Abstract
Human microbial communities are characterized by their taxonomic, metagenomic, and metabolic diversity, which varies by distinct body sites and influences human physiology. However, when and how microbial communities within each body niche acquire unique taxonomical and functional signatures in early life remains underexplored. We thus sought to assess the taxonomic composition and potential metabolic function of the neonatal and early infant microbiota across multiple body sites, and assess the impact of mode of delivery and its potential confounders or modifiers. A cohort of pregnant women in their early 3rd trimester (n=81) were prospectively enrolled for longitudinal sampling through 6 weeks post-delivery, and a second matched cross-sectional cohort (n=81) was additionally recruited for sampling once at delivery. Samples were collected for each maternal-infant dyad across multiple body sites, including stool, oral gingiva, nares, skin and vagina. 16S rRNA gene sequencing analysis and whole genome shotgun sequencing was performed to interrogate the composition and function of the neonatal and maternal microbiota. We found that the neonatal microbiota and its associated functional pathways were relatively homogenous across all body sites at delivery, with the notable exception of neonatal meconium. However, by 6 weeks, the infant microbiota structure and function had significantly expanded and diversified, with body site serving as the primary determinant of the bacterial community composition and its functional capacity. Although minor variations in the neonatal (immediately at birth) microbiota community structure were associated with Cesarean delivery in some body sites (oral, nares, and skin; R2 = 0.038), this was not true in neonatal stool (meconium, Mann-Whitney p>0.05) and there was no observable difference in community function regardless of delivery mode. By 6 weeks of age, the infant microbiota structure and function had expanded and diversified with demonstrable body site specificity (p<0.001, R2 = 0.189), and no discernable differences in neither community structure nor function by Cesarean delivery were identifiable (p=0.057, R2 = 0.007). We conclude that within the first 6 weeks of life, the infant microbiota undergoes significant reorganization that is primarily driven by body site and not by mode of delivery.
Introduction
The human microbiome comprises a rich ecosystem of microbes that are essential to human health and physiology. In the adult, the microbiota inhabiting each body site is characterized by a distinct microbial community structure and function1–4. For instance, in the gastrointestinal tract, enteric microbes produce and modify numerous biologically active compounds that support host metabolism, including bile acids, vitamins and other macromolecules, while Lactobacillus species in the vagina support a low vaginal pH5,6. Indeed, the importance of microbiota to human health is underscored by the observation that dysbiotic shifts in these microbial communities have been associated with a number of human diseases, including obesity, inflammatory bowel disorders, autoimmune disease, and gastrointestinal cancer7–10. However, in order to understand how our microbiota may contribute to disease progression later in life, the mechanisms by which host-microbial symbiosis is established and maintained in early life requires further exploration.
Evidence from germ-free (GF) murine models has demonstrated that normal development is dependent on the presence of commensal microbiota, particularly in the gastrointestinal tract11. For example, GF mice exhibit an altered immune phenotype with deficits in both innate and adaptive immune components of the gut mucosa12,13. Reintroducing microorganisms post-natally (after birth) partially corrects many of these defects, though even a brief GF neonatal period can induce immunological changes that persist into adulthood12,14. Importantly, different bacterial species have been shown to distinctly modulate the host immune system, indicating that the presence of specific bacteria within a given developmental window is important for normal patterning of host immunity15–17.
Work by the Human Microbiome Project Consortium and others have demonstrated that in the adult, distinct microbial communities uniquely inhabit each body site1,4. Even sites in close proximity (e.g. supragingival plaque vs. tongue) have discernable differences in microbial composition1 that may be due to differences in microenvironment conditions such as oxygen exposure and nutrient availability. A previous study in a small cohort of primarily preterm, very low birth-weight infants demonstrated that the microbiota of skin, saliva and stool rapidly diverged within the first three weeks of life18. However, more precise approximations of when body site differentiation is achieved have not been explored in larger cohorts of healthy term infants. Furthermore, how this assembly process is altered by exogenous factors, such as mode of delivery and breastfeeding practices, is not fully understood.
In recent years, the impact of mode of delivery (i.e. vaginal vs. Cesarean) on the infant microbiota has been subjected to scrutiny due to the increased rate of Cesarean deliveries worldwide and their potential association with allergic and autoimmune disease19–22. However, the clinical decision to deliver via Cesarean is often indicated by the underlying maternal or fetal medical diagnoses or co-morbidities, and is accompanied by varying use of medications, including antibiotics and anti-inflammatory pain analgesics23. Several previous studies identified distinct microbiota initially colonizing the neonatal microbiome differentially associated with either Cesarean or vaginal deliveries, but many of these were limited by small cohort sizes, potential for significant confounders, or a lack of longitudinal infant sampling24–26. A subsequent study by Azad et al. reported differences in the infant gut microbiota by virtue of the type of Cesarean delivery performed (emergent versus non-labored/elective), suggesting that the differences associated with Cesarean births may be due to the underlying medical indication, rather than the surgical procedure per se27. Indeed, the overwhelming majority of Cesarean births in the United States are not by maternal request but are rather indicated (e.g. arrest of active labor, fetal malpresentation, severe preeclampsia & HELLP syndrome (Hemolysis, Elevated Liver Enzymes, Low Platelets), fetal macrosomia, and/or prior Cesarean delivery28) and yet the impact of such potential confounders on the offspring microbiota have not been established and remains underexplored23. In addition, while Cesarean deliveries can be classified based on whether or not the mother was in active labor prior to the Cesarean surgery (potentially indicating descent of the fetus into the vaginal canal or exposure to vaginal microbes), the impact of labored versus unlabored Cesarean on the offspring microbiota has been initially considered, but remains underexplored and often overlooked by many studies. Finally, since diet is known to be a potent and persistent modifier of the microbiota in both adults and children29,30, and human milk modifies the infant microbiota31–33, infant feeding practices in association with mode of delivery have been accounted for in some studies, but need to be more thoroughly evaluated
Given the complexities of the clinical context surrounding the decision to deliver an infant via Cesarean surgery, we conducted a large, population-based prospective cohort study of maternal-infant pairs to (1) assess the taxonomic composition and potential metabolic function of the early neonatal microbiota across multiple body sites and up to 6 weeks of age, then (2) determine the impact of Cesarean mode of delivery and its potential confounders or modifiers on the neonatal and infant microbiota structure and function. Mothers and their infants were sampled at two time points in early life across multiple body sites, and 16S rRNA gene sequencing and whole genome shotgun (WGS) sequencing was performed on collected samples to interrogate its bacterial composition and function.
Results
We enrolled a population-based cohort of mother-infant dyads for longitudinal sampling (delivery and 4 to 6 weeks postpartum) across multiple body sites (skin, oral cavity, nares, stool, posterior fornix, vaginal introitus; Fig. S1). An endpoint of approximately 4–6 weeks postpartum (hereon out referred to simply as 6 weeks) was chosen because at this age, infants still consume a relatively homogenous diet of human milk and/or formula, have limited person-to-person contact, and are not yet exposed to a wide variety of environmental sources of microbes (e.g. not yet attending daycare and not yet crawling). In all, a cohort of mother-neonate dyads (n=81) was prospectively enrolled from our county hospital (Ben Taub General Hospital, Harris Health System, Houston, Texas) and sampled starting in the early third trimester. The Dirichlet-multinomial distribution was used to power our study to detect a difference in the infant microbiota by virtue of mode of delivery at the 6 week time point. As subjects were enrolled, preliminary sequencing efforts demonstrated lower read counts in neonatal samples taken at delivery as opposed to postpartum (Table S1). Therefore, to increase our power to detect a difference based on mode of delivery at birth, we enrolled a matched cross-sectional cohort of gravidae (n=82) that was sampled once at the time of delivery (Fig. S1A). Of the longitudinally sampled cohort, two withdrew and four delivered at another site and thus were excluded. An additional fifteen subjects were lost to follow up. No losses were incurred in the cross-sectional cohort. In all, 157 mother-neonate dyads were sampled at delivery, with 60 dyads longitudinally sampled (Fig. S1A). As shown in Table S2 and consistent with our delivery population, the enrolled cohort consisted of primarily Hispanic women (90.0%) who delivered singleton pregnancies (96.2%) at term (88.5%). The rate of Cesarean delivery was similar to the U.S. national incidence (33.1% vs. 32.7%, respectively, p=0.91)20. The baseline maternal characteristics of the two cohorts were similar, except for a greater proportion of gestational diabetes in mothers enrolled in the longitudinal cohort (44.0% vs. 17.1%, p<0.001, Table S2). Furthermore, the baseline maternal characteristics were similar between mothers who delivered vaginally (n=105) or by Cesarean (n=52) (all p>0.05), except for a greater proportion of twin pregnancies undergoing Cesarean delivery (Student’s t-test, p=0.008; Table S3). Finally, the rate of gestational diabetes (GDM) and occurrence of Class I-Class III obesity, mean pre-pregnancy BMI, and incidence of fetal macrosomia did not vary among women who underwent Cesarean surgery when compared to those who delivered their infants vaginally (Tables S2 and S3). Overall, the characteristics of the enrolled cohort reflect the typical demographic of patients seen in most Maternal-Fetal Medicine clinics. However, as evidenced by the nationally comparable Cesarean delivery rate, intrapartum management of these subjects’ pregnancies was likely in line with the standard of care provided to the general population.
Neonates and mothers were sampled across five different body habitats (antecubital fossa, retro-auricular crease, keratinized gingiva, anterior nares and stool) representing four major body sites (skin, oral cavity, nares and gut). Samples from the maternal vagina (introitus and posterior fornix) were additionally obtained (Fig. S1B). A second set of samples was obtained for all subjects enrolled in the longitudinal cohort at a follow up visit (mean 43.6 days, s.d. of 17.6 days post-delivery). After quality control and 16S rRNA gene sequencing analysis by 454 pyrosequencing, 1429 high quality samples were available for downstream analysis (mean 8579, s.d. 5681 filtered sequences per sample; Table S1). A summary of filtered sequences per sample by age, body habitat and time point is provided in Table S1.
Early life microbiota community structure
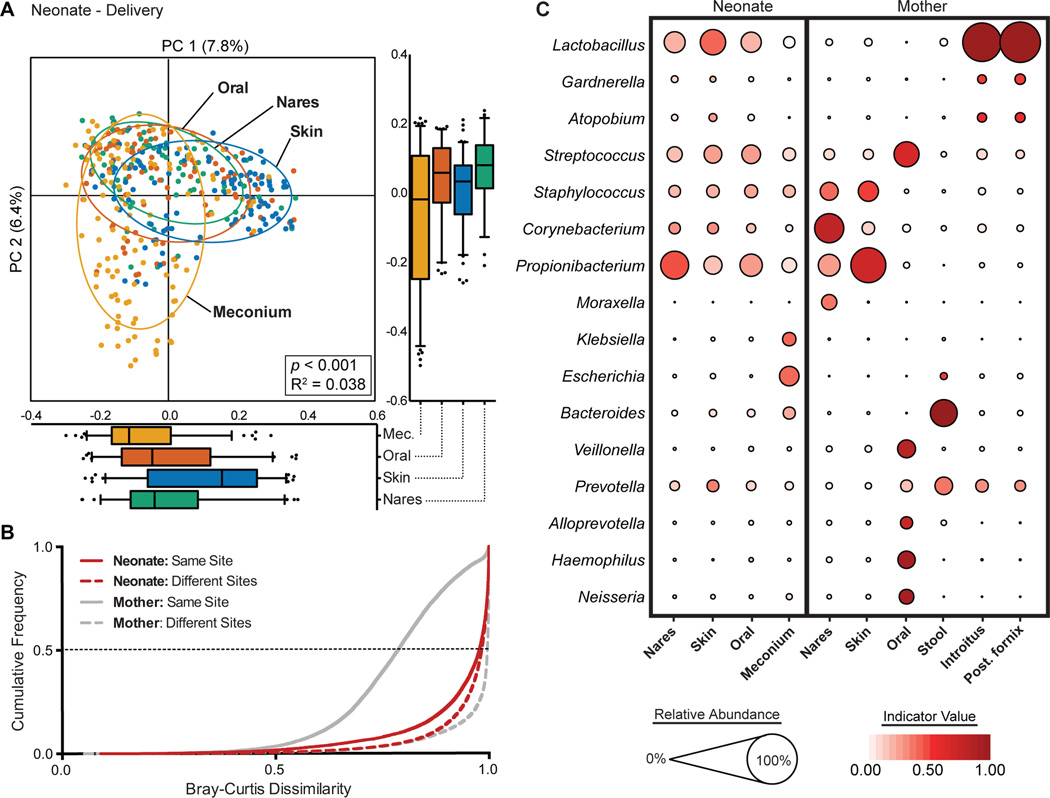
We first sought to examine how the neonatal microbiota varied across different body sites at the time of delivery and then at 6 weeks of age, using the maternal microbiota as a reference of the adult microbiota. Unlike the maternal microbiota, whose community structure was driven primarily by the major body site groupings (Adonis, p<0.001, R2=0.192, Fig. S2), the neonatal microbial community structure at delivery did not demonstrate strong body site differentiation (Fig. 1A). Only the neonatal meconium appeared to cluster separately from the skin, nares and oral cavity (Adonis p<0.001), with a small R2 value (0.038) suggestive of minimal body site variation. Pairwise Bray-Curtis dissimilarity comparisons between samples among subjects (beta diversity) further corroborated these observations. In maternal subjects, the beta diversity between maternal samples obtained from the same body site was significantly lower than that between different body sites (Fig. 1B, gray solid line vs. gray dotted line). Conversely, among neonatal subjects, Bray-Curtis distances were as dissimilar among samples from the same body site as they were between different body sites (Fig. 1B, red solid line vs. red dotted line).
Fig. 1. Neonatal (at birth) microbial community structure.
(A) Principal coordinate analysis (PCoA) on unweighted UniFrac distances between the neonatal microbiota is shown along the first two principal coordinate (PC) axes. Boxplots shown along each PC axis represents the distribution of samples along the given axis, representing the median and interquartile range with whiskers determined by Tukey’s method. Each point represents a single sample and is colored by body site: Meconium (Mec.), Orange; Skin, blue; Oral Cavity, red; Nares, green. Ellipses represent a 95% confidence interval around the cluster centroid. Clustering significance by virtue of body site was determined by Adonis (p<0.001). (B) Cumulative distribution of Bray-Curtis dissimilarity distances calculated pairwise between samples of the same body site (solid lines), and between different body sites (dashed lines). Distance comparisons for neonates and maternal samples are shown in red and gray, respectively. Smaller values indicate a greater similarity between samples. (C) The average relative abundance (circle size) of the most prevalent genera (y-axis) in each body site (x-axis) is plotted for neonates and mothers at delivery. The indicator value index (related to circle color shade darkness) represents the strength of association between a taxa and a given body site, with larger values indicating greater specificity.
Although many different taxa were present in neonates at the time of delivery, few were characteristic of a given body site. To measure the specificity of a taxa to a given site, we determined its Indicator Value (IndVal) Index, which considers the relative abundance of a taxa in a given site and its relative frequency of occurrence across all sites34. A maximum IndVal value represents taxa that are found in only a single site and are found within all individuals. Thus, taxa with a large IndVal can be thought of as a signature taxa for a given body site. Linear Discriminate Analysis Effect Size (LEfSe) analysis was additionally performed to further corroborate representative taxa (Fig. S3). For example, in the maternal subjects, Lactobacillus was both highly abundant and highly specific for the vagina (avg. abundance 64.7%, IndVal = 0.922; Fig. 1C; Table S4), while Bacteroides was prevalent and highly specific for the maternal stool (avg. abundance 27.8%, IndVal = 0.943; Fig. 1C; Table S4), which is consistent with previous observations by ourselves and others1,6,35. In contrast, few signature taxa were detected in the neonatal microbiota at delivery. Consistent with previous studies6,24, predominant members of the vaginal and skin microbiota of the adult, namely Lactobacillus, Propionibacterium, Streptococcus and Staphylococcus, were also the most abundant genera in the neonates across all body sites (Fig. 1C; Table S4). However, none of these taxa were specific to any body site (all IndVal < 0.5). Of interest, many neonatal meconium samples harbored Escherichia and Klebsiella (avg. abundance 14.3% and 6.4%, respectively), which was not seen in any other body site (all < 0.03%). Notably, these taxa are known facultative anaerobes typical of the early gastrointestinal tract30,36, which have been previously detected in the placenta and amniotic fluid by both 16S rRNA gene and WGS sequencing37–39.
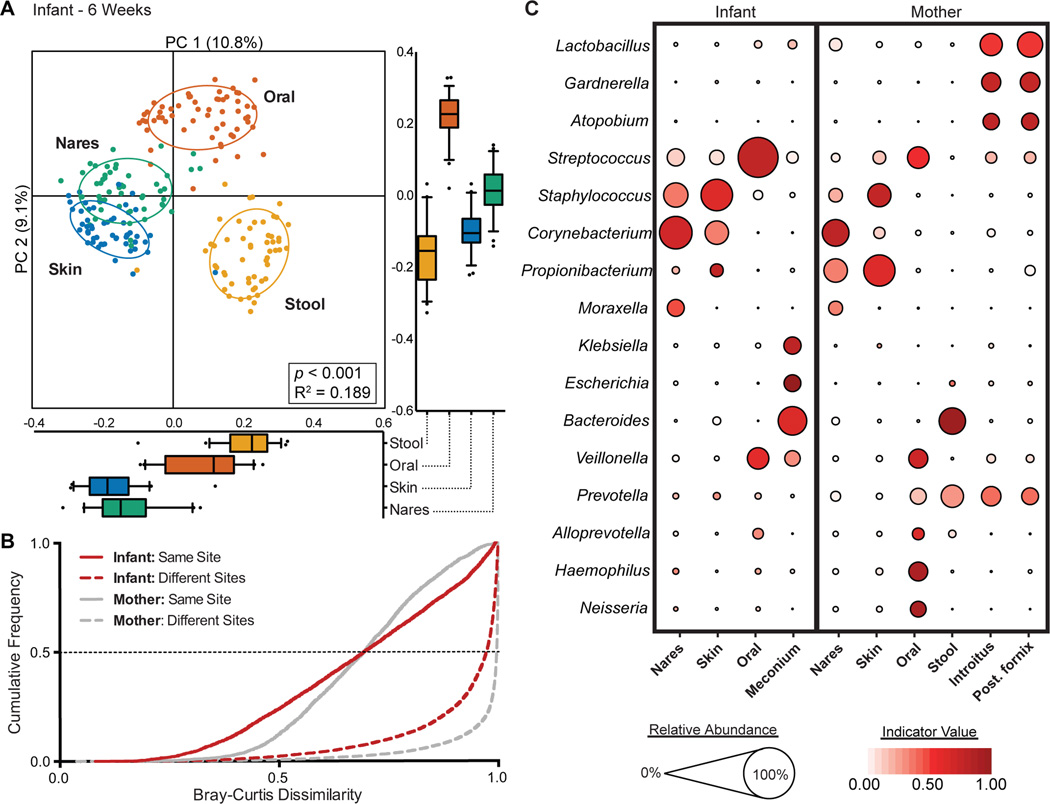
By 6 weeks of age, however, the community structure of the infant microbiota appeared to be primarily driven by body site differences, similar to the maternal microbiota. The stool, oral and skin microbiota clustered distinctly with the nares bridging the oral and skin body sites (Adonis p<0.001, R2=0.189; Fig. 2A). Similarly, patterns of beta diversity (Fig. 2B) and signature taxa (Fig. 2C) were similar to maternal communities (Fig. 2B,C). For instance, the infants’ oral gingiva was similarly characterized by Streptococcus (60.7%, IndVal = 0.73), while the infant’s skin and nares were similarly characterized by Staphylococcus and Corynebacterium (Table S5). More appreciable differences could be detected between the infant and maternal stool. As has been previously published35, the maternal stool was dominated by either Bacteroides or Prevotella, though only Bacteroides was considered a signature taxa for the maternal gut (Bacteroides IndVal = 0.92). In contrast, Escherichia and Klebsiella were both prominently abundant (~10% on average) and highly specific for the infant gut (IndVal, 0.95 and 0.75, respectively), which conforms to previous observations of the typical microbial constituents of infant stool at this age30,40 (Fig. 2C; Table S5). Although the infant and maternal microbiota as a whole shared similar community structure and genus level membership (Fig. 2), the microbial communities of the infant within most body habitats remained distinct at the OTU level. The infant nares, oral cavity and gut clustered distinctly from its maternal counterpart, while no difference could be detected between the infant and maternal skin (Fig. S4). Furthermore, measurements of taxa diversity within a sample revealed that at 6 weeks, with the exception of skin, the infant tended to harbor more simple communities than in mothers (Fig. S5). Thus, while body site differences appear to drive the reorganization of the infant microbiota across body habitats within the first 6 weeks of life, these site-specific communities are generally less ecologically rich and harbor unique communities from the mother.
Fig. 2. The infant microbiota at 6 weeks demonstrates body site specificity.
(A) PCoA on unweighted UniFrac distances between the infant microbiota is shown along the first two PC axes. Boxplots shown along each PC axis represents the distribution of samples along the given axis, representing the median and interquartile range with whiskers determined by Tukey’s method. Each point represents a single sample and is colored by body site: Stool, orange; Skin, blue; Oral cavity, red; Nares, green. Ellipses represent a 95% confidence interval around the cluster centroid. Clustering significance by virtue of body site was determined by Adonis (p<0.001). (B) Cumulative distribution of Bray-Curtis dissimilarity distances calculated pairwise between samples of the same body site (solid lines), and between different body sites (dashed lines). Distance comparisons for infant and maternal samples are shown in red and gray, respectively. Smaller values indicate a greater similarity between samples. (C) The average relative abundance (circle size) of the most prevalent genera (y-axis) in each body site (x-axis) is plotted for infants and mothers at 6 weeks. The indicator value index (related to circle shade darkness) represents the strength of association between a taxa and a given body site, with larger values indicating greater specificity.
Impact of Mode of Delivery on Microbiota Community Structure
Although patterns of microbial composition were influenced by mode of delivery at birth, these differences were absent at 6 weeks of age. By PCoA, we observed marginal significant clustering in the neonatal microbiota by virtue of mode of delivery only in the oral cavity, nares, and skin (Adonis R2 0.038), but not the meconium (Mann-Whitney p>0.05; Fig. 3A). Alpha diversity was similarly reduced in neonates at the time of birth born by Cesarean delivery in the oral cavity and skin microbiota (p<0.001), but not the nares or meconium (p>0.05; Fig. S6). These marginal differences appeared to be driven by an increased association of Propionibacterium and Streptococcus with Cesarean-born neonates while Lactobacillus was associated with those born vaginally (Fig. S7). Reflecting this observation, pairwise beta diversity comparisons between mother-neonate dyads revealed that neonates born vaginally tended to be more similar to the maternal vagina (Fig. S8), which was predominantly comprised of Lactobacillus at the time of delivery (Fig. 1C). However, at 6 weeks, the infant microbiota did not significantly cluster by mode of delivery at any body site (Fig. 3B), nor did alpha diversity significantly differ (Fig. S6). Hierarchical clustering of taxa at the genus level further demonstrated primary clustering by body site, but without appreciable clustering by mode of delivery (Fig. S9).
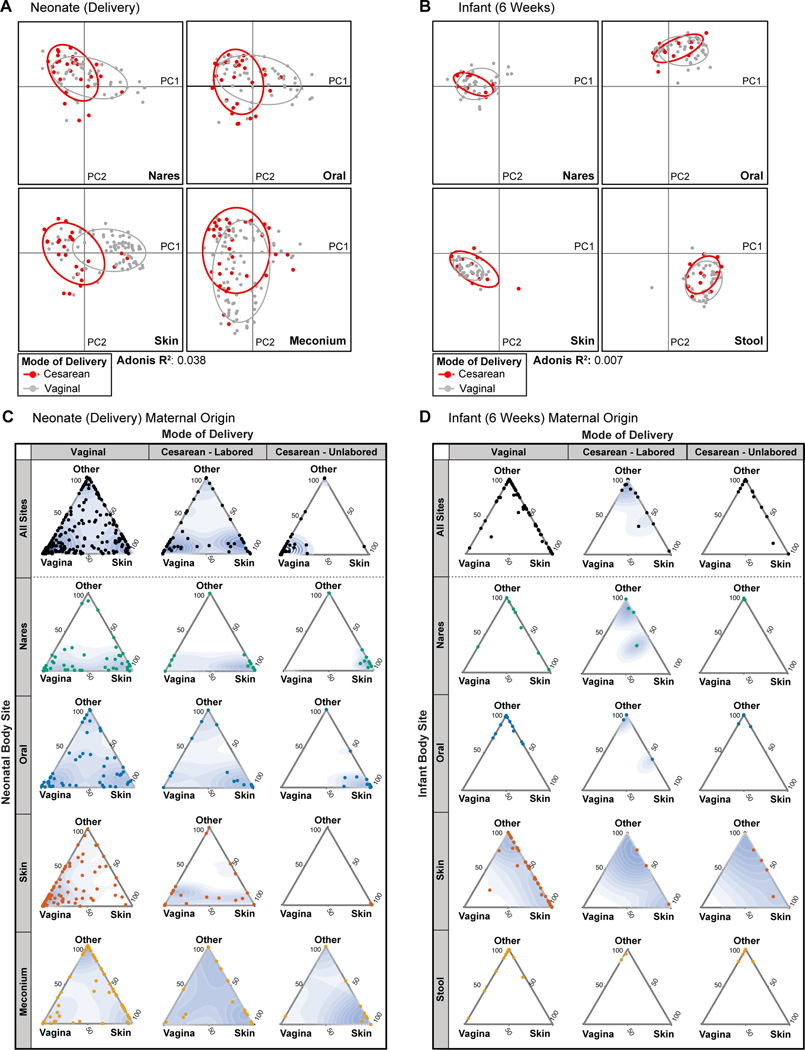
Fig. 3. Failure to demonstrate a significant impact of mode of delivery on the infant microbiota across body sites and time.
(A and B) PCoA on unweighted UniFrac distances between the neonatal microbiota at delivery (A) and 6 weeks (B). Data are stratified by body site, with mode of delivery indicated by color (Cesarean, red; vaginal, gray). Ellipses represent the 95% confidence interval around the cluster centroid. Significance of clustering, which was seen among neonatal (A, delivery) nasal, oral, and skin communities, but not meconium (Mann-Whitney test of PC1 values: p>0.05), was determined by an Adonis test by virtue of mode of delivery, stratified by body site (R2=0.038). Conversely, among infants (B,6 weeks), significant clustering was not observed (Adonis p=0.057, R2=0.007). (C and D) Three axes ternary plots indicating the proportion of OTUs within a neonatal sample (each point) predicted to originate from a maternal body site (indicated by the triangle vertices). Each point represents a neonatal sample while its position indicates the predicted relative contribution from either the maternal vagina (posterior fornix or introitus), maternal skin (retroauricular crease or antecubital fossa) or from another maternal site (supragingival plaque, anterior nares, stool, unknown). Points closer to the vertices indicate that a greater proportion of the samples OTUs are predicted to originate from the microbiota of the indicated maternal body site. Data for samples obtained at delivery (C) and at 6 weeks (D) are stratified by body site and by mode of delivery (Vaginal, Cesarean-Labored, or Cesarean-Unlabored). A 2D point density topography map (blue shading) for each plot is given to indicate the point density.
Cesarean mode of delivery has been demonstrated to impact patterns of maternal transmission of microbiota at delivery, though to what degree of significance and extent this is modified by other maternal or infant clinical co-variates or comorbities remains underexplored. Having extensively sampled multiple maternal body sites in parallel with equivalent neonatal and infant samples, we thus sought to perform SourceTracker analysis41 to predict the most probable maternal origin of the neonatal microbiota. Samples from different maternal body sites were considered to be potential sources of OTUs for each neonatal sample. From this analysis, we obtained an estimated proportion of OTUs of a given neonatal sample predicted to originate from either the maternal skin, vagina, stool, nares, oral cavity or an unknown source (Fig. S10, S11). Because neonates delivered by Cesarean are thought to be populated by maternal skin microbiota rather than vaginal microbiota24,40, data were plotted on a ternary plot to highlight these two maternal body sites (Fig. 3C,D). Moreover, because differences in the neonatal microbiota have previously been shown by virtue of Cesarean indication (emergent vs. non-emergent)27, comparisons were further stratified by whether or not the mother had labored before the Cesarean procedure (here on out referred to as a labored or unlabored Cesarean delivery). For neonates delivered vaginally, most body sites demonstrated a bimodal pattern of maternal origin – while many sites were predominantly populated by microbiota originating of the mother’s vagina, a substantial number were predominantly populated by microbiota of the maternal skin (Fig. 3C). Notably, meconium samples additionally harbored OTUs predicted to originate from the maternal stool (Fig. S10). Thus, except for the infant gut, the maternal vagina and skin appear equally contribute to a majority of the early taxa of the vaginally delivered neonate. This bimodal pattern among vaginally born neonates was similarly seen in neonates delivered by a labored Cesarean, but not by an unlabored Cesarean (Fig. 3C). In the latter case, a majority of samples from neonates delivered by an unlabored Cesarean were primarily populated by microbiota found in the maternal skin. Thus, whether or not labor had occurred prior to delivery, rather than the Cesarean procedure itself, appeared to have the greatest impact on the maternal origin of microbiota.
By 6 weeks of age, however, infant samples were predominantly populated by microbiota from its corresponding maternal body site, irrespective of mode of delivery (Fig. 3D, Fig. S11). Although the maternal skin and vagina represented a substantial portion of taxa of the neonatal microbiota at delivery, most appeared to be replaced in favor of taxa more typical of the given body niche. For example, while either Propionibacterium or Lactobacillus largely comprised the neonatal oral microbiota at the time of delivery (Fig. S7), by 6 weeks of age, Streptococcus was the most prominent genera of nearly all infant oral samples (Fig. S9).
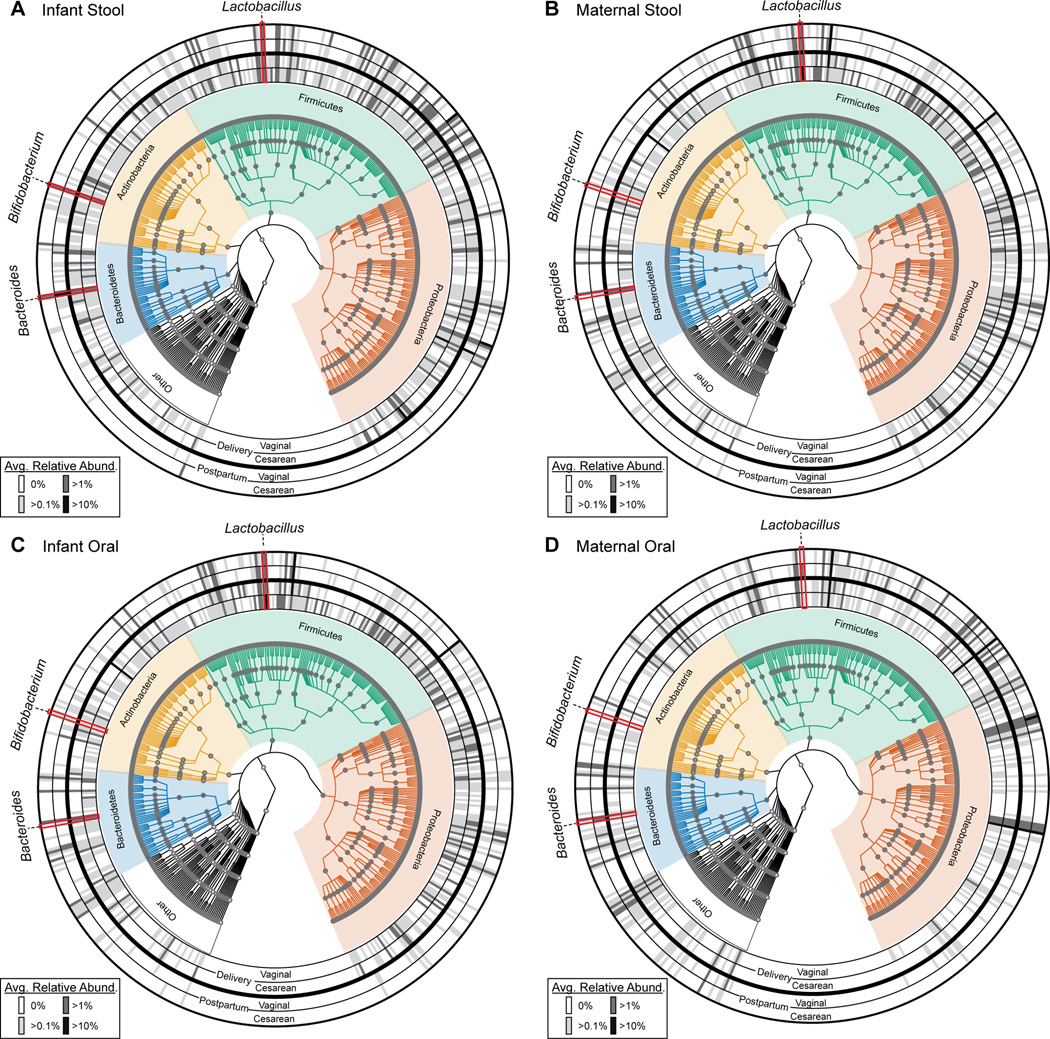
Previous studies24,40,42–44 have indicated that mode of delivery has the greatest influence on the presence and abundance of several notable taxa of the early infant gut microbiome, namely Bacteroides, Lactobacillus and Bifidobacterium. We next sought to examine how these three genera were impacted by mode of delivery and other clinical variables at 6 weeks of age. The abundance of these three taxa were classified as either being undetectable (0%), sparsely present (>0.1%), appreciably present (>1%) or moderately abundant (>10%). Comparisons of the gut microbiota at delivery revealed that in all Cesarean and the majority of vaginally delivered infants, Lactobacillus, Bifidobacterium and Bacteroides were appreciably (i.e., not undetectable at greater than 0%) present at the time of delivery (Fig. 4A–D). By 6 weeks of age, Lactobacillus and Bifidobacterium continued to be appreciably present and similar by virtue of mode of delivery in the neonatal stool (Fig. 4A). By shotgun sequencing, Bacteroides were undetectable in just three infant stool samples, all of which belong to infants born not by Cesarean but rather by vaginal delivery (Fig. S12). To further interrogate if mode of delivery had a significant impact on the relative abundance of these three genera while controlling for other factors, we employed a generalized linear model. Based on previous literature, breastfeeding practices26,32,42, gestational age36, maternal and infant obesity/macrosomia45,46, maternal diet (% fat intake)47 and maternal gestational diabetes48 all have been reported to impact the very early infant microbiome; thus these were used in model creation. Of the imputed factors, breastfeeding practices was the only significant contributor to the abundance of Bacteroides in the infant stool at 4–6 weeks of age, with exposure to formula associated with increased levels of Bacteroides (Fig. S13, Table S6). We have recently published47 that increased maternal fat intake during gestation is significantly associated with decreased abundance of Bacteroides, but only after a potential outlier was excluded from analysis. No factors were significantly associated with altered Bifidobacterium or Lactobacillus abundance (Fig. S13, Table S6).
Fig. 4. Taxonomic profiles of infant and maternal stool and oral microbiomes according to mode of delivery and time.
A phylogenetic representation of the taxonomic composition in the infant stool (A), maternal stool (B), infant oral (C), and maternal oral (D) samples at delivery (inner two rings) and 6 weeks postpartum (outer two rings). Each time point is further divided by mode of delivery (indicated by the label at the bottom of the rings. The average relative abundance of each genera are plotted within the concentric rings, represented by the shaded cells, with higher relative abundance indicated by a darker shade. The phyla to which each taxa belongs is indicated by the phylogenetic tree. Three notable taxa are indicated by the red outline, namely Bacteroides, Bifidobacterium and Lactobacillus.
Expansion and Diversification of Microbial Community Function
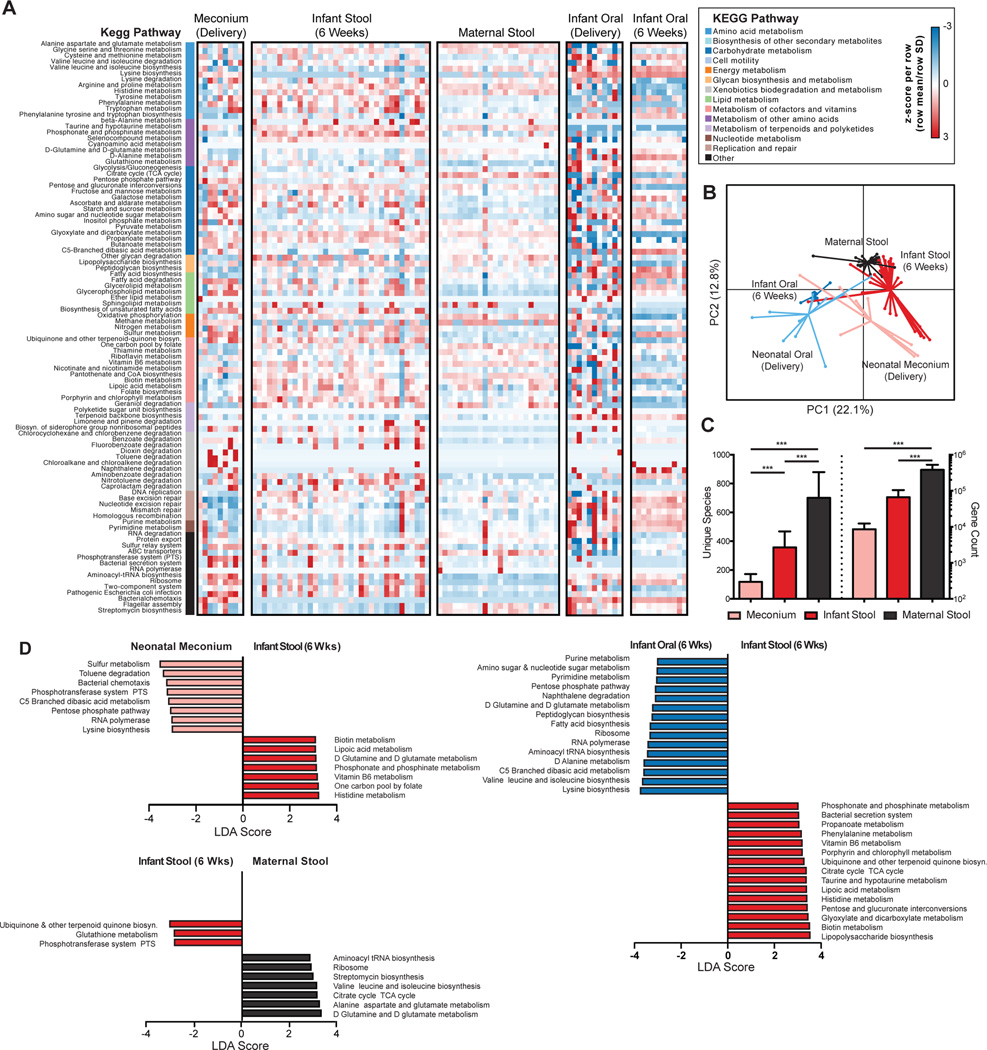
We next sought to examine how the microbial metabolic and functional pathways of the early neonatal metagenome changed from delivery to 6 weeks across different body sites. Overall, the metagenomic profiles clustered by virtue of age and by body site (Fig. 5A and 5B), indicating overall similarities in site-specific microbial activities. However, while the maternal gut metagenome appeared much more consistent and evenly diverse than taxonomic abundances (Fig. 5A, see also Table S5), a significant amount of heterogeneity was seen in the neonatal oral and gut metagenomes at delivery (Fig. S14). This was particularly apparent in the neonatal oral cavity. For example, the relative abundance of alanine, aspartate and glutamate metabolism in neonatal oral samples were undetectable in half of the sequenced samples (0% relative abundance), but present at greater than 2.3% in the other half (median abundance across all samples 2.4%, Table S7). However, at 6 weeks, both the infant oral and stool metagenomes were significantly more similar between body sites of different individuals (Fig. S14), indicating convergence toward a shared set of microbial metabolic pathways and activities unique to each body site. Comparisons of the infant stool and oral cavity metagenomes revealed that each was enriched for pathways that potentially reflected a selective advantage of specific microbiota to its local environment (Fig. 5D). For example, lipopolysaccharide biosynthesis is a key feature of gram-negative enteric bacteria and was fittingly increased in the infant stool metagenome compared to that of the oral cavity. Additionally, taurine and hypotaurine metabolism, which is critical for bile acid metabolism and conjugation49, was also increased in the infant stool metagenome compared to that of the infant oral cavity. In contrast, amino acid biosynthesis and metabolism was seen to be a predominant feature of the infant oral metagenome.
Fig. 5. Expansion and diversification of the infant microbial community structure and function by 6 weeks of age.
(A) Heatmap showing distinct microbial gene (KEGG pathway) profiles of the infant stool and oral metagenome at delivery and 6 weeks of age. The microbial gene profile for maternal stool is shown as a comparison. The relative abundance of a pathway in a given sample is colored by its row z-score ((value – row mean)/row standard deviation). The vertical color bar represents the higher order KEGG module to which each pathway belongs. (B) PCoA of Bray-Curtis distances based on pathway relative abundances demonstrates primarily clustering by body site and time point (PERMANOVA, p<0.001). (C) Enumeration of the unique number of genes and species within the neonatal meconium (delivery), stool (6 weeks) and maternal stool (ANOVA, *** p<0.001 by post-hoc Tukey’s tests). (D) Results from Linear discriminate analysis Effect Size (LEfSe) analysis conducted to identify pathways that differentiated the infant stool metagenome at delivery and at 6 weeks (top left), the infant and maternal stool metagenomes at 6 weeks (top right) and the infant oral and stool metagenomes at 6 weeks (bottom). Only significant pathways with an LDA score >3.0 are shown.
From delivery to 6 weeks, the number of unique species and microbial genes found within the infant gut metagenome was also significantly increased, indicating an overall expansion and diversification within this early time period (Fig. 5C). The neonatal meconium was enriched for several microbial pathways including the pentose phosphate pathway and the phophotransferase system (PTS), which participate in sequestering and utilizing glucose for anabolism of amino acids and cell wall components. In comparison, the infant gut metagenome at 6 weeks was significantly enriched for pathways related to co-factor metabolism and biosynthesis, including folate, biotin and vitamin B6, which are known to be important for growth, metabolism and neurodevelopment50. However, the infant gut metagenome was still greatly outnumbered by the maternal in both gene counts and unique species present (Fig. 5C). Furthermore, several pathways discriminated the infant and maternal gut metagenomes (Fig. 5D), including an enrichment of TCA cycle and amino acid metabolism in the maternal gut. Thus, while the infant gut metagenome was significantly expanded and diversified from birth, both its taxonomic composition and metabolic capability remained distinct from maternal gut.
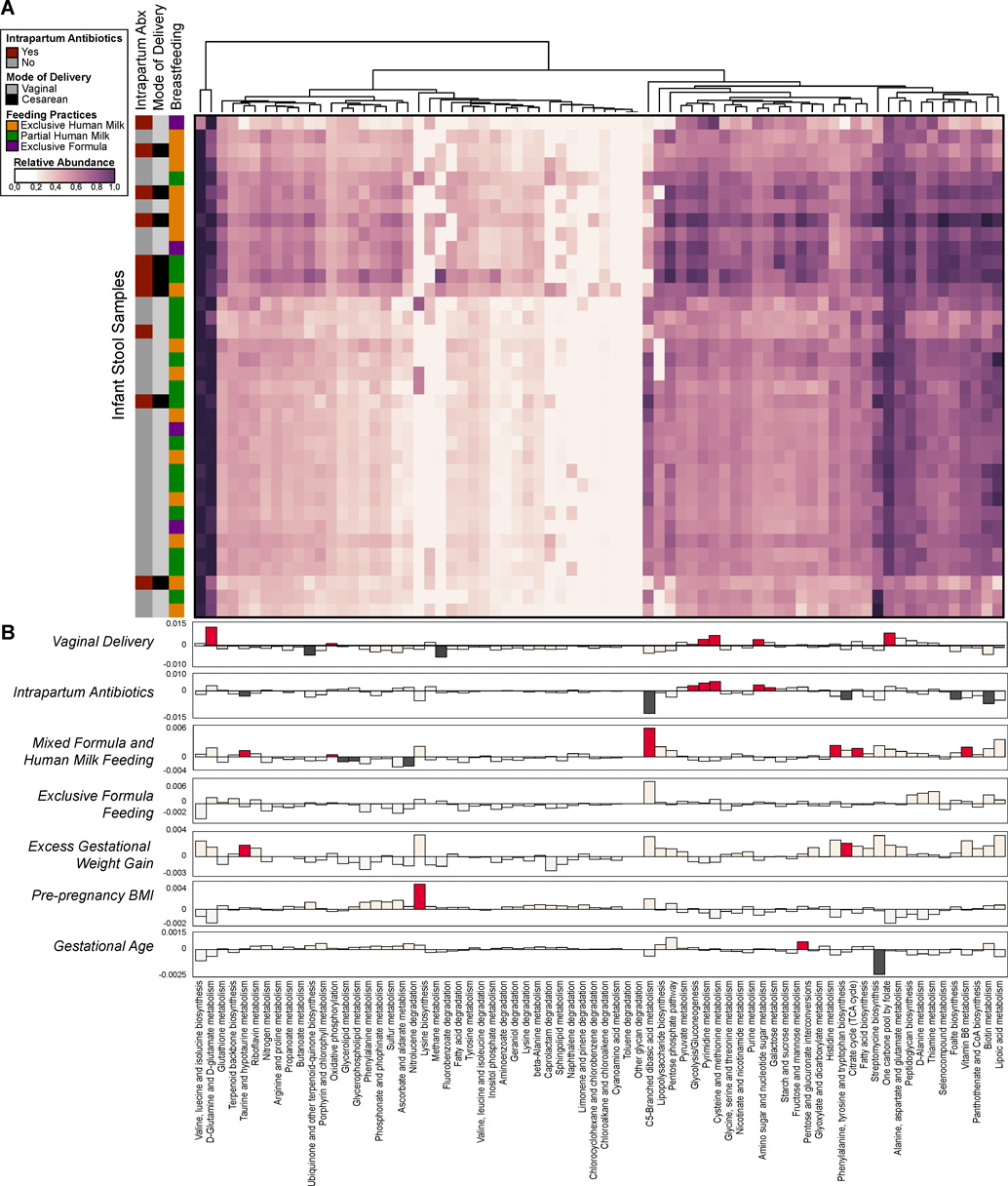
In order to assess how the metabolic and functional pathways of the infant stool metagenome varied with Cesarean or vaginal mode of delivery, breast feeding practices and other clinical metadata, we fitted a generalized linear mixed model to infant stool samples to identify significantly differing pathways. This approached allowed us to quantify the contribution of each variable to the pathway abundances, while controlling for possible confounding between clinical metadata. Again based on previous literature, we considered mode of delivery, intrapartum antibiotics51, breastfeeding practices at 6 weeks of age32, gestational weight gain38, maternal gestational diabetes or pre-pregnancy BMI46, and gestational age36 at the time of delivery as possible fixed effects in our model. Of all of the pathways considered, no single pathway correlated with any clinical variable considered in our model, indicating that the relative frequency of most functions throughout the infant stool microbiota were consistent across individuals and robust to exogenous perturbations including Cesarean delivery (Fig. 6A). Similarly, few species in the infant stool identified by WGS sequencing were significantly altered in our model (Fig. S15). Intrapartum antibiotics appeared to have the greatest effect on pathway variation, as ten distinct pathways were either positively or negatively correlated with intrapartum antibiotic usage (Fig. 6B). In contrast, gestational age and pre-pregnancy BMI had little impact on pathway variation within the infant stool. Of particular interest, given co-linearity with antibiotic usage and breastfeeding practices, Cesarean mode of delivery per se did not bear a differential effect on the infant metagenome nor its function when subjected to robust linear mixed modeling controlling for covariates as fixed effects.
Fig. 6. Infant microbial community function with clinical metadata in a generalized linear model.
(A) Heatmap of the relative abundances of KEGG pathways found in infant stool samples as determined by WGS sequencing. The vertical bar on the left indicates mode of delivery, breastfeeding practices and intrapartum antibiotic usage. Dendrograms represent hierarchical clustering on Euclidean distances using average linkage. (B) A generalized linear mixed model was fitted for each pathway to identify pathways whose abundances differed significantly between individuals by virtue of mode of delivery, antibiotic usage, breastfeeding, gestational weight gain, BMI and gestational age. The strength of the linear model predictions for each pathway is represented by bar height. Significant correlations are indicated by the darker color (dark red or dark gray). Labels correspond to the following comparisons: Vaginal Delivery - pathways enriched in infants born vaginally (red, up) or by Cesarean (gray, down); Intrapartum Antibiotics - pathways enriched in infants exposed to intrapartum antibiotic usage (red, up) as opposed to no antibiotics (gray, down); Mixed Formula and Human Milk Feeding - pathways increased in partially breast fed (human milk and formula) infants (red, up) as opposed to exclusive human milk only (gray, down); Exclusive Formula Feeding - pathways higher in exclusively formula fed infants (red, up), as opposed to human milk only (gray, down); Excess Gestational Weight Gain - pathways higher in cases of excess maternal gestational weight gain (red, up), as opposed to normal weight gain (gray, down); Pre-pregnancy BMI & Gestational age - pathways positively (up, red) or negatively (down, gray) correlated with pre-pregnancy BMI or gestational age.
Discussion
In this large cohort of term infants reflective of characteristics common to the contemporary U.S. birth population, we found significant reorganization of the neonatal microbiota across multiple body sites within the first 6 weeks of life that was primarily shaped by the major body site groupings as seen previously in the adult1. Although the neonatal microbiota at the time of delivery was sparsely populated and predominantly comprised of taxa of the maternal skin and vaginal microbiota, by 6 weeks of age, the major patterns of community variation was most strongly associated with body site grouping with concomitant coalescence of functional and metabolic microbial pathways reflective of probable selection within or adaptations to a given body niche. However, while the infant skin most resembled the maternal microbiota, the stool, nares and oral cavity harbored distinct microbial communities from their maternal counterparts. These differences likely reflect age related physiological differences between the maternal and infant body habitats, including nutrient availability and oxygen exposure.
Overall, our data reach similar conclusions to those derived from observations of low birth weight infants18, which found that changes to microbiota composition within the first weeks of life is primarily shaped by body site. We similarly found that after several weeks, each body niche was enriched for taxa that were characteristic of their adult counterparts, such as Streptococcus in the oral cavity, likely indicating a common maturation process. However, our study further extends these observations though the generation of rich WGS data, which provides the benefit of interrogating both taxonomic abundances and community function within this early time period. Our results suggest that similar to microbiota abundance, site-specific functional pathways emerge very early on in life, before extensive contact to environmental sources which may influence community membership and consequent function.
There are several distinctions between the characteristics of our study cohort and those of other studies which have reported a variation in the neonatal or infant microbiome by mode of delivery24,40,42,43,52. First, our cohort was ethnically homogenous (mostly Hispanic) and was comprised of primarily term infants; thus specific observations of the predominant taxonomic and pathway abundances of the infant metagenome may not be generalizable to preterm neonates or other ethnic populations. Previous observations have indicated that in adult and infant populations43, there are large differences between individuals of different ethnic populations residing in different countries and societal structures. Thus the specific taxa or functions that become enriched early on may vary depending on the environmental context. Nevertheless, considering that the adult microbiota is driven to a larger extent by body site habitat and not ethnicity, we anticipate that gross reorganization of the infant microbiota by body site within this early time period is generalizable across populations, though additional studies are needed to confirm this.
Second, reflective of our clinical population from which these women were recruited for our longitudinal cohort, we observed a high prevalence of gestational diabetes. However, the rate of gestational diabetes did not vary among women who later delivered their infants via Cesarean or vaginal birth (Table S2) and gestational diabetes (as well as other potential or likely confounders) were included in our GLM analysis. Since other studies have not reported maternal gestational nor Type II diabetes as characteristics of their cohort, we cannot directly compare nor speculate as to whether their findings might be a result of confounding or co-linearity24,40,42–44,52. This would necessarily include those characteristics and comorbidities which render risk or indication for Cesarean delivery, such as poorly controlled diabetes with resultant fetal macrosomia. As one such example, it is worthwhile noting that the landmark study of Dominquez-Bello et al does not report at-risk maternal characteristics such as diabetes nor BMI, but does describe infant birth weight ranges of up to 5.2 kg24. Thus, in their study comprised of 11 neonates from Venezuela (6 born via Cesarean surgery), at least one neonate was macrosomic (“Babies weighed between 2 and 5.2 kg (the smallest baby was the twin in second order of birth, after his 3-kg brother)”)24. A 5.2 kg infant would be 1.0 kg heavier than more than 98% of the birth population by WHO standards. While there are multiple underlying causes of fetal macrosomia, common causes include poorly controlled maternal diabetes. Additional causes include genetic and epigenetic overgrowth disorders, chronic caloric excess, and maternal obesity. These were excluded or controlled for in our study analysis, and our mean infant birthweight was 3.237 kg (±0.68 kg) with no significant difference in birthweight observed when comparing infants born vaginally to those delivered by Cesarean (p 0.85; Table S2 and S3). Notably, in our entire cohort of 162 infants, only one exceeded 5 kg (birthweight 5.085 kg; Table S8).
Our observations of the early neonatal microbiota are consistent with the growing body of evidence that is challenging the notion of a sterile in utero environment. Previous studies have detected bacteria associated with the placenta and amniotic fluid of preterm and healthy, term pregnancies37,39,53–55 while work in murine models have demonstrated maternal transmission of bacteria to the fetal gut during gestation, consistent with microbial colonization of the mouse fetus occurring prior to delivery56. More recently, the neonate’s first intestinal discharge (meconium) has been shown to harbor a microbial community similar to that of the amniotic fluid and placenta39,57,58. Interestingly, the meconium microbiota was shown to vary with maternal glycemic control, but not mode of delivery, indicating that establishment of the neonatal microbiota can be potentially altered by gestational exposures48. Consistent with these findings, we observed that the meconium microbiota was markedly different from other neonatal body sites at the time of delivery, indicating a potentially different maternal origin. Additionally, unlike the skin, oral cavity or nares, the neonatal gut microbiota at the time of delivery did not significantly vary by mode of delivery. The content of the first meconium is hypothesized to reflect the in utero environment (whereby the infant is swallowing amniotic fluid continuously from mid to late gestation), and thus we speculate that these microbes were similarly transmitted from the mother to the fetus during gestation, suggesting that seeding of the early microbiota may occur earlier than was previously thought. However, additional studies are needed to evaluate potential mechanisms of transmission and its potential impact on fetal programming and the establishment of the infant gut microbiome long-term.
In agreement with previous observations24,25, mode of delivery was associated with differences in the neonatal microbiota immediately after delivery only within the nares, skin and oral cavity. However, this was not true of the gut microbiota, which appeared to have a distinct maternal origin from the rest of the body sites. A number of studies have indicated that the early composition of the gut may be influenced by maternal exposures or health status, which may account for the lack of separation by mode of delivery in our study. Our previously published work in a non-human primate model demonstrated that maternal diet during gestation and lactation has a persistent impact on the gut microbiota of the offspring at least up to one year of age. In conjunction with these findings, we’ve similarly seen in human cohorts that the composition of the early infant gut microbiome associated with maternal diet in the last trimester of pregnancy, independent of mode of delivery and maternal obesity47. In addition to maternal gestational diet, maternal pre-pregnancy obesity, and maternal glycemic control us have also been attributed to differences in the early infant gut microbiome. Notably, these maternal health states also increase the risk that the pregnancy will be delivered by Cesarean28. However, accounting for these potential confounders in our study, we did not find significant differences in infant gut microbiome which could be reliably nor significantly attributed to Cesarean nor vaginal mode of delivery.
We additionally sampled multiple body sites of both the neonate and mother, which allowed for an in depth interrogation of the potential maternal origin of neonatal microbiota. Division of Cesarean deliveries by indication (labored vs. unlabored) revealed that the differences in the neonatal microbiota at birth associated with mode of delivery was most pronounced when comparing unlabored Cesarean to vaginal deliveries. In our cohort, these unlabored Cesarean deliveries were generally among women with a history of prior Cesarean and electing for a repeat Cesarean, rather than a trial of labor with attempted vaginal birth after Cesarean. This is in agreement with the observations of Azad et al.27, which identified differences in the infant gut microbiome at 4 months of age when Cesarean deliveries were classified as either being emergent (typically labored) or elective (typically unlabored). This is not surprising, as Cesarean delivery is not a randomly allocated procedure but rather is indicated for an underlying maternal or fetal medical condition23. Ergo, prior small studies24,25 which do not account for underlying indication for the Cesarean surgery, may be prone to both Type I and Type II error resulting in misclassification of the attribution of Cesarean delivery per se as a driving force in any dysbiosis in the infant microbiota.
A number of perinatal and postnatal factors have been associated with relatively low neonatal and infant stool abundance of Bacteroides. These include: Cesarean delivery40,43,52, exclusive breastfeeding26,42,59, maternal high fat diet47, maternal obesity46, transition to solid foods30, and other yet unknown factors52. Based on the work of Bakhed et al.40, Bacteroides species can be acquired postnatally, with a notable increase at 4 months of age among both Cesarean and vaginally delivered infants. Ergo, any supposition relating relative abundance and less abundance to Cesarean delivery per se must be threaded with caution. This may include introduction of antibiotics postnatally51,52,60, formula feeding42,59, as well as solid foods30. In the current study, we focused on early exposures since they are most proximal to the intrauterine and intrapartum period of exposure, and least likely to be subjected to exogenous postnatal influences. We have now demonstrated with WGS data that both vaginally and Cesarean delivery infants can have low Bacteroides counts. In our cohort, and distinct from that of Bäcked40, no Bacteroides at birth at 6 weeks of age was found only among vaginally delivered infants (n=3) and every Cesarean delivered infant had detectable Bacteroides (Fig. S12). These findings are consistent with a recent publication by Yassour et al.60 which similarly observed low Bacteroides in 20% of vaginally delivered infants. Even when we extend our definition of significantly variant taxa to include those identified by Dominquez-Bello as being perturbed in Cesarean and only restored with vaginal wiping44, we failed to see significantly altered taxonomic profiles. Specifically, sequential definitions of low Bifidobacterium or high Lactobacillus were equally probable in vaginally and Cesarean delivered infants (Fig. S12). Finally, when we examined 4–6 week old infants with either absent or relatively low Bacteroides and assigned relative risk estimated for a number of antenatal factors, low Bacteroides were most likely to be delivered by vaginal birth (Fig. S12).
In sum, we have undertaken the largest study to date utilizing whole genome shotgun analysis to analyze both the composition and function of the neonatal and infant microbiota with paired maternal-infant across multiple body sites. We have observed that by 6 weeks of age, the microbial community structure and function has significantly expanded and diversified. We further demonstrate that there is no discernable impact of Cesarean mode of delivery on the early microbiota beyond the immediate neonatal period (and never inclusive of meconium nor stool), and that early taxonomic distinctions are not recapitulated by functional pathway analysis. These findings underscore the likely importance of interrogating not only the relative abundance of members of the microbiota, but their potentially overlapping metabolic pathways, when considering the impact of an intervention on diversity of the microbiota.
Methods
Experimental Design
The intent of this prospective cohort study was to characterize the neonatal microbiota in early life and subsequently compare the neonatal microbiota by virtue of mode of delivery. An overview of the study design is shown in Fig. S1A. An initial cohort of gravidae and their infants (n=81) were prospectively enrolled in the 3rd trimester from the Ben Taub County Hospital in Houston, Texas. Assuming a Cesarean rate equivalent to the U.S. national average (32%)20, this cohort size achieved a power of at least 0.80 to detect a difference in the taxonomic composition by virtue of mode of delivery at the 6 week time point, based on an estimated read count of at least 5000 and a small effect size (Φ=0.07 at an α=0.05). Power calculations were based on the Dirichlet-multinomial distribution as described in La Rosa et al.61. A second cross-sectional cohort (n=82) was additionally enrolled to increase the power to detect differences by virtue of mode of delivery at the time of delivery. This second cohort increased enrollment of Cesarean deliveries, thus capturing a larger number of types of Cesarean indications (labored vs. unlabored, n=26 for both, Table S9). All subjects were enrolled under Baylor College of Medicine Institutional Review Board (IRB) H-27393. Subjects included in this study had a viable pregnancy >28 weeks gestation, were 18 years of age or older, and willing to consent to all aspects of the protocol. Subjects were excluded if there was known HIV or Hepatitis C infection, known immunosuppressive disease, known use of cytokines or immunosuppressive agents within the last 6 months, a history of cancer except squamous or basal cell carcinoma of the skin managed by local excision, treatment of suspicion of ever having toxic shock syndrome, or major surgery of the GI tract except cholecystectomy or appendectomy in the past five years. During the consent process, subjects were informed of the potential risks of participation, including the minimal physical risks associated with specimen collection, and the possibility that protected health information or de-identified project data could be accidentally released. Several precautions described in the protocol and consent form were taken to reduce these risks.
Clinical Metadata
Clinical metadata for all subjects are listed in Supplementary Tables 6 and 7. The cohort that each subject was recruited is listed under Study as either longitudinal or cross-sectional. Gestational age at delivery is provided as weeks and days. Standard definitions of “neonate” (28 days or less) and “infant” (29 days or greater) were employed, albeit neonates were sampled within 1 hour of birth. Neonates were considered preterm if delivered <37 weeks gestational age. Twin births (multiples) are indicated. The indication for Cesarean delivery is provided as either being primary or repeat (first Cesarean or repeat Cesarean). Whether the mother was in labor prior to Cesarean delivery was also recorded (Labored vs. Unlabored). Antibiotic usage was noted if given antepartum or intrapartum (around the time of delivery). Breastfeeding practices were assessed by careful chart review and classified as either being exclusively formula fed, exclusively fed human milk, or fed both formula and human milk (partial human milk) within the first 6 weeks of life. Birth weight was provided in grams and percentile given as a function of gestational age and gender. Maternal diet during pregnancy was determined for the one month period prior to enrollment using the National Health and Examination Survey Dietary Screener Questionnaire as described previously47.
Sample Collection and Processing
Specimens were sterilely and uniformly collected by trained personnel according to a standardized protocol as previously described62. A summary of specimens collected at each time point from each maternal-infant pair is shown in Fig. S1B. All specimens were stored at 4°C and processed in the lab within 24 hours. In a decontaminated, sterile environment, genomic DNA was isolated from each specimen using the MOBIO PowerSoil DNA Isolation Kit (MOBIO) with the standard manufacturer’s protocol. Extracted DNA was prepared for sequencing according to the HMP consortium outlined protocol1 for 16S rRNA gene sequencing.
16S rRNA Gene Sequencing Analysis
To characterize the microbial composition of each specimen, the hypervariable regions (V) of the 16S ribosomal gene were sequenced from the extracted DNA as described in the Human Microbiome Project1,62. The V5-V3 region of the 16S rRNA gene was amplified by polymerase chain reaction using bar-coded universal primers 926R (R, Reverse Primer) and 357F (F, Forward Primer). Amplicons were quantified by PicoGreen and pooled in equimolar amounts. Multiplex sequencing was performed on a 454-FLX Titanium Sequencer (Roche) at the Human Genome Sequencing Center (HGSC) at Baylor College of Medicine (BCM). In total, all samples were sequenced across 32 sequencing runs (Table S10). To reduce bias related to sequencing in different runs, samples from both mother and infant of all body sites were sequenced as they were obtained, blinded to subject clinical metadata. Given the nature of enrollment, samples from either the cross sectional and longitudinal cohort tended to be sequenced in different pools; however, as shown in Fig. S16, the data did not appear to strongly vary by virtue of cohort enrollment.
Raw 16S rRNA gene sequence data was processed using the QIIME platform63 (v 1.9.0) using the default parameters on the suggested pipeline (http://qiime.org/tutorials/tutorial.html) with notable changes as described below. Sequences were preprocessed to remove sequences less than 200 nucleotides, greater than 700 nucleotides, or sequences with a minimum average quality score less than 25. Remaining sequences were trimmed of reverse primer sequences and trimmed at the first ambiguous base call. Sequences below 200 nucleotides after trimming were subsequently removed. Retained high quality sequences were subsequently assigned to the sample of origin using the uniquely identifying barcode sequence and thereafter trimmed of the barcode and 5’ primer. To remove human contamination, high quality reads were mapped to the human genome (GRCh38) using DeconSeq (v 0.4.3) and highly similar sequences to the human genome reference were subsequently removed from the dataset64. Remaining sequence data and associated flowgram data were aggregated by body site and denoised to reduce sequencing errors using the QIIME denoising pipeline (denoise_wrapper.py). All denoised sequences were aggregated and de novo operational taxonomic unit (OTU) were identified. Sequences were clustered into distinct OTUs at 97% similarity using the UCLUST65 method. Chimeric sequences were identified and removed using ChimeraSlayer on the QIIME workflow script (v 1.9.0, identify_chimeric_seqs.py)66. The RDP classifier67 (v2.2) retrained against the May 8th 2013 version of the GreenGenes68 taxonomic database was used to assign taxonomy for each OTU at a confidence greater than 50% at the lowest level of assignment. Read counts for each OTU were tabulated for downstream analysis. In all, 2581 samples were collected from the maternal-infant pairs and used for 16S rRNA gene sequencing. After filtering of singletons and removal of samples with less than 100 sequences, 1429 samples were retained for downstream analysis (mean 8578.5 s.d. 5680.9 filtered sequences per sample (Table S1).
The specificity of certain taxa to a given body site was assessed by calculating the indicator value index as previously described34. Each body grouping (skin, nares, oral cavity, stool, vagina) within the infant or mother was considered a different site. For each genera in each site, we computed the mean abundance of each taxa compared to all sites, multiplied by the frequency of appearance of that taxa (abundance > 0.0%) across all samples within the site. Bubble plots of relative taxonomic abundances were generated using the R package ggplot. Linear Discriminant Analysis Effect Size (LEfSe v1.0) was performed with a LDA cut off of 2.0 with a significance value (p) of 0.0569. Diversity within samples (alpha diversity) and between samples (beta diversity) were evaluated on the operational taxonomic unit (OTU) table (16S) or KEGG pathway table (WGS) as indicated. For alpha diversity, samples were first rarefied at 2000 sequences before diversity indexes were calculated. Significance was determined by Mann Whitney U tests. To evaluate OTU discovery as a function of sequencing depth, reads per sample were plotted against the number of unique OTUs using PRISM (GraphPad Software, La Jolla, CA) and regression lines were fitted. Significance between linear regression slopes was determined by Student’s t-test. For beta diversity measurements, both phylogenetic (UniFrac) and non-phylogenetic (Bray-Curtis, Jaccard) distance matrixes were determined. Cumulative distribution plots of beta diversity distances were generated using PRISM. Principal component analysis (PCoA) was performed on distance matrices as indicated with significance of clustering determined by PERMANOVA or Adonis with 999 permutations. PCoA plots and confidence ellipses were generated by the R package ggplot. Heatmaps were generated using the R package pheatmap. Heirarchical clustering was performed using complete linkage on Jaccard similarity coefficients or Bray-Curtis dissimilarity distances using the R package vegan as indicated. Phylogenetic trees were created with PhyloPhlAn (v0.99). Association of the metadata categories with the hierarchical clustering in the dataset was determined by a Chi-squared test. A generalized linear model with a normal distribution according to a log link function was fit by maximum likelihood using with JMP (v12.2.0). All statistical analysis was performed on the QIIME platform (v 1.9) or R (v 3.2.2) as indicated.
SourceTracker Analysis to Predict the Maternal Origin of OTUs in Neonatal Samples
SourceTracker analysis was performed on the QIIME platform to predict the likely origin of microbiota in the neonatal microbiota using the maternal microbiota as potential sources41. OTUs present in less than 1% of samples were first filtered, and the resultant OTU table was imputed using default parameters, with the neonatal samples identified as the “sink” and maternal samples identified as the “source”. Results were aggregated into 3 categories: skin (retroauricular crease and antecubital fossa), vagina (posterior fornix and vaginal introitus) and other (stool, nares, supragingival plaque). Data was visualized as ternary plots generated by the R package ggtern.
Whole Genome Shotgun (WGS) Sequencing and Analysis
Due to limiting sample volumes, whole genome shotgun sequencing and analysis was constrained to a subset of all samples. We focused our sequencing efforts on neonatal meconium (n=9), infant stool (n=36), maternal stool (n=24), neonatal oral samples obtained at delivery (n=11), and infant oral samples obtained at 6 weeks (n=11). A list of samples is provided in Table S10. Isolated genomic DNA was sheared and Illumina adapters ligated to the resultant fragments. The DNA was pooled and paired-end sequenced on the Illumina HiSeq 2500 platform at the HGSC at BCM. DNA was then processed and analyzed through in-house pipelines as described previously37. In brief, human contamination was identified and removed with Best Match Tagger according to the HMP Human Sequence Removal Standard Operating Procedure. The resultant filtered high-quality sequences were deposited in the NCBI Sequence Read Archive (SRA) database with Bio project ID SRP078001. Identification and quantification of taxa in each sample was determined using Metagenomic Phylogenic Analysis (MetaPhlAn) v1.7.770 while the relative abundance of microbial pathways was determined by The HMP Unified Metabolic Analysis Network (HUMAnN) v 0.9971. The total counts of mapped reads by each algorithm are provided in Table S11. Gene counts and unique species counts were determined as described in the MetaHit project72. After quality filtering and removal of human contamination, reads were aligned against the MetaHIT gene catalog by using SOAPalign version 2.21 with parameters ‘-m 200 -x 600’. As long as the read could be aligned to a gene, the aligned gene was taken as one gene count. In order to assess the number of unique species, the collection of genes for each sample where then checked against the origin of species using the MetaHIT gene catalog taxonomic reference. Species were tabulated if any gene was represented in the data set.
Statistical modeling of Infant Gut Metagenome Pathway and Species Relative Abundance
Heatmaps for WGS based KEGG pathways and species were generated using Python seaborn package. Hierarchical clustering was performed using average linkage on Euclidean distance. To identify factors with a significant impact on either species relative abundance or gene pathway abundance while controlling for other factors, we fitted a generalized linear model for each pathway or taxa with a normal distribution and a linear link function by residual maximum likelihood using the lme4 package in R. The KEGG pathway or taxa abundance was treated as the dependent variable as a function of mode of delivery, feeding method (any formula vs. exclusive breastfeeding), gestational weight gain category (excess vs. normal), maternal age, pre-pregnancy BMI, GDM, antibiotic usage and gestational age as fixed effects. The categorical variables (mode of delivery, feeding and GWG) were represented using dummy variables. The base case for linear regression was set as vaginal delivery, exclusive breastfeeding, normal gestational weight gain and no antibiotic usage.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the support of the NIH Director’s New Innovator Award (K.A., DP2 DP21DP2OD001500), the NIH/NINR (K.A., NR014792-01), the NIH National Children’s Study Formative Research (N01-HD-80020), the Burroughs Welcome Fund Preterm Birth Initiative (K.A.), the March of Dimes Preterm Birth Research Initiative (K.A.), the Baylor College of Medicine Medical Scientist Training Program (D.C. and K.M.A., NIH NIGMS T32 GM007330), the National Institute of General Medical Sciences (D.C., T32GM088129), Baylor Research Advocates for Student Scientists (D.C.), and the Human Microbiome Project funded through the NIH Director’s Common Fund at the National Institutes of Health (as part of NIH RoadMap 1.5). All sequencing and adaptation of protocols for whole genome shotgun sequencing were performed by the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC), which is funded by direct support from the National Human Genome Research Institute (NHGRI) at NIH (U54HG004973 (BCM), Dr. Richard Gibbs, P.I.). The authors also thank the staff directly involved in clinical recruitment and specimen processing (Michelle Moller, Brigid Boggan, Renata Benjamin, Jia Chen, Claire Cook, and Dr. Diana Racusin). The authors are grateful to Drs. Michael Belfort, James Versalovic, Tor Savidge, Ruth Ann Luna, Diana Racusin, Melissa Suter, and Kristen Meyer for critical review of the manuscript.
Footnotes
Author Contributions
D.M.C. and K.M.Aa. designed and conceived the study. K.M.Aa. and K.M.A. assembled the cohort and developed the infrastructure to obtain swabs, samples and clinical metadata from all samples. K.M.Aa, and K.M.A. recruited and sampled all subjects. A.L.P., D.M.C. and M.D.S. prepared samples for 16S rRNA gene sequencing and whole genome shotgun sequencing. D.M.C., J.M. and K.M.Aa. performed and supervised all analysis and statistical modeling. D.M.C. and K.M.Aa. wrote the manuscript, with contributions from J.M., A.L.P., K.M.A., and M.S.
Competing Financial Interest Statement
The authors declare no conflicts of interest.
Accession Codes
NCBI Sequence Read Archive (SRA) database: Bio project ID PRJNA322188, Bio project ID SRP078001
Data Availability
All specimens and associated sequence data were assigned a de-identified code and stored in controlled-access repositories. 16S rRNA gene sequence data was deposited to the NCBI Sequence Read Archive (SRA) database with Bio project ID PRJNA322188. WGS sequence data was deposited in the NCBI Sequence Read Archive (SRA) database with Bio project ID SRP078001.
Main text References
- 1.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aagaard K, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PloS One. 2012;7:e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 6.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U. S. A. 2011;(108 Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 9.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulz MD, et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature. 2014;514:508–512. doi: 10.1038/nature13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon HA, Pesti L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol. Rev. 1971;35:390–429. doi: 10.1128/br.35.4.390-429.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wesemann DR, et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. 2013;501:112–115. doi: 10.1038/nature12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez de Aguero M, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 15.Ivanov II, et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith PM, et al. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atarashi K, et al. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costello EK, Carlisle EM, Bik EM, Morowitz MJ, Relman DA. Microbiome Assembly across Multiple Body Sites in Low-Birthweight Infants. mBio. 2013;4:e00782–13. doi: 10.1128/mBio.00782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng XL, Xu L, Guo Y, Ronsmans C. Factors influencing rising caesarean section rates in China between 1988 and 2008. Bull. World Health Organ. 2012;90:30–39. doi: 10.2471/BLT.11.090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osterman MJK, Martin JA. Trends in low-risk cesarean delivery in the United States, 1990–2013. Natl. Vital Stat. Rep. Cent. Dis. Control Prev. Natl. Cent. Health Stat. Natl. Vital Stat. Syst. 2014;63:1–16. [PubMed] [Google Scholar]
- 21.Almqvist C, Cnattingius S, Lichtenstein P, Lundholm C. The impact of birth mode of delivery on childhood asthma and allergic diseases--a sibling study. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2012;42:1369–1376. doi: 10.1111/j.1365-2222.2012.04021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black M, Bhattacharya S, Philip S, Norman JE, McLernon DJ. Planned Repeat Cesarean Section at Term and Adverse Childhood Health Outcomes: A Record-Linkage Study. PLoS Med. 2016;13:e1001973. doi: 10.1371/journal.pmed.1001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.American College of Obstetricians and Gynecologists (College) et al. Safe prevention of the primary cesarean delivery. Am. J. Obstet. Gynecol. 2014;210:179–193. doi: 10.1016/j.ajog.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 24.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biasucci G, et al. Mode of delivery affects the bacterial community in the newborn gut. Early Hum. Dev. 2010;(86 Suppl 1):13–15. doi: 10.1016/j.earlhumdev.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Fallani M, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J. Pediatr. Gastroenterol. Nutr. 2010;51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 27.Azad MB, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. Can. Med. Assoc. J. 2013;185:385–394. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barber EL, et al. Indications Contributing to the Increasing Cesarean Delivery Rate. Obstet. Gynecol. 2011;118:29–38. doi: 10.1097/AOG.0b013e31821e5f65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. 2011;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 2014;16:2891–2904. doi: 10.1111/1462-2920.12238. [DOI] [PubMed] [Google Scholar]
- 32.Azad M, et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG Int. J. Obstet. Gynaecol. 2015 doi: 10.1111/1471-0528.13601. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 33.Ardeshir A, et al. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci. Transl. Med. 2014;6:252ra120. doi: 10.1126/scitranslmed.3008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dufrene M, Legendre P. Species Assemblages and Indicator Species: The Need for a Flexible Asymmetrical Approach. Ecological Monographs. 1997;67:345–366. [Google Scholar]
- 35.Koren O, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.La Rosa PS, et al. Patterned progression of bacterial populations in the premature infant gut. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12522–12527. doi: 10.1073/pnas.1409497111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aagaard K, et al. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antony KM, et al. The preterm placental microbiome varies in association with excess maternal gestational weight gain. Am. J. Obstet. Gynecol. 2015;212:653.e1–16. doi: 10.1016/j.ajog.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016;6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bäckhed F, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:852. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 41.Knights D, et al. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods. 2011;8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penders J, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 43.Vatanen T, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dominguez-Bello MG, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016;22:250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galley JD, Bailey M, Kamp Dush C, Schoppe-Sullivan S, Christian LM. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PloS One. 2014;9:e113026. doi: 10.1371/journal.pone.0113026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mueller NT, et al. Birth mode-dependent association between pre-pregnancy maternal weight status and the neonatal intestinal microbiome. Sci. Rep. 2016;6:23133. doi: 10.1038/srep23133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu DM, et al. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 2016;8:77. doi: 10.1186/s13073-016-0330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu J, et al. Diversified Microbiota of Meconium Is Affected by Maternal Diabetes Status. PLoS One. 2013;8:e78257. doi: 10.1371/journal.pone.0078257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014;30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kennedy D. B Vitamins and the Brain: Mechanisms, Dose and Efficacy—A Review. Nutrients. 2016;8:68. doi: 10.3390/nu8020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibson MK, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat. Microbiol. 2016;1:16024. doi: 10.1038/nmicrobiol.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bokulich NA, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016;8:343ra82–343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DiGiulio DB, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PloS One. 2008;3:e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steel JH, et al. Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatr. Res. 2005;57:404–411. doi: 10.1203/01.PDR.0000153869.96337.90. [DOI] [PubMed] [Google Scholar]
- 55.Jiménez E, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr. Microbiol. 2005;51:270–274. doi: 10.1007/s00284-005-0020-3. [DOI] [PubMed] [Google Scholar]
- 56.Jiménez E, et al. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008;159:187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 57.Ardissone AN, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PloS One. 2014;9:e90784. doi: 10.1371/journal.pone.0090784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansen R, et al. First-Pass Meconium Samples from Healthy Term Vaginally-Delivered Neonates: An Analysis of the Microbiota. PLOS One. 2015;10:e0133320. doi: 10.1371/journal.pone.0133320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fallani M, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157:1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 60.Yassour M, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 2016;8:343ra81–343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 61.La Rosa PS, et al. Hypothesis Testing and Power Calculations for Taxonomic-Based Human Microbiome Data. PLoS One. 2012;7:e52078. doi: 10.1371/journal.pone.0052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aagaard K, et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2013;27:1012–1022. doi: 10.1096/fj.12-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmieder R, Edwards R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PloS One. 2011;6:e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinforma. Oxf. Engl. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 66.Haas BJ, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cole JR, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abubucker S, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Chatelier E, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.