Abstract
Macrocycles are appealing drug candidates due to their high-affinity, specificity, and favorable pharmacological properties. In this study, we explored the effects of chemical modifications to a natural product macrocycle upon its activity, 3D geometry, and conformational entropy. We chose thiocillin as a model system, a thiopeptide in the ribosomally-encoded family of natural products that exhibits potent antimicrobial effects against gram-positive bacteria. Since thiocillin is derived from a genetically-encoded peptide scaffold, site-directed mutagenesis allows for rapid generation of analogs. To understand thiocillin’s structure-activity relationship, we generated a site-saturation mutagenesis library covering each position along thiocillin’s macrocyclic ring. We report the identification of eight unique compounds more potent than WT thiocillin, the best having an 8-fold improvement in potency. Computational modeling of thiocillin’s macrocyclic structure revealed a striking requirement for a low entropy macrocycle for activity. The populated ensembles of the active mutants showed a rigid structure with few adoptable conformations while inactive mutants showed a more flexible macrocycle which is unfavorable for binding. This finding highlights the importance of macrocyclization in combination with rigidifying post-translational modifications to achieve high potency binding.
Graphical abstract
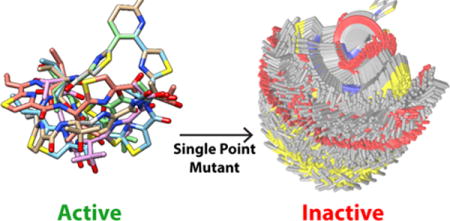
Natural products are critical modulators of microbial and multicellular biology. Roughly one-third of the pharmacopeia are derived from small molecule natural products. Many of these exceed Lipinski’s rules with sizes in the range of 500–1500 Da but retain favorable pharmacokinetic properties, enabling oral dosing in many cases. Macrocycles are conformationally constrained by cyclization, which has been suggested to reduce their apparent size and pre-organize the compound into a low entropy state, facilitating permeation and target binding. However, macrocyclic rings of the same size can vary dramatically in their conformational flexibility, due to the presence or absence of rigidifying elements such as double bonds or backbone rings. Here we assess the effect of ring entropy on binding and activity in a model natural product system, by combining systematic mutational analysis with computational modeling of ring entropy.
We chose to study the natural product thiocillin, a thiopeptide in a class called RiPPs, ribosomally synthesized and post-translationally modified peptides. Thiocillin undergoes a cascade of post-translational modifications (PTMs) to form a mature macrocyclic natural product.1 Thiopeptide antibiotics inhibit the growth of gram-positive bacteria including MRSA and VRE at nanomolar concentrations.2 Like many other thiopeptides, thiocillin targets the interface between ribosomal protein L11 and the 23S rRNA.3 Thiocillin’s prepeptide contains a C-terminal core peptide and an N-terminal leader sequence, which is removed once modifications are completed.4 Common thiopeptide PTMs include thiazole (from Cys), oxazole (from Ser), methyloxazole (from Thr), dehydroalanine (Dha) (from Ser), and dehydrobutyrine (Dhb) (from Thr); all of these modifications rigidify the peptide.5 The modified core peptide undergoes an enzyme-catalyzed [4+2] cycloaddition reaction to close the macrocycle and forms a pyridine core.6,7 One of our goals was to understand the importance of rigidifying modifications in the peptide to macrocyclization and potency.
We present a systematic SAR analysis of thiocillin by saturation mutagenesis and computational modeling. Thiocillin has previously been subjected to alanine scanning, cysteine-to-serine scanning, ring-size variants, incorporation of noncanonical amino acids, and various point mutants.8–11 In this work, we conducted saturation mutagenesis on macrocycle residues 2–9, the 8 residues not involved in macrocycle linkage, thus producing a total of 152 single point mutants. This provided a comprehensive understanding of tolerated amino acid replacements, including variants with enhanced antibacterial activity. Importantly, we noted sharp SAR that separated active and inactive analogs. Molecular mechanics modeling combined with NMR studies showed the steep loss of activity seen with certain mutants was the result of dramatic increases in ring entropy. Thus, conformational constraints beyond macrocyclization are critical for the activity of this natural product.
To rapidly generate mutants of thiocillin, we used a plasmid complementation strategy (Figure 1B). By expressing the prepeptide gene, tclE, in Bacillus cereus ΔtclE-H, a strain lacking the endogenous prepeptide gene, we demonstrate the rescue of thiocillin production (Figure S1).
Figure 1.
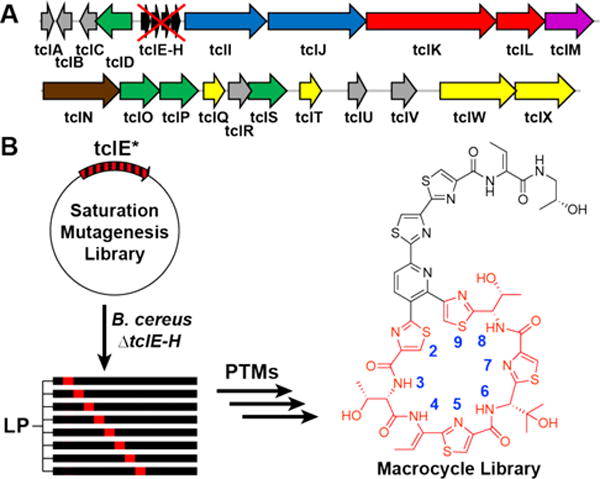
A) Thiocillin gene cluster with genomic deletion of endogenous prepeptide genes tclE-H, B. cereus ATCC 14579 ΔtclE-H. B) A plasmid complementation method to generate site-saturation mutagenesis libraries at residues 29 of the macrocycle. LP = leader peptide.
The mature form of thiocillin contains a large macrocycle closed via a pyridine ring which absorbs light at 350 nm. To screen for macrocycle formation, methanolic extracts from small-scale 1.5 mL cultures were analyzed by LC/MS. Presence of a 350 nm peak and a mass consistent with the mutation indicated the successful macrocyclization of 25 mutants (Figure 2). Our results suggest mutations to non-thiazole forming residues 3, 4, 6, and 8 are tolerant to mutations, with 6 and 8 being the most tolerant without loss of activity. Only residue 8 was able to accept large aromatic side chains such as phenylalanine and tyrosine. Mutating thiazole-forming Cys residues was poorly tolerated, consistent with a previous publication.8 These mutants were not detectable in our small scale high-throughput assay, which deliberately focused on identifying highly-expressing and well- tolerated mutants.
Figure 2.
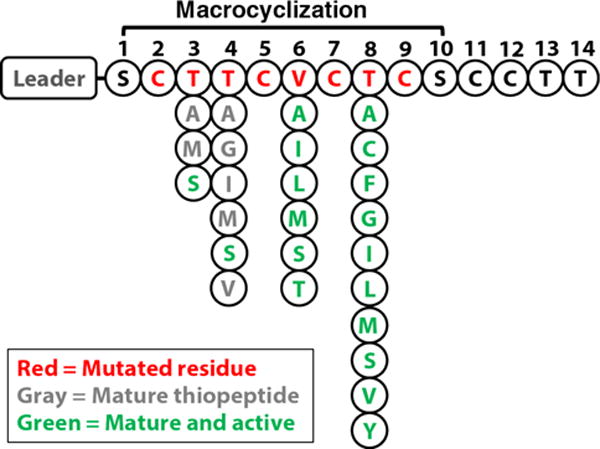
Thiocillin prepeptide shown as a drop-down diagram designating tolerated mutations leading to macrocyclization (gray and green) and activity (green).
To screen for active mutants, an overlay assay was used against Bacillus subtilis 168, a representative grampositive bacterium. Engineered B. cereus strains producing an active thiopeptide create a zone of inhibition when overlaid with B. subtilis (Figure S3). This screen identified 18 thiocillin mutants with antibiotic activity (Figure 2). Production of these 18 active mutants and an inactive negative control mutant were scaled up and purified for quantitative minimal inhibitory concentration (MIC) determination. Since each mutant produces multiple unique compounds due to auxiliary PTMs on residues 6 and 8, we isolated and screened all unique compounds with yields greater than 0.1 mg/L. In total, 33 unique analogs were purified from the active mutants and 2 from the negative control. The MIC assay resulted in 7 compounds more active than WT (Table 1). The most potent compound was V6A2 with an MIC of 0.06 μg/mL, an 8-fold increase in potency. This particular mutant was previously discovered to have a 4-fold increase in potency when assayed as a mixture of compounds.8 Interestingly, we identified mutants such as T8F, which showed activity on solid media, but not in liquid culture. This may be due to the hydrophobic Phe side-chain limiting its solubility.
Table 1.
MIC table of thiocillin analogs.
| Mutanta | Yieldb | MICc | Modificationsd |
|---|---|---|---|
| WT | 3.9 | 0.5 | All variants |
| T3S1 | 1.3 | >8 | V6-OH |
| T3S2 | 1.9 | >8 | |
| T3S3 | 0.3 | >8 | T8-CH3 |
| T3S4 | 0.3 | >8 | T3-Dha |
| T4A1(−) | 0.7 | >8 | V6-OH |
| T4A2(−) | 1.1 | >8 | |
| T4S1 | 0.6 | 2 | T4-Dha; V6-OH |
| T4S2 | 0.3 | 1 | T4-Dha; V6-OH; T8-CH3 |
| T4S3 | 0.1 | >8 | T4-Dha |
| V6A1 | 0.5 | 1 | |
| V6A2 | 1.9 | 0.06 | T8-CH3 |
| V6I1 | 2.1 | 0.5 | I6-OH |
| V6I2 | 0.3 | 0.5 | |
| V6L1 | 0.3 | 0.5 | L6-OH; T8-CH3 |
| V6L2 | 0.5 | 2 | T8-CH3 |
| V6M1 | 1.1 | 0.25 | M6-OH; T8-CH3 |
| V6M2 | 2.4 | 0.13 | T8-CH3 |
| V6S1 | 0.6 | 0.5 | T8-CH3 |
| V6T1 | 1.6 | 0.13 | T8-CH3 |
| T8A1 | 1.2 | 0.5 | V6-OH |
| T8C1 | 0.4 | 8 | V6-OH; C8-CH3 |
| T8F1 | 1.0 | >8 | |
| T8G1 | 0.9 | 2 | V6-OH |
| T8G2 | 0.4 | 0.5 | |
| T8I1 | 0.7 | 0.13 | V6-OH |
| T8L1 | 1.3 | 1 | V6-OH |
| T8L2 | 0.7 | >8 | |
| T8M1 | 1.4 | 1 | V6-OH |
| T8M2 | 1.3 | 2 | |
| T8S1 | 0.9 | >8 | V6-OH |
| T8S2 | 0.1 | 1 | V6-OH; S8-Dha |
| T8V1 | 2.0 | 0.13 | V6-OH |
| T8V2 | 0.3 | 1 | |
| T8Y1 | 1.6 | 0.25 | V6-OH |
| T8Y2 | 2.1 | 1 |
Compound numbering is in order of chromatographic retention time. (−) indicates an inactive negative control.
mg/L.
μg/mL.
Modifications are predicted based on high-resolution MS, retention time, and NMR (structural characterizations are included in the SI).
To examine the influence of each single point mutation on conformational entropy, we sampled the potential energy landscape for every analog using BRIKARD.12 BRIKARD applies inverse kinematics to enable efficient generation of low-energy conformations of macrocycles. We initially validated our computational approach by reproducing near-native states extracted from crystallography data for similar thiopeptides, including one bound to ribosomal protein L11.3 Detailed methods are available in the SI. The conformations generated for each thiopeptide by BRIKARD were then clustered using a stringent Cartesian RMSD metric (0.25 Å) to eliminate redundant (nearly identical) conformations. While this collection of low-energy states cannot be interpreted as a true thermodynamic ensemble, the number of such states varied dramatically among the thiocillin variants (Table 2), reflecting the rigidity of the macrocycle and hence its entropy.
Table 2.
Populated ensembles of conformations resulting from clustering the BRIKARD sampling results.
| Mutant | Conformations | Mutant | Conformations |
|---|---|---|---|
| WTa | 5 | V6Ia | 5 |
| C2A | 600 | V6La | 4 |
| C2S | 591 | V6Sa | 5 |
| T3A | 5 | C7A | 253 |
| T3M | 5 | C7S | 264 |
| T3Sa | 5 | T8Aa | 5 |
| T4A | 355 | T8Ga | 7 |
| T4L | 380 | T8Ia | 5 |
| T4M | 442 | T8La | 4 |
| T4S | 384 | T8Ma | 4 |
| T4Dhaa | 7 | T8Sa | 5 |
| C5A | 290 | T8Va | 5 |
| C5S | 340 | C9A | 501 |
| V6Aa | 5 | C9S | 486 |
Active mutant.
Our results activity data, with a clear division between the highly rigidified active analogs having a maximum of 47 conformational states compared to the >250 conformational states accessible to the inactive mutants (Table 2). This finding suggests that, within the thiocillin family, mutations that dramatically increase the flexibility of the macrocycle ablate binding, due at least in part to the increased entropic loss required for binding. We cannot, of course, rule out other effects of the mutations also impacting binding affinity and activity. Conversely, conformational rigidity does not guarantee activity, i.e., the data suggests that rigidity is necessary but not sufficient for activity.
One dramatic example with mutant T4A shows that breaking the planar character of the Dhb residue at position 4, which resulted in a mature macrocycle with no detectable activity. In this case, simply changing a planar sp2 alpha carbon to a tetrahedral sp3 geometry leads to a dramatic increase in backbone entropy (Figure 3). These striking changes in macrocycle entropy exemplify the need for computational sampling of ring entropy.
Figure 3.
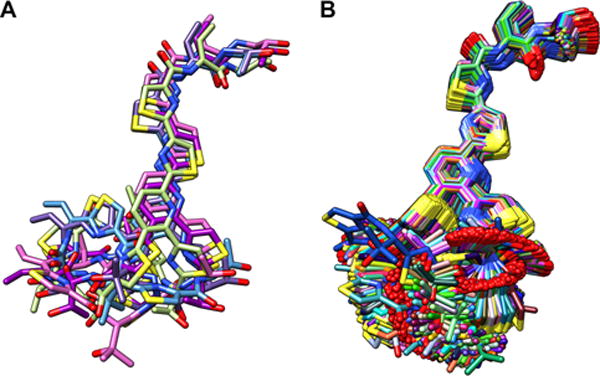
BRIKARD simulations identified the populated ensembles of low energy conformations for A) WT thiocillin (low entropy) and B) T4A mutant (high entropy).
Following our computational study of the potential energy landscape of thiocillin and our mutant analogs, NMR was used to determine the structure of WT thiocillin in DMSO. The 3D HNHA NMR experiment was used for structural determination because it is an accurate method for measuring homonuclear three-bond 3JHNHa coupling constants and was used to help elucidate the structure of lassomycin.13 The 3JHNHa values obtained from 15N-labelled thiocillin were used to estimate dihedral phi angles for Thr-3, Val-6, and Thr-8 according to the Karplus equation (Table S5).14 These values were used as restraints in BRIKARD to generate an ensemble of conformations consistent with the NMR data (Figure S4A). Five similar conformations were obtained with and without experimental restraints, further supporting the use of BRIKARD for conformational sampling. Our predominant NMR structure (Figure S4B) (31% occupancy) showed a similar folded conformation to a previous NMR structure obtained using ROESY data.8
Based on our experimental and computational SAR, we developed a second generation combinatorial thiocillin library randomizing residues 6 and 8, the most tolerated sites. 1200 colonies were screened by overlay assay, observing many previously known single mutants as well as 16 new double mutants. Six double mutants that were combinations of potent single mutants were purified to produce 8 unique compounds and subjected to MIC testing. Only the V6A-T8V double mutant showed a 2-fold increase in potency over WT (Table S2). These double mutants did not show the expected synergy, suggesting that other global molecular properties such as solubility or permeability may be limiting their activity.
The three PTMs characteristic of thiopeptides are formation of (1) five-membered heterocyclic thiazoles and oxazoles, (2) sp2 side chains of Dha/Dhb, and (3) a macrocycle enclosing pyridine/dehydropiperidine ring. All are rigidifying modifications. The conformational analyses in this study suggest the entropy restricting property of these planar Dha/Dhb side chains may be as important as their previously appreciated chemical reactivity, as electrophiles in lanthionine residue formation and as dual electrophiles and nucleophiles in pyridine/dehydropiperidine ring formation. If generalizable, the Dha/Dhb-forming PTM may reveal yet another layer of chemical logic used in nature to create high affinity molecular scaffolds.
Our studies suggest that a rigid macrocycle is a requirement for binding in this system and that very small chemical changes can lead to substantial increases in entropy as measured by computational modelling. The formation of a macrocycle greatly reduces backbone entropy, however, macrocyclization alone appears insufficient for preorganization of the compound. Particularly in thiopeptides, the presence of rigidifying PTMs such as heterocyclization and dehydrations appears to lower the entropic barrier for binding. In other macrocycle systems that are not heavily modified, entropy reduction can be achieved by intramolecular hydrogen bonding. Entropy is an often overlooked parameter in macrocycle design and the principles learned here can be applied to other natural and synthetic macrocycle systems.
Supplementary Material
Acknowledgments
We thank Professors Michael Fischbach and Susan Miller for useful discussion, Dr. Evangelos Coutsias for advice on BRIKARD, Dr. Hector Huang for help with HRMS, and Drs. Mark Kelly and Greg Lee for help with NMR. Molecular graphics and analyses were performed with the UCSF Chimera package. This research was supported by the NIH (AI111662 and GM100619).
Footnotes
Supporting Information. Supporting figures, detailed experimental procedure, and computational methods. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
References
- 1.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. Proc Natl Acad Sci U S A. 2009;106:2549. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li C, Zhang F, Kelly WL. Mol Biosyst. 2011;7:82. doi: 10.1039/c0mb00129e. [DOI] [PubMed] [Google Scholar]
- 3.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. Mol Cell. 2008;30:26. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Oman TJ, van der Donk WA. Nat Chem Biol. 2010;6:9. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh CT, Malcolmson SJ, Young TS. ACS Chem Biol. 2012;7:429. doi: 10.1021/cb200518n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowers AA, Walsh CT, Acker MG. J Am Chem Soc. 2010;132:12182. doi: 10.1021/ja104524q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wever WJ, Bogart JW, Baccile JA, Chan AN, Schroeder FC, Bowers AA. J Am Chem Soc. 2015;137:3494. doi: 10.1021/jacs.5b00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowers AA, Acker MG, Koglin A, Walsh CT. J Am Chem Soc. 2010;132:7519. doi: 10.1021/ja102339q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowers AA, Acker MG, Young TS, Walsh CT. J Am Chem Soc. 2012;134:10313. doi: 10.1021/ja302820x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo X, Zambaldo C, Liu T, Zhang Y, Xuan W, Wang C, Reed SA, Yang PY, Wang RE, Javahishvili T, Schultz PG, Young TS. Proc Natl Acad Sci U S A. 2016;113:3615. doi: 10.1073/pnas.1602733113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Just-Baringo X, Albericio F, Alvarez M. Angew Chem Int Ed Engl. 2014;53:6602. doi: 10.1002/anie.201307288. [DOI] [PubMed] [Google Scholar]
- 12.Coutsias EA, Lexa KW, Wester MJ, Pollock SN, Jacobson MP. J Chem Theory Comput. 2016;12:4674. doi: 10.1021/acs.jctc.6b00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gavrish E, Sit CS, Cao S, Kandror O, Spoering A, Peoples A, Ling L, Fetterman A, Hughes D, Bissell A, Torrey H, Akopian T, Mueller A, Epstein S, Goldberg A, Clardy J, Lewis K. Chem Biol. 2014;21:509. doi: 10.1016/j.chembiol.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vuister GW, Bax A. J Am Chem Soc. 1993;115:7772. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


