Abstract
Objective:
To investigate the origin of the HIV-1 viremia induced by the latency-reversing agent romidepsin.
Design:
Six individuals on suppressive antiretroviral therapy received romidepsin administered intravenously once weekly for 3 consecutive weeks. CD4+ T cells were obtained at baseline, following the second and third romidepsin infusion, and 10 weeks after the final romidepsin treatment. Plasma samples were collected 24 and 72 h after romidepsin infusions.
Methods:
Single-genome sequencing of the env and p24-RT region was used to genetically characterize the virus from proviral DNA, the transcribed cell-associated RNA and the plasma RNA pool.
Results:
In three of six participants with available plasma samples we identified plasma HIV-1 RNA sequences that were identical to DNA and/or cell-associated RNA sequences from peripheral blood CD4+ T cells. In two participants, plasma RNA sequences contained expansions of identical sequences, corresponding to 62 and 100% of the total sequences, respectively. Plasma HIV-1 RNA had very low amounts of defective viruses compared to cell-associated RNA (odds ratio 20.85, P < 0.001) and to DNA (odds ratio 7.07, P = 0.011) during romidepsin therapy.
Conclusions:
Romidepsin induced transcription from proviruses in peripheral blood cells, which contributed to viremia in patients on suppressive therapy. The intermingling of these cell-associated HIV-1 RNA with DNA sequences indicates transcription from a diverse range of proviruses, but the expansions of identical viral plasma sequences with few defects indicate that the romidepsin-induced viremia arises from intact proviruses with highly similar or identical genetic backgrounds.
Keywords: histone deacetylase inhibitor, HIV latency, HIV persistence, HIV reservoirs, latency-reversing agent, romidepsin, single-genome sequencing
Introduction
The integration of HIV-1 DNA into the host genome of long-lived CD4+ T cells establishes a latent infection that is not eliminated by antiretroviral therapy (ART). The administration of latency-reversing agents (LRAs) like histone deacetylase inhibitors (HDACis) alone or in combination with immunotherapy may facilitate the clearance of replication-competent HIV-1. The goal is, through induction of HIV-1 transcription, cell surface expression of viral proteins, viral antigen presentation, and ultimately viremia, to facilitate immune or virus-mediated killing of latently infected cells. Several clinical studies have shown that HDACis are generally safe, well tolerated, and capable of enhancing HIV-1 transcription measured as increases in cell-associated RNA [1–4]. Sequencing of HIV-1 DNA and cell-associated RNA from individuals treated with the HDACis panobinostat [5] and vorinostat [2] demonstrated that both drugs induced broad reactivation of latent viruses [6]. Interestingly, the study by Barton et al. showed that 29–45% of cell-associated HIV-1 RNA detected following HDACi administration are defective in the env region, suggesting that reactivated latent proviruses may not contribute to the replication-competent HIV-1 reservoir. Characterization of plasma viruses from these studies could further inform the development of LRAs since reactivation of cells infected with replication-competent virus is a prerequisite for the success of ‘shock and kill’ strategies. However, due to limited sample material from the panobinostat trial and lack of plasma viremia in the vorinostat trials, it has not been possible to characterize the nature of plasma HIV-1 RNA species being released during HDACi treatment.
To investigate the origin and nature of LRA-induced plasma HIV-1 RNA, we turned our attention to the potent HDACi, romidepsin. In a recent trial, romidepsin was administered once weekly for 3 consecutive weeks to individuals on suppressive ART, leading to quantifiable increases of cell-associated HIV-1 RNA in all six participants and quantifiable increases of plasma HIV-1 RNA in five of six individuals [7]. Careful characterization of HIV-specific T-cell responses – which remained unaffected through the romidepsin dosing period – allowed us to conclude that the effect of romidepsin on plasma viremia was due to direct drug effect on HIV transcription and not due to loss of immune control by cytotoxic T lymphocytes [7]. In the present study, we compared HIV-1 DNA and cell-associated RNA sequences from peripheral blood CD4+ T cells to HIV-1 RNA sequences obtained from the plasma during romidepsin treatment.
Materials and methods
Study design
The study is a follow-on study to the REDUC Part A clinical trial. In this trial, six HIV-1-infected individuals on long-term suppressive ART received romidepsin administered intravenously once weekly for three consecutive weeks while maintaining ART. Detailed clinical trial design and patient characteristics have been published previously [7].
Participants and samples
From all six study participants, we analysed DNA and cell-associated HIV-1 RNA from CD4+ T cells from baseline, two to three time points during romidepsin therapy and a follow-up visit. Baseline samples were obtained immediately prior to the first romidepsin infusion. CD4+ T-cell samples obtained during romidepsin administration were designated time point 1, 2 and 3. For participants 1, 2, 3, 5 and 6, time point 1 was obtained 4 h after the second romidepsin infusion. For participant 7, time point 1 was obtained 4 h after the third romidepsin infusion. For all participants, time point 2 was obtained 7 days after the third romidepsin infusion. Participant 5 had an additional time point during romidepsin treatment, which was obtained 4 h after the third romidepsin infusion and was designated time point 3. Follow-up samples were obtained 10 weeks after the third romidepsin infusion. From three study participants, we obtained plasma from two to three time points during romidepsin therapy with quantifiable viral loads (participants 1, 2 and 7). Plasma samples obtained following the first, second and third romidepsin infusion were designated time points 1, 2 and 3, respectively. Plasma sample days are listed in Supplemental Digital Content 7. The plasma sample time points were chosen individually for each participant to prioritize samples with high viral loads to ensure high numbers of sequences. From two study participants, we also obtained sample material prior to initiation of ART. From participant 1, we analysed peripheral blood mononuclear cells (PBMCs) obtained 1 year prior to ART initiation. From participant 7, we analysed PBMCs and plasma obtained 4 months prior to ART initiation.
Ethics statement
The study was approved by the Danish Health and Medical Authorities, and also the Danish Data Protection Agency. Ethics committee approval was obtained in accordance with the principles of the Helsinki Declaration. Each patient provided written informed consent prior to any study procedures. The trial is registered at http://clinicaltrials.gov (NTC02092116). Pre-ART samples were obtained under protocol no. M 2007-0135 approved by Central Region of Denmark Committee on Health Research Ethics.
Single copy assay
The single copy assay (SCA) was performed on 1–2 ml of plasma to quantify HIV-1 RNA levels and rule out plasma contamination by cellular debris. This method has been described in detail previously [8].
Nucleic acid extraction and cDNA synthesis
Plasma was obtained following centrifugation of blood EDTA collection tubes, and PBMCs were separated by gradient centrifugation. CD4+ T cells were isolated from PBMCs using a CD4+ T-cell isolation kit and magnetic-activated cell sorting (MACS) columns (Miltenyi Biotec, Teterow, Germany; purity >95%). One million CD4+ T cells were lysed, and the lysates were stored at −80°C until the RNA and DNA were extracted using the QIAgen AllPrep DNA/RNA Mini Kit. RNA columns were treated twice with QIAgen RNase-Free DNase. Plasma HIV-1 RNA for sequencing was extracted from 2 to 6 ml of plasma by ultracentrifugation and a guanidinium-based method, including a prespin to sediment potential cell remnants [8]. From cell-associated HIV-1 RNA and plasma HIV-1 RNA, we generated cDNA using the Superscript III (Thermo Fisher Scientific, Australia, Cat. no. 18080044) cDNA synthesis kit and a gene-specific primer for env/p24-RT according to the manufacturer's instructions. See Supplemental Digital Content 1 for primers and PCR conditions.
Single-genome/proviral sequencing
We performed single-genome/proviral sequencing of HIV-1 env (V1–V3) as previously described for all six participants [6,9]. For participants 5, 6, and 7, we also sequenced a 2.1-kb region from p24 to reverse transcriptase [10]. See Supplemental Digital Content 1 for primers and PCR conditions. Single HIV-1 molecules were sequenced using Sanger sequencing (Australian Genome Research Facility, Sydney, Australia).
Phylogenetic analysis
Contigs were generated from the raw sequencing data using an in-house computer program written in Perl scripting language (available upon request). Vigorous automated and manual quality-control parameters were used to eliminate low-quality sequences prior to and following the generation of the contigs. Multiple alignment files were created for each participant using MUSCLE [11]. Defective viruses were characterized using the Los Alamos HIV-1 Database Hypermut tool (http://www.hiv.lanl.gov) to screen for sequences containing G-A hypermutations and by screening the amino acid sequences for premature stop codons. Sequences assigned with a P value <0.05 by the Hypermut tool were defined as hypermutated. Defective viruses were excluded, and the remaining sequences were used to construct maximum likelihood phylogenetic trees using MEGA 6.0 [12]. An appropriate model for nucleotide substitution was determined for each phylogenetic tree using MEGA model finder. The models used to generate the phylogenetic trees in this study included the following: generalized-time reversible model, Tamura three-parameter model, Hasegawa-Kishino-Yano model, and Tamura Nei model incorporating gamma distributed (gamma category 4) and/or invariant sites, when appropriate. Our heuristic tree search strategy used the nearest neighbour interchanges branch swapping algorithm. Branch support was inferred using 1000 bootstrap replicates. Measurements of genetic HIV-1 diversity (average pair-wise distance) of HIV-1 DNA or RNA sequences were calculated after excluding defective sequences using the Ape package version 3.4 in R programming language [13]. Samples with fewer than five sequences were excluded from analyses to reduce bias. Identical and similar sequences were identified using the number of differences model in MEGA 6.0. Sequences with one to two nucleotide differences equating to above 99.7% similarity were included. All intact sequences from the study are available in GenBank (env: KX841174-KX842073 and p24-RT: KX840928-KX841173).
Statistics
Only time points with five or more intact sequences were included in the analysis of average pair-wise distance, and time points with five or more total sequences were included in the hypermutation analysis. Env and p24-RT sequences from all time points during romidepsin treatment were pooled for analysis. To account for the longitudinal nature of the data, the difference in average pair-wise difference between cell-associated RNA and DNA was investigated using a mixed-effects model using function lme in package nlme in R. To investigate the percentage of dead end viruses, a logistic mixed-effects model using function glmer in package lme4 in R was fitted. P values were calculated using a chi-square likelihood ratio tests, with Wald confidence intervals (CIs) reported. P values less than 0.05 were considered significant.
Results
Broad reactivation of quiescent HIV-1 proviruses by romidepsin
Cell-associated HIV-1 RNA and DNA extracted from peripheral CD4+ T cells were sequenced immediately prior to, two to three times during, and once following romidepsin administration. A total of 1087 env sequences (187 defective and 900 intact) and 310 p24-RT sequences (64 defective and 246 intact) were analyzed (see Table, Supplemental Digital Content 2. Phylogenetic analysis of the sequences from all six participants formed individual clades with no inter-participant mixing (see figure, Supplemental Digital Content 3. HIV-1 DNA sequences were obtained to establish a baseline level of HIV-1 genetic diversity to which the induced cell-associated HIV-1 RNA sequences could be compared. We quantitated the genetic diversity of cell-associated HIV-1 RNA and DNA sequences for each participant by calculating the average pair-wise distance (Fig. 1 and figure, Supplemental Digital Content 4a and b. Phylogenetic trees were constructed for all participants. We found that cell-associated HIV-1 RNA sequences were distributed throughout the tree and intermingled with their corresponding HIV-1 DNA sequences (Figs. 2 and 3, and figures, Supplemental Digital Content 5 and 6. We found no significant difference in average pair-wise distance within each participant between the HIV-1 DNA and cell-associated RNA sequences collected at baseline, on romidepsin, and at follow-up (percentage difference DNA to cell-associated RNA 0.02, 95% CI −0.54 to 0.49, P = 0.96, mixed-effects model). Furthermore, although DNA showed increased genetic diversity to cell-associated RNA during romidepsin administration, this difference was below any meaningful amount (percentage difference on romidepsin DNA to cell-associated RNA 0.24%, 95% CI −0.04% to 0.53%, P = 0.04, mixed-effects model). Less than five HIV-1 DNA sequences were obtained from participant 3 during romidepsin treatment, and this data point was excluded from analysis. The inclusion of this data point changes the P value for average pair-wise difference on romidepsin treatment between DNA and cell-associated RNA from 0.04 to 0.75.
Fig. 1.
Romidepsin non-selectively activates transcription from latent HIV-1 proviruses.
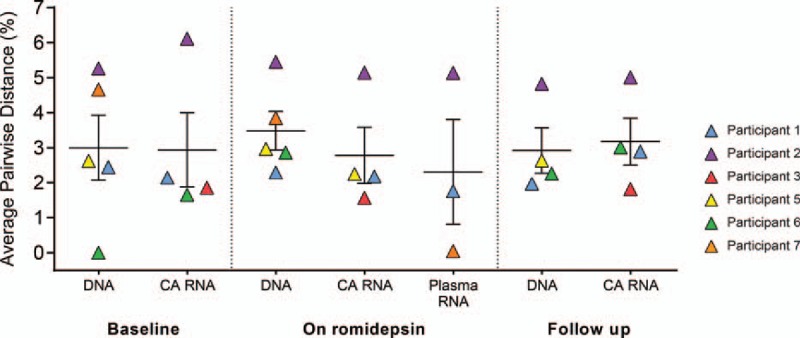
Average pair-wise distance of env sequences from HIV-1 DNA, cell-associated (CA) RNA and plasma RNA at study baseline, during romidepsin administration and at follow-up. Error bars show mean ± standard error of the mean. On romidepsin data points represent the average pair-wise distance of the pooled sequences for all time points during romidepsin treatment.
Fig. 2.
Proviruses in peripheral CD4+ T cells contribute to romidepsin-induced viremia.
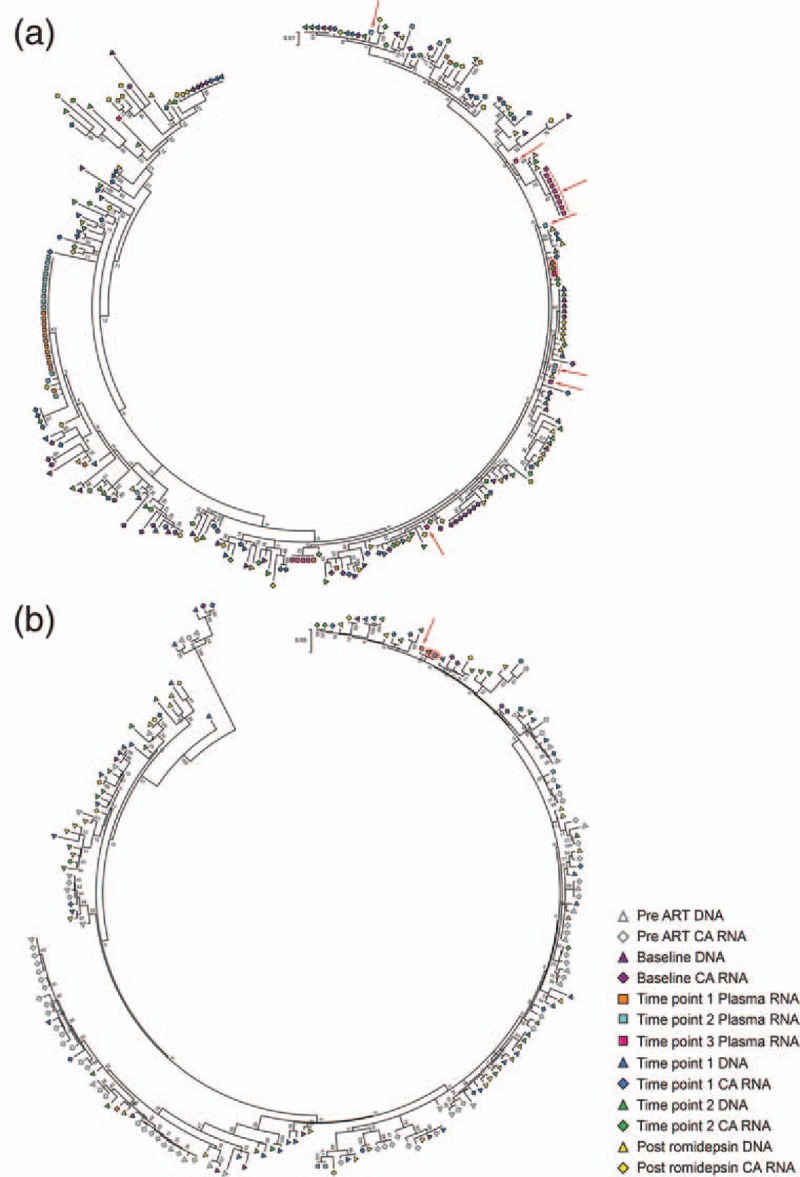
Maximum likelihood phylogenetic tree of HIV-1 env sequences from (a) participant 1 and (b) participant 2. Highlighted in red are romidepsin-induced plasma sequences that are identical to a DNA and/or a cell-associated (CA) RNA sequence. Red arrows point to plasma HIV-1 RNA sequences that are above 99.7% similar to HIV-1 DNA and/or CA RNA sequences.
Fig. 3.
Romidepsin-induced viremia contains identical viral sequences.
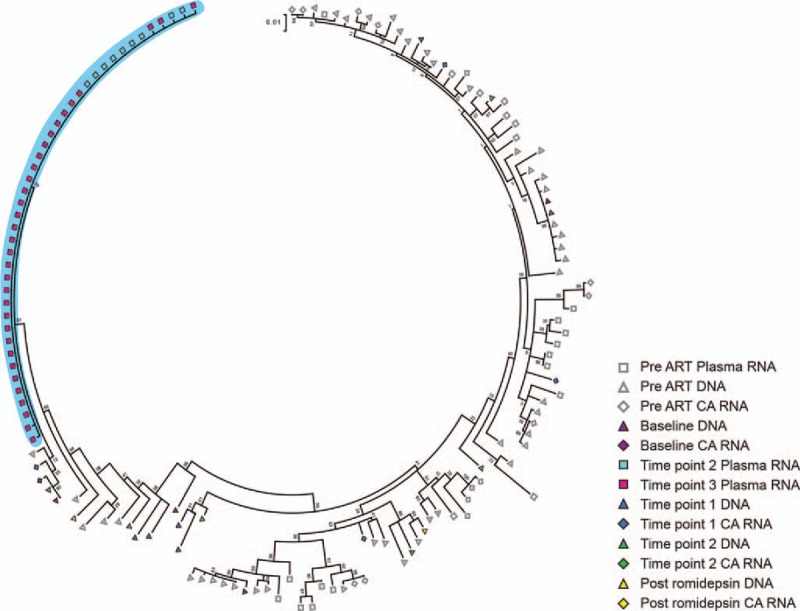
Maximum likelihood phylogenetic tree of HIV-1 env sequences from participant 7. Highlighted in blue is a cluster containing all plasma HIV-1 RNA sequences obtained during romidepsin therapy.
The similarity in the genetic diversity of cell-associated HIV-1 RNA and DNA during romidepsin therapy indicates that romidepsin non-selectively induced transcription from proviruses in HIV-1-infected individuals on long-term suppressive therapy.
Viremic plasma samples do not contain HIV-1 DNA
To quantify romidepsin-induced plasma viremia, we used the SCA on multiple samples (two to three time points) from participants 1, 2, and 7 who experienced elevated plasma viremia at least twice after romidepsin infusion. As shown in table (Supplemental Digital Content 7, HIV-1 RNA values ranged from 3 to 70 copies/ml. We added replication competent avian sarcoma–leukosis retroviral vector RCAS BP(A) (RCAS) to each plasma sample as an internal control and confirmed that their values were in the assay dynamic range. To rule out the presence of HIV-1 DNA from lysed cells in the plasma, one reaction mixture for each sample was processed and amplified without the addition of reverse transcriptase. All the non-reverse transcriptase controls were negative, demonstrating that none of the plasma samples contained HIV-1 DNA and confirming that the source of the detected romidepsin-induced viremia was HIV-1 RNA.
Romidepsin-induced plasma HIV-1 RNA contains identical viral sequences
We obtained plasma HIV-1 RNA sequences from the three participants with the highest viral loads during romidepsin therapy (participants 1, 2 and 7). For participant 1, we obtained 55 plasma HIV-1 RNA sequences from three time points during romidepsin therapy. Of these, 39 sequences grouped into three clusters of sequences that were identical or above 99.7% similar. Two of these clusters contained identical sequences from several time points. Considering all participants had undetectable viral loads between romidepsin infusions [7], this indicates that identical proviruses can contribute to viremia following subsequent doses of romidepsin. The remaining 16 sequences were unique and intermingled throughout the phylogenetic tree. This indicates that romidepsin therapy resulted in broad induction of viremia from genetically diverse proviruses, and also activation of multiple proviruses with a highly similar genetic background (Fig. 2a).
For participant 2, we obtained five plasma HIV-1 RNA sequences from two time points during romidepsin therapy. Four of these sequences fell into two groups of identical/highly similar sequences. Despite a relatively lower level of romidepsin-induced viremia, the pattern was similar to that observed in participant 1 (Fig. 2b).
All of the 42 env plasma-derived HIV-1 RNA sequences from the two time points obtained from participant 7 were above 99.7%, similar to each other, and no corresponding expansions of DNA were detected, indicating that either a few cells produced a large number of virions or that there was activation of multiple proviruses with a highly similar genetic background (Fig. 3). Similar to what was observed for participant 1, this cluster contained sequences from several time points, demonstrating that identical proviruses can contribute to viremia following subsequent doses of romidepsin. For participant 7, we also obtained five p24-RT plasma HIV-1 RNA sequences that were above 99.7%, similar to each other (Supplemental Digital Content 6C, which supports the finding from the env region, indicating that the env and p24-RT sequences likely represent the same single cluster of identical plasma HIV-1 RNA sequences.
Proviruses in CD4+ T cells from peripheral blood contribute to viremia
In all three participants from whom we obtained plasma samples during romidepsin therapy, we identified plasma HIV-1 RNA sequences that were identical to DNA sequences from peripheral blood CD4+ T cells. Participant 1 had one plasma HIV-1 RNA sequence that was identical to both a HIV-1 DNA and a cell-associated RNA sequence. An additional 15 plasma HIV-1 RNA sequences from this participant were above 99.7%, similar to HIV-1 DNA and cell-associated RNA sequences. Of these 15 plasma HIV-1 RNA sequences, 5 sequences were unique and the remaining 10 sequences belonged to two clusters of two and eight identical sequences each (Fig. 2a). In participant 2, one plasma HIV-1 RNA sequence was identical and another was highly similar (>99.7%) to an HIV-1 DNA sequence (Fig. 2b). Participant 7 had two p24-RT plasma HIV-1 RNA sequences that were identical to two HIV-1 DNA sequences (see figure, Supplemental Digital Content 6c. An additional two p24-RT plasma RNA sequences contained only a single-nucleotide mismatch (>99.7% similar) relative to the same two DNA sequences. These results strongly indicate that proviruses from the peripheral blood contributed to the plasma viremia. We did not detect any romidepsin-induced env plasma RNA sequences that were identical to pre-ART env plasma RNA (Fig. 3).
Romidepsin-induced plasma HIV-1 RNA contains few defective viruses
Defective viruses are defined as sequences containing hypermutation and/or stop codons in the region that is sequenced rendering them replication incompetent. At every time point during romidepsin administration we found a significantly higher number of defective env/p24-RT sequences in cell-associated HIV-1 RNA compared to DNA [Fig. 4, DNA to cell-associated RNA [odds ratio (OR) 2.95, 95% CI 1.71–5.07, P < 0.001, mixed-effects model]. We observed no differences in proportions of defective sequences between baseline, during romidepsin administration, and follow-up (DNA P = 0.29, cell-associated RNA 0.91, mixed-effects model), indicating that treatment with romidepsin does not selectively activate transcription from hypermutated or non-hypermutated sequences, but rather uniformly increases transcription from all proviruses (see figures, Supplemental Digital Content 4c and d). In a subset of participants, we sequenced HIV-1 DNA and cell-associated RNA from cells collected prior to the initiation of ART. In contrast to the high level of defective cell-associated HIV-1 RNA sequences during ART, there were far fewer defective sequences in these pre-ART peripheral blood cell samples, and the frequencies of defective sequences were similar between cell-associated HIV-1 RNA and DNA (Supplemental Digital Content 4C, mean 4.0% in pre-ART DNA and 4.5% in pre-ART cell-associated RNA). This difference in the proportion of defective sequences likely reflects the predominance of replication-competent viruses prior to ART and a clearance of these intact viruses after the initiation of ART, whereas defective viruses persist as they might not trigger immune or virus-mediated cell lysis. Notably, the plasma HIV-1 RNA sequences obtained from three participants during romidepsin therapy revealed significantly fewer defective viral sequences at all time points compared to cell-associated HIV-1 RNA and DNA (Fig. 4, plasma RNA to cell-associated RNA, OR 20.85, 95% CI 4.64–93.72, P < 0.001; plasma RNA to DNA, OR 7.07, 95% CI 1.56–32.06, P = 0.011, mixed-effects model). This indicates that plasma sequences are much more likely to originate from infection-competent proviruses than cell-associated HIV-1 RNA sequences.
Fig. 4.
Romidepsin-induced plasma viremia contains low amounts of defective envelope.
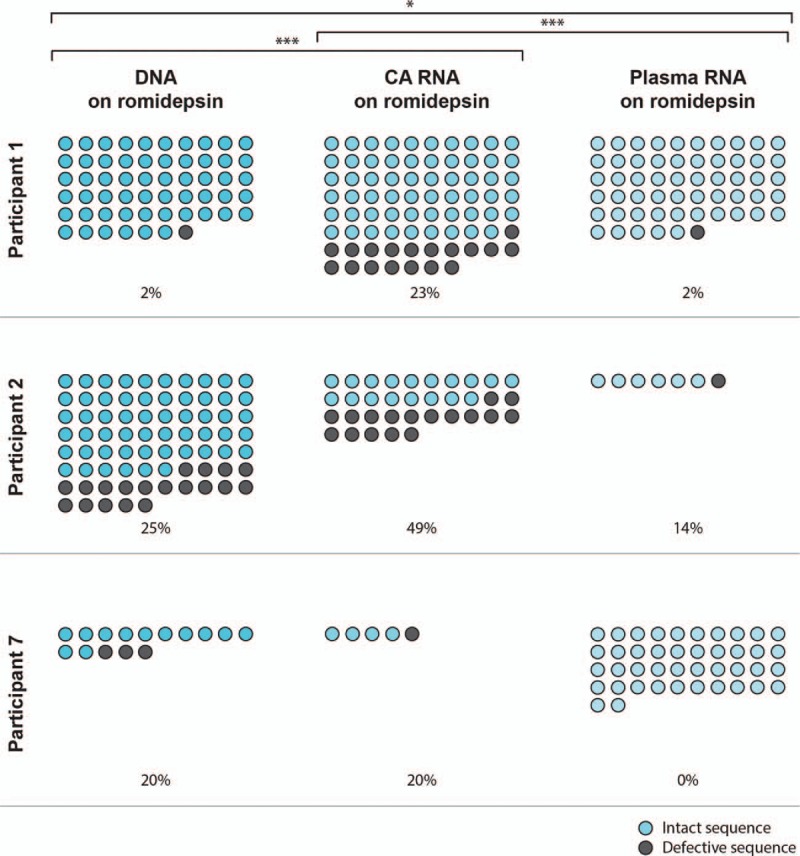
Absolute numbers of all env DNA, cell-associated (CA) RNA and plasma RNA sequences obtained from participants 1, 2 and 7 during romidepsin administration. Intact sequences are shown as blue circles. Defective sequences, defined as containing hypermutation and/or stop codons in the region sequenced, are shown as grey circles. Percentages of defective viruses of the total number of sequences are shown. ∗P < 0.05, ∗∗∗P < 0.001, mixed-effects model.
Discussion
This is the first study to sequence plasma HIV-1 RNA induced during HDACi treatment in vivo. Herein, we show that proviruses contributing to romidepsin-induced viremia can be identified in CD4+ T cells obtained from the periphery. Furthermore, we report activation of transcription from an extensive range of proviruses by romidepsin; however, clusters of identical viral sequences containing low amounts of deleterious mutations were isolated from the plasma during romidepsin therapy.
The intermingling of plasma-derived HIV-1 RNA sequences with intracellular HIV-1 RNA and DNA sequences indicates activation of proviral expression from multiple infected cells. In two participants, however, the finding of clusters of identical plasma HIV-1 RNA sequences indicates that a specific subset of transcriptionally activated proviruses contributed to the majority of viremia. The phylogenetic pattern showing clusters of identical plasma HIV-1 sequences in participants 1 and 7 resembles rebound virus after ART cessation [6,14,15]. These expansions most likely represent activation of multiple proviruses with a highly similar genetic background rather than high numbers of virions arising from a few cells [16]. Of note, several of the clusters of identical viral sequences from participants 1 and 7 contained sequences from more than one time point, indicating that identical proviruses can contribute to viremia following subsequent doses of romidepsin. Furthermore, the clusters of identical plasma HIV-1 RNA sequences did not match any expansions of intracellular HIV-1 DNA sequences from peripheral blood CD4+ T cells, which could be due to induction of genetically similar proviruses from cells located in other anatomical sites. This corresponds with the observation that many romidepsin-induced plasma HIV-1 RNA sequences did not match proviral sequences from the peripheral blood, similar to what has been shown for sequences of low-level viremia on suppressive ART [17]. However, it should be stressed that this could also be attributed to the number of peripheral blood cells included in our analyses. Notably, we did not find any similarities between HIV-1 DNA, cell-associated RNA or pre-ART plasma HIV-1 RNA sequences and romidepsin-induced plasma HIV-1 RNA sequences, suggesting that either the HDACi-inducible reservoir was not present at the pre-ART sample time point or conversely that activation by romidepsin was required to induce the production of virions from these latent proviruses.
Similar to findings following induction by vorinostat and panobinostat in vivo, the intracellular RNA transcripts observed following romidepsin administration were genetically diverse [6]. These findings indicate that romidepsin induced transcription from a broad range of proviruses, as the majority of HIV-infected cells contain one HIV-1 DNA molecule [18]. However, in contrast to the genetically diverse cell-associated HIV-1 RNA sequenced during romidepsin therapy, plasma HIV-1 RNA sequences during romidepsin therapy were less genetically diverse and contained clusters of identical viruses, indicating that only a proportion of the reactivated proviruses were capable of producing virions. It is possible that more potent latency reversal, either with increased dosing of romidepsin or a combination of LRAs, could result in the release of more genetically diverse virions from several different latent reservoirs, but it is also likely that the diversity of plasma virus demonstrated here is representative of the replication-competent, inducible reservoir.
Intriguingly, we observed a high percentage of defective virus in the cell-associated HIV-1 RNA compared to the DNA sequences, whereas the percentage of defective virus pre-ART and in the romidepsin-induced plasma HIV-1 RNA was much lower. This indicates that cells containing defective transcripts are not cleared during ART possibly because they seemingly cannot translate into functional virus proteins. The low amounts of defective romidepsin-induced sequences in plasma together with the clusters of identical plasma viruses indicate a selection against incorporation of mutated RNA into virions and further suggest that the virions detected are replication competent. It should be noted, however, that we sequenced p24-RT and env of HIV-1, and, thus, are not able to fully determine the replication-competency of the virions released into the plasma.
In summary, this study shows that romidepsin caused a broad activation of proviruses in vivo. HIV-infected cells in the blood are important reservoirs of HIV-1 during effective therapy and harbour proviruses capable of contributing to viremia during romidepsin therapy. Importantly, the viremia induced by romidepsin contained very low amounts of deleterious mutations. The low genetic diversity of viral RNA isolated from viremic plasma samples during romidepsin therapy suggests that a specific subset of cells containing HIV-1 proviruses contribute to this viremia. Additional trials with more potent LRAs, potentially in combination, are warranted to further assess the potential to induce HIV-1 RNA transcription and virion production from a greater number of cells containing replication-competent virus.
Acknowledgements
We acknowledge the participation and commitment of study participants, which made the study possible.
This work was supported by the Delaney AIDS Research Enterprise (DARE) to Find a Cure 1U19AI096109 and the National Health and Medical Research Council (NHMRC) of Australia (grant APP1061681). The clinical study from which these samples were derived was sponsored by Bionor Pharma.
A.W. designed and performed experiments, analysed data and wrote the manuscript. K.B. and B.H. designed and performed experiments, analysed data and assisted with manuscript preparation. W.S. prepared sequences for analysis. T.E.S. performed the mixed-effects model statistical analyses and contributed to manuscript editing. T.A.R., M.T., L.O. and O.S.S. designed the study, provided samples and assisted with manuscript preparation. S.P. designed the study, supervised the work performed and edited the manuscript. All authors have read and accepted the final manuscript.
T.A.R., L.Ø., M.T. and O.S.S. were supported by a grant from the Danish Council for Strategic Research (grant #0603–00521B). The funders had no role in data collection and analysis, decision to publish, or preparation of the manuscript. Bionor Pharma contributed to the design of the clinical trial.
Conflicts of interest
The authors declare no competing financial interests.
Supplementary Material
References
- 1.Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 2014; 1:e13–e21. [DOI] [PubMed] [Google Scholar]
- 2.Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 2014; 10:e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012; 487:482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archin NM, Bateson R, Tripathy MK, Crooks AM, Yang KH, Dahl NP, et al. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis 2014; 210:728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 2015; 1:e13–e21. [DOI] [PubMed] [Google Scholar]
- 6.Barton K, Hiener B, Winckelmann A, Rasmussen TA, Shao W, Byth K, et al. Broad activation of latent HIV-1 in vivo. Nature Commun 2016; 7:12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sogaard OS, Graversen ME, Leth S, Olesen R, Brinkmann CR, Nissen SK, et al. The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog 2015; 11:e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 2003; 41:4531–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Stockenstrom S, Odevall L, Lee E, Sinclair E, Bacchetti P, Killian M, et al. Longitudinal genetic characterization reveals that cell proliferation maintains a persistent HIV type 1 DNA pool during effective HIV therapy. J Infect Dis 2015; 212:596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Josefsson L, von Stockenstrom S, Faria NR, Sinclair E, Bacchetti P, Killian M, et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci USA 2013; 110:E4987–E4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24:1596–1599. [DOI] [PubMed] [Google Scholar]
- 13.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics (Oxford, England) 2004; 20:289–290. [DOI] [PubMed] [Google Scholar]
- 14.Kearney MF, Spindler J, Shao W, Yu S, Anderson EM, O'Shea A, et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog 2014; 10:e1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Boni J, et al. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci USA 2008; 105:16725–16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci USA 2016; 113:1883–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol 2006; 80:6441–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Josefsson L, King MS, Makitalo B, Brannstrom J, Shao W, Maldarelli F, et al. Majority of CD4+ T cells from peripheral blood of HIV-1-infected individuals contain only one HIV DNA molecule. Proc Natl Acad Sci USA 2011; 108:11199–11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


