterial antigens. American Journal of Perinatology, 26, 647–657. doi:10.1055/s-0029-1220788 [DOI] [PubMed] [Google Scholar]
- Pérez A., Gurbindo M. D., Resino S., Aguarón Á., & Muñoz-Fernández M. Á. (2007). NK cell increase in neonates from the preterm to the full-term period of gestation. Neonatology, 92, 158–163. doi:10.1159/000101567 [DOI] [PubMed] [Google Scholar]
- Remington J. S., Klein J. O., Wilson C. B., Nizet V., & Maldonado Y. (2001). Infectious diseases of the fetus and newborn infant. Philadelphia, PA: Elsevier. [Google Scholar]
- Remy S., Govarts E., Bruckers L., Paulussen M., Wens B., Hond E. D., … Schoeters G. (2014). Expression of the sFLT1 gene in cord blood cells is associated to maternal arsenic exposure and decreased birth weight. PLOS ONE, 9, e92677 doi:10.1371/journal.pone.0092677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R., Dey S. K., & Fisher S. J. (2014). Preterm labor: One syndrome, many causes. Science, 345, 760–765. doi:10.1126/science.1251816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafidou T., Kahl C., Martinez-Garay I., Mangelsdorf M., Gesk S., Baker E., … European Collaborative Consortium for the Study of ADLTE. (2004). Folate-sensitive fragile site FRA10A is due to an expansion of a CGG repeat in a novel gene, FRA10AC1, encoding a nuclear protein. Genomics, 84, 69–81. doi:10.1016/j.ygeno.2003.12.017 [DOI] [PubMed] [Google Scholar]
- Segel G. B., & Palis J. (1995). Hematology of the newborn. In Williams W. J. (Ed.), Williams hematology (5th ed pp. 57–72). New York, NY: MacGraw-Hill. [Google Scholar]
- Thilaganathan B., Nicolaides K. H., Mansur C. A., Levinsky R. J., & Morgan G. (1993). Fetal B lymphocyte subpopulations in normal pregnancies. Fetal Diagnosis and Therapy, 8, 15–21. doi:10.1159/000263742 [DOI] [PubMed] [Google Scholar]
- Wang D., Liu X., Zhou Y., Xie H., Hong X., Tsai H. J., … Wang X. (2012). Individual variation and longitudinal pattern of genome-wide DNA methylation from birth to the first two years of life. Epigenetics, 7, 594–605. doi:10.4161/epi.20117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zuckerman B., Pearson C., Kaufman G., Chen C., Wang G., … Xu X. (2002). Maternal cigarette smoking, metabolic gene polymorphism, and infant birth weight. JAMA, 287, 195–202. doi:10.1001/jama.287.2.195 [DOI] [PubMed] [Google Scholar]
- Ygberg S., & Nilsson A. (2012). The developing immune system—From foetus to toddler. Acta Paediatrica, 101, 120–127. doi:10.1111/j.1651-2227.2011.02494.x [DOI] [PubMed] [Google Scholar]


