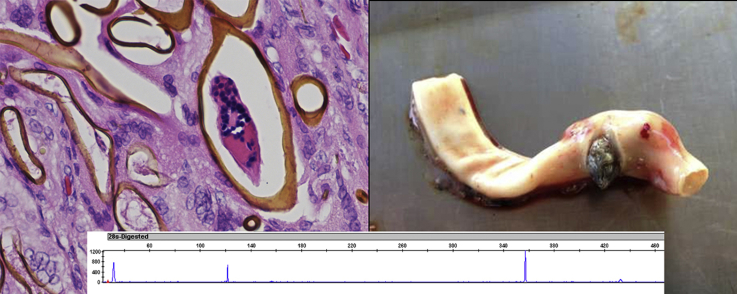
Abstract
Spirorchiid blood fluke infections affect endangered turtle populations globally, and are reported as a common cause of mortality in Queensland green sea turtles. Both the flukes and their ova are pathogenic and can contribute to the stranding or death of their host. Of particular interest are ova-associated brain lesions, which have been associated with host neurological deficits. Accurate estimations of disease frequency and the relative effect of infection relating to different spirorchiid species are made difficult by challenges in morphological identification of adults of some genera, and a lack of species-level identifying features for ova. A new specifically designed molecular assay was used to detect and identify cryptic spirorchiids and their ova in Queensland green sea turtle tissues collected from 2011 to 2014 in order to investigate epidemiology, tissue tropisms and pathology. Eight spirorchiid genotypes were detected in 14 distinct tissues, including multiple tissues for each. We found no evidence of a characteristic pathway of the eggs to the exterior; instead the results suggest that a high proportion of eggs become lost in dead-end tissues. The most common lesions observed were granulomas affecting most organs with varying severity, followed by arteritis and thrombi in the great vessels. The number of spirorchiid types detected increased with the presence and severity of granulomatous lesions. However, compared with other organs the brain showed relatively low levels of spirorchiid diversity. An inverse relationship between host age and spirorchiid diversity was evident for the liver and kidneys, but no such relationship was evident for other organs. Molecular data in this study, the first of its kind, provides the first species-level examination of spirorchiid ova and associated pathology, and paves the way for the future development of targeted ante-mortem diagnosis of spirorchiidiasis.
Keywords: Spirorchiidiasis, Chelonia mydas, Epidemiology, Pathology, Tissue tropisms, Terminal restriction fragment length polymorphism
Graphical abstract
Highlights
-
•
First species-level molecular study of spirorchiidiasis.
-
•
Eight genotypes detected across fourteen tissue types.
-
•
Species investigated in terms of tissue tropisms and pathology.
-
•
Granulomas and arteritis/thrombosis most common lesions.
-
•
Number of species present correlated with presence and severity of lesions.
1. Introduction
Each year, hundreds of marine turtles are reported stranded or dead on the east coast of Queensland, Australia (Flint et al., 2015a, Queensland Department of Environment and Heritage Protection, 2015). From 2009 to 2014, an annual average of 1152 strandings or mortalities were recorded; the majority of these were green sea turtles (Flint et al., 2015a), which are currently listed as Endangered by the International Union for the Conservation of Nature. Disease is among the most commonly recorded causes of stranding or mortality (Flint et al., 2009a, Meager and Limpus, 2012), but infectious causes of turtle mortality are poorly understood. Parasites, particularly spirorchiid blood flukes and the coccidian Caryospora cheloniae, are noted for their capacity to cause disease (Gordon et al., 1998, Santoro et al., 2007, Flint et al., 2010, Stacy et al., 2010) and mortality (Gordon et al., 1993, Flint et al., 2010, Chapman et al., 2016a). Of the two, spirorchiids are the more common and widespread.
Spirorchiid flukes infect all major organs, and both adults and ova can have deleterious effects that may contribute to the stranding or death of their host (Glazebrook et al., 1989, Glazebrook and Campbell, 1990, Aguirre et al., 1998, Gordon et al., 1998, Work et al., 2004, Work et al., 2005, Santoro et al., 2007, Flint et al., 2010, Flint et al., 2015b, Stacy et al., 2010). In Queensland, spirorchiidiasis is considered the most significant infectious disease among sea turtles (Flint et al., 2010). The earliest studies in the region found that spirorchiids were present in 40.9%–72.2% of wild sea turtles (Glazebrook et al., 1989, Glazebrook and Campbell, 1990) and were associated with a range of lesions and general debilitation. In 1998, Gordon et al. (1998) found spirorchiids to be the primary cause of death in 10% of locally stranded green sea turtles and a severe problem in a further 30%, with an overall infection rate of 98% on the basis of histopathology. Spirorchiids were also found to be the most common cause of death (41.8%) in Queensland green sea turtles between 2006 and 2009 (Flint et al., 2010). Spirorchiids maintain a high prevalence in turtles globally, including Hawaii (Graczyk et al., 1995, Aguirre et al., 1998, Work et al., 2005, Work et al., 2015), the eastern United States (Stacy et al., 2010) and South America (Santoro et al., 2007). Incidental, asymptomatic infections are common (Gordon et al., 1998, Santoro et al., 2007, Flint et al., 2010, Stacy et al., 2010, Work et al., 2015). However, the occurrence of severe disease appears to vary spatially; while spirorchiidiasis is often a primary cause of death in Australian sea turtles (Flint et al., 2010), it is less frequently fatal in turtles from the south-eastern United States (Stacy et al., 2010) and Hawaii (Work et al., 2015). The reasons behind these geographic variations are unexplained.
Accurate estimations of prevalence and the relative impact of individual spirorchiid species are constrained by two factors. First, the adults of some species are microscopic and show an apparent predilection for small blood vessels, making them very difficult to detect and collect intact (Gordon et al., 1998, Stacy et al., 2010). Secondly, ova can only be categorised into one of three broad morphological types, and identification to species level is not possible. Given the limitations of traditional gross and microscopy based methods in this field, molecular techniques may contribute substantially to our knowledge of the parasite and the disease. Molecular tools have superior sensitivity and specificity relative to traditional microscopic/histologic identification (McManus and Bowles, 1996, Ndao, 2009, Chapman et al., 2016b), and the additional advantage of having potential for diagnostic applications in live turtles. Such approaches can therefore increase capacity to detect and identify parasites and explore their connection with pathology. Reports of pathology associated with ova are so far restricted to generalised comments on associated pathology and rarely attempt to associate lesions with particular species. Given that spirorchiid ova are almost universally present and often present in the apparent absence of adult flukes (Flint et al., 2010, Stacy et al., 2010), they require detailed investigation of their impacts and association with pathology. Recently, a new molecular assay was developed and validated for the identification of spirorchiids and their ova in green sea turtle tissues (Chapman et al., 2016b) enabling the collection of data of a kind that has, to date, been unavailable.
This paper aims to improve understanding of spirorchiidiasis in turtle populations of Australia and other regions around the globe by examining and quantifying infection rates, tissue tropisms, and predisposing host factors using the newly developed test (Chapman et al., 2016b).
2. Materials and methods
2.1. Study population
Green sea turtle carcasses were obtained from the wildlife rehabilitation facilities Sea Life Underwater World (Mooloolaba) and Australia Zoo Wildlife Hospital (Beerwah) as well as government agencies (Queensland Parks and Wildlife Service – QPWS) between 2011 and 2015. Prior to necropsy turtles were stored in refrigeration for a maximum of 2 days, or otherwise frozen. Turtles were mainly from two broad locations: the central Queensland region from Gladstone Port (23.8251°S, 151.2975°E), nearby islands (Quoin, Facing and Boyne) south to the town of 1770 (24.1594°S, 151.8658°E), and the southern Queensland area between Hervey Bay (25.2538°S, 152.8605°E) and Ormiston (27.5112°S, 153.2675°E). One further turtle was collected from the Townsville area (19.2372°S, 146.8985°E) in northern Queensland. These areas encompass three of several ‘hotspots’ for marine turtle strandings on Australia's east coast (Queensland Department of Environment and Heritage Protection, 2013) and therefore provided an opportunity to investigate the role of diseases in strandings. Turtles were classified into age groups using the size criteria utilised by Flint et al. (2010) and body condition scores were estimated visually and assigned using the criteria described by Flint et al. (2009b) to provide an overall post mortem health profile.
2.2. Parasitological methods
Turtles were necropsied between 2011 and early 2015 using standard methods for sea turtle post mortem examination (Flint et al., 2009b). During necropsy, adult flukes were collected and identified using molecular and morphological methods as described by Chapman et al. (2015). Samples of turtle tissues were collected for molecular analysis. For tubiform organs such as the gastrointestinal tract and major blood vessels, a cross section was taken to include all cell layers. When sampling the gastrointestinal tract, samples were collected from the stomach or small intestine, depending on evidence of spirorchiid infection i.e. visible egg deposition. Samples were stored in 70% ethanol at 4 °C before analysis using PCR and T-RFLP as described by Chapman et al. (2016b). This method involved an initial multiplex PCR to detect and identify spirorchiid genera based on the 28S gene, followed by a second round of singleplex reactions using fluorescently tagged genus specific primers to produce PCR products for species level analysis through restriction nuclease digestion and capillary electrophoresis. A total of twelve genotypes were tested for. Many of these genotypes corresponded with known species, though several were not able to be matched to an existing morphological or molecular identification; regardless, they were interpreted as distinct species based on the level of genetic variation observed between types (Chapman et al., 2016b). Negative results from first round multiplex PCR were validated by assessing the presence of amplifiable DNA with universal eukaryote 18S primers (Fajardo et al., 2008, Chapman et al., 2016b). For unfrozen carcasses with minimal decomposition, further samples were collected and fixed in 10% neutral buffered formalin for histological examination to correlate turtle health with parasitic effect. These samples were embedded in paraffin wax prior to being sectioned at 5 μm and stained with haematoxylin and eosin (HE). Sections were examined by a specialist veterinary pathologist.
2.3. Histopathological methods
The presence of spirorchiid ova in tissues was assessed by histology, along with the presence of associated inflammatory lesions (e.g. granulomas). Granulomas were graded based on relative severity, accounting for size and number of lesions as well as disruption to surrounding cellular architecture. Grading was based on the methodology described by Flint et al. (2010), however, a five point scale was used with a score of 1 designated for mild, 3 for moderate and 5 for severe lesions; scores of 2 and 4 were used in cases where lesions did not clearly meet the criteria either side, or varied in severity across the section.
2.4. Statistical analyses
The proportion of organs infected with each spirorchiid genus was estimated from PCR data, and genotypes from T-RFLP results. Samples with PCR results but without species level T-RFLP were omitted from species level calculations. For the purpose of this study, Hapalotrema, Learedius and Amphiorchis were grouped together due to their genetic and morphological similarity and comparable reported site tropisms, with genus level proportions calculated as one.
We compared the occurrence of granulomas within each organ type by age, sex, body condition and infection type (single or multi species infections) using the Fisher's exact test (95% confidence interval).
We investigated the association between granuloma presence in brain samples (outcome) and spirorchiid infection type (exposure of interest) using multivariable generalised linear model (GLM). The model was adjusted for the effect of age, sex (female – 0, male – 1), body condition and Bernoulli distributed residuals (binomial family) with a logit link function. Each sample was categorised as either single infection (i.e. one spirorchiid genotype present - 0) or multiple infection (i.e. two or more genotypes - 1). Mature and large immature classes were combined for analysis, resulting in two age groups of <65 cm CCL (small immature – assigned 0) and >65 cm (mature and large immature - 1). Turtles judged to be in poor or very poor body condition were combined into one category (0) representing turtles with deemed chronic debility, while those assessed as good or fair formed the second category (1). Analyses of spirorchiid occurrence were undertaken at both the genus and species level. The effect size of each of the predictor variables were expressed as odds ratios with 95% confidence intervals. The Mann-Whitney U— test was used to compare the average number of spirorchiids in relation to age, sex and body condition in other organs. All analyses were performed using STATA version 13.1 (StataCorp, Texas, USA).
2.5. Ethical standards
All work described above was completed under approval SVS/037/11/ARC/DERM/AUSTZOO issued by the University of Queensland Animal Ethics Committee. Approval to undertake marine research activities was granted through Queensland State Government Marine Parks permit QS2011/CVL1414 and Scientific Purposes permit WISP09021911.
3. Results
3.1. Dataset for analysis
Necropsies were completed on a total of 51 turtles. Various tissue samples from 39 turtles were examined histologically, while tissue samples from 44 animals were collected for molecular characterisation of spirorchiids. A summary of host characteristics and tissue samples collected is provided in Table 1.
Table 1.
Tissue samples collected from turtles, summarised by age group and body condition.
| Total turtles | Adrenal gland | Aorta | Bladder | Brain | Cornea | Fibropapilloma | Gall bladder | GIT | Gonads | Heart | Kidney | Liver | Lung | Pancreas | Parathyroid | Salt gland | Spleen | Thyroid | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Body Condition | |||||||||||||||||||
| Good - Fair | 20 | 4, 0 | 6, 2 | 1, 2 | 13, 20 | 0, 0 | 1, 2 | 1, 1 | 14, 10 | 2, 3 | 11, 9 | 14, 10 | 11, 8 | 14, 10 | 11, 5 | 0, 0 | 6, 0 | 12, 8 | 12, 4 |
| Poor - Very Poor | 31 | 12, 0 | 13, 0 | 4, 0 | 23, 23 | 1, 0 | 0, 0 | 0, 1 | 21, 6 | 9, 0 | 21, 3 | 19, 4 | 23, 3 | 22, 4 | 18, 3 | 1, 0 | 14, 0 | 22, 4 | 8, 3 |
| Age group | |||||||||||||||||||
| Small immature | 34 | 13, 0 | 14, 1 | 5, 1 | 22, 28 | 0, 0 | 1, 2 | 1, 1 | 21, 9 | 8, 1 | 20, 6 | 20, 7 | 22, 5 | 23, 7 | 19, 4 | 1, 0 | 15, 0 | 20, 6 | 10, 3 |
| Large immature | 9 | 1, 0 | 2, 1 | 0, 1 | 7, 9 | 0, 0 | 0, 0 | 0, 0 | 7, 4 | 1, 2 | 7, 3 | 6, 3 | 6, 3 | 6, 4 | 5, 3 | 0, 0 | 1, 0 | 7, 3 | 5, 2 |
| Mature | 8 | 2, 0 | 3, 0 | 0, 0 | 7, 6 | 1, 0 | 0, 0 | 0, 1 | 7, 3 | 2, 0 | 5, 3 | 7, 4 | 6, 3 | 7, 3 | 5, 1 | 0, 0 | 4, 0 | 7, 3 | 5, 2 |
| Total | 51 | 16, 0 | 19, 2 | 5, 2 | 36, 43 | 1, 0 | 1, 2 | 1, 2 | 35, 16 | 11, 3 | 32, 12 | 33, 14 | 34, 11 | 36, 14 | 29, 8 | 1, 0 | 20, 0 | 34, 12 | 20, 7 |
Total turtles refers to the total number of turtles in each category that underwent gross post mortem examination. The first number in each case denotes the number of samples examined histologically, while the second number indicates samples tested by T-RFLP to detect and characterise spirorchiid species. Samples that failed to amplify either spirorchiid or Eukaryote 18S DNA have been omitted. Abbreviations: GIT = gastrointestinal tract.
3.2. Parasitological findings
3.2.1. Adult flukes
Adult flukes were found in 16/51 turtles (Table 2). The majority of flukes found were larger worms, i.e. species of Hapalotrema and Learedius. The most common was H. pambanensis, which was found in the heart of six turtles, with additional flukes found in the aortic vessels (left or pulmonary) in two of these cases. In two additional turtles, they were recovered from the body cavity, presumably after having been released from the circulatory system during necropsy. Hapalotrema postorchis showed a particular affinity for the major vessels (five turtles) while L. learedi was found in the heart of two turtles, with further specimens also present in the lung of one. In most cases multiple flukes were collected, with over 80 recovered in severe infections (Table 2).
Table 2.
Summary of adult flukes collected.
| Host characteristics (age, body condition) |
Aorta | Brain | Body cavity | Heart | Liver | Lung | Pancreas | Notes |
|---|---|---|---|---|---|---|---|---|
| Small immature, poor | 10 | H. postorchis | ||||||
| Adult, poor | 23* | 23* | H. pambanensis (A, H) | |||||
| Small immature, very poor | N | 2 | 1 | 5 | H. postorchis (A), H. pambanensis (BCx1, Hx1), Carettacola sp. (BCx1), C. hawaiiensis (P) | |||
| Small immature, very poor | N | H. postorchis (A) | ||||||
| Small immature, poor | N | H. postorchis (A) | ||||||
| Small immature, fair | 2 | 1 | 1 | Carettacola sp. (BC, Li), Neospirorchis Gen. 2 (H) | ||||
| Small immature, very poor | 1 | 1 | 1 | C. hawaiiensis | ||||
| Adult, fair | 3 | H. pambanensis (x2), Novel species (x1) | ||||||
| Small immature, poor | 3 | H. pambanensis (H) | ||||||
| Small immature, very poor | 1 | Neospirorchis Gen. 2 | ||||||
| Small immature, very poor | 2 | C. hawaiiensis | ||||||
| Small immature, poor | 4 | H. pambanensis x1, L. learedi x2, Neospirorchis Gen. 2 x1 | ||||||
| Large immature, poor | 84 | H. pambanensis | ||||||
| Adult, poor | 7 | Carettacola sp. | ||||||
| Small immature, poor | 1 | Neospirorchis Gen. 3 | ||||||
| Small immature, good | 6 | 5 | 2 | 5 | 4 | H. postorchis (Ax2), H. pambanensis (Ax4, Hx4), L. learedi (Hx1, Lu), Carettacola sp. (BCx1), Neospirorchis Gen 1. (BCx1, Bx5) |
All totals are estimates based on counts of intact worms and fragments. * indicates combined total between two organs. Column headed ‘notes’ provides details of species present in each organ and an estimate of the total number of adult flukes recovered. Abbreviations: N = numerous, A = aorta, B = brain, BC = body cavity, H = heart, Li = liver, Lu = lung, P = pancreas.
Adult flukes confirmed as Carettacola sp. were found in six turtles, and were recovered from sites including the liver (three turtles), pancreas (one turtle), and body cavity (four turtles). In some instances, these flukes were unable to be identified to species level on account of the samples being broken or in poor condition. However, flukes from three turtles were identified as C. hawaiiensis based on either morphological or molecular characteristics. Others were genetically distinct but did not match limited sequences available in Genbank. One fluke retrieved from the body cavity was initially identified as a Carettacola sp. but sequence data indicates that it was likely to relate to an unknown spirorchiid genus (Genbank KU600078.1).
Difficulties were also experienced in identifying Neospirorchis spp. that were retrieved from the heart, brain, lung and body cavity of five turtles, due to damaged and broken samples and a lack of genetic data available in public databases. Flukes from Genotype 1 were collected from the brain and body cavity, Genotype 2 from the heart, and Genotype 3 from the lungs.
3.2.2. Molecular detection - tissue samples
Spirorchiid infections were detected in 142/148 tissue samples, encompassing 43/44 turtles from which samples were tested. Of six that failed to give a positive PCR result, four were validated as genuine negative results, while the remaining two were discarded owing to failure of the generic eukaryote 18S PCR assay to amplify genomic host DNA. Full molecular characterisation was achieved for 129 samples; the remainder were limited to initial multiplex PCR characterisation or, in some cases, multiplex PCR plus T-RFLP results for one primer set only, most likely due to DNA degradation.
A total of eight distinct spirorchiid genotypes (interpreted as representing distinct species) were detected. The occurrence of each spirorchiid genotype as detected by PCR and T-RFLP is summarised in Table 3. Overall, Neospirorchis was the most common genus. Of samples that were successfully characterised to species level, between 80% and 100% of each organ (excepting the gall bladder) were positive for Neospirorchis Genotype 2, making this the most common spirorchiid genotype. Genotype 1 was also recorded in the majority of tissue types. Genotype 3 showed a more restricted range, being detected only in gastrointestinal, kidney, lung and spleen tissues. It appeared to show a predilection for lung tissue, where it was found in 50% of samples tested.
Table 3.
Occurrence of spirorchiids in each organ, by genera and by species.
| Aorta | Bladder | Brain | FP | GB | GIT | Gonads | Heart | Kidney | Liver | Lung | Pancreas | Spleen | Thyroid | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total samples | 2 | 2 | 43 | 2 | 2 | 16 | 3 | 12 | 14 | 11 | 14 | 8 | 12 | 7 |
| PCR positive (%) | ||||||||||||||
| Hapalotrema | 100 | 0 | 37 | 100 | 100 | 56 | 0 | 50 | 29 | 55 | 57 | 75 | 75 | 71 |
| Carettacola | 0 | 0 | 5 | 0 | 0 | 13 | 0 | 0 | 0 | 18 | 0 | 0 | 17 | 0 |
| Neospirorchis | 100 | 100 | 93 | 100 | 50 | 88 | 100 | 100 | 100 | 82 | 93 | 100 | 100 | 100 |
| Total T-RFLP tested - Hapalotrema | 2 | 2 | 39 | 2 | 2 | 13 | 3 | 12 | 14 | 11 | 13 | 8 | 12 | 6 |
| T-RFLP Positive (%) | ||||||||||||||
| Hapalotrema postorchis | 50 | 0 | 13 | 0 | 50 | 46 | 0 | 25 | 21 | 36 | 15 | 50 | 50 | 33 |
| Hapalotrema pambanensis | 50 | 0 | 21 | 50 | 50 | 31 | 0 | 33 | 14 | 27 | 31 | 50 | 58 | 33 |
| Hapalotrema synorchis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Learedius learedi | 0 | 0 | 15 | 50 | 50 | 8 | 0 | 17 | 7 | 9 | 23 | 25 | 17 | 0 |
| Hapalotrema mistroides | 0 | 0 | 0 | 0 | 50 | 0 | 0 | 8 | 0 | 9 | 15 | 0 | 25 | 33 |
| Amphiorchis sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total T-RFLP tested - Carettacola | 2 | 2 | 41 | 2 | 2 | 16 | 3 | 12 | 14 | 11 | 14 | 8 | 11 | 7 |
| T-RFLP Positive (%) | ||||||||||||||
| Carettacola hawaiiensis | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 18 | 0 | 0 | 9 | 0 |
| Carettacola Gen. 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total T-RFLP tested - Neospirorchis | 2 | 2 | 43 | 2 | 2 | 16 | 3 | 10 | 14 | 10 | 14 | 8 | 11 | 7 |
| T-RFLP Positive (%) | ||||||||||||||
| Neospirorchis Gen. 1 | 50 | 0 | 30 | 50 | 0 | 19 | 33 | 50 | 43 | 20 | 21 | 50 | 27 | 43 |
| Neospirorchis Gen. 2 | 100 | 100 | 95 | 100 | 50 | 88 | 100 | 100 | 100 | 80 | 93 | 100 | 100 | 100 |
| Neospirorchis Gen. 3 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 21 | 0 | 50 | 0 | 9 | 0 |
| Average no. Hapalotrema spp. present | 1 | 0 | 0 | 1 | 2 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 2 | 1 |
| Average no. Carettacola spp. present | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Average no. Neospirorchis spp. present | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 2 | 1 | 1 |
| Average total species present | 2.5 | 1 | 1.8 | 2.5 | 2.5 | 2.2 | 1.3 | 2.1 | 2.1 | 2 | 2.5 | 2.8 | 3.1 | 2.5 |
Total samples refers to total samples available for molecular testing. Total PCR positive refers to number of samples that were positive for each genus/group of genera based on first round multiplex PCR results. 'Total T-RFLP results' columns refer to total number of samples that were successfully characterised by T-RFLP, or had a validated negative result on multiplex PCR. Abbreviations: GIT - Gastrointestinal tract, GB = Gall bladder, FP = Fibropapilloma.
Four species from the Hapalotrema group (incorporating Learedius and Amphiorchis) were detected. Of these, the most common were H. pambanensis and H. postorchis, though the remaining two species, L. learedi and H. mistroides, were also regularly found. The group as a whole was common across most tissues, with the exception of the gonads and bladder where they were not detected from limited samples. In the remaining organs, H. postorchis recorded its lowest infection rate in the brain (13%) and the lung (15%), while H. pambanensis was least common in the kidneys (14%) and in the brain (21%). Hapalotrema mistroides had a more restricted distribution among tissues than other species, and was not found in any brain, gastrointestinal, kidney or pancreas tissues. Neither H. synorchis nor Amphiorchis spp. were detected in any sample.
Carettacola was less common than Neospirorchis or the Hapalotrema group, and was restricted to the gastrointestinal tract, liver and spleen. The only species detected was C. hawaiiensis, although characterisation was only achieved to genus level in some samples.
Based on molecular results, the greatest average species diversity was observed in the spleen (3.1 spirorchiid species per sample) followed by the pancreas (2.8 species) and lung (2.5 species) (Table 3). An average of 2.5 species each were found in aorta/thrombus, gall bladder and fibropapilloma samples, but only low numbers of samples were available for these tissues. The brain showed a relatively low diversity, with an average of 1.8 species detected. For the liver (Mann-Whitney U test, z = −2.124, N1 = 5, N2 = 5, P > 0.0336) and kidney (Mann-Whitney U test, z = −2.320, N1 = 7, N2 = 7, P > 0.0204), the average number of spirorchiid types in each sample was significantly greater in small immature turtles than in large immature and mature turtles. No effect of body condition or sex on the occurrence of granulomas was observed.
3.3. Pathological findings
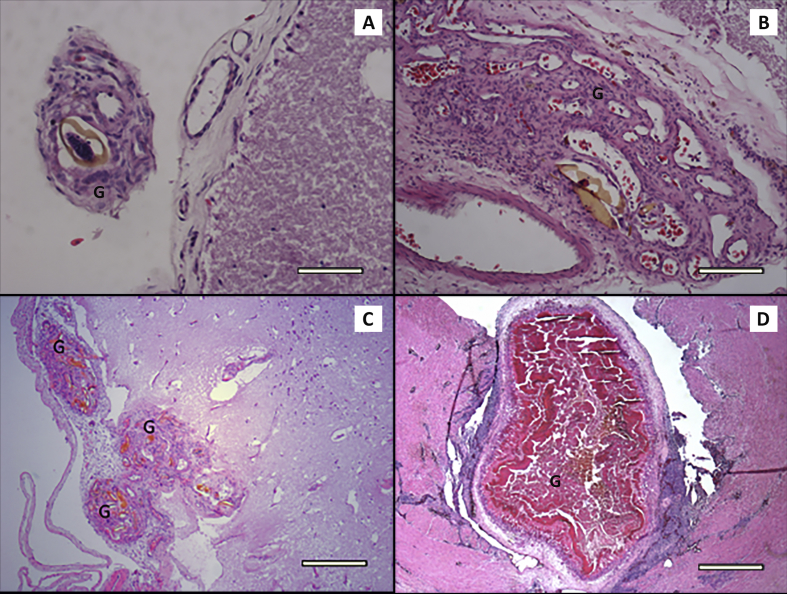
Ova were observed in 242 of 364 (66.5%) tissue samples examined by histology. In a large proportion of infected tissues (169 - 70%) a granulomatous response to the ova was observed. The majority of granulomas were scored as 1 (mild – 82, 48%), 2 (mild-moderate – 37, 22%) or 3 (moderate – 33, 20%) in severity, with remaining lesions being classed in the range of 4 (moderate-severe – 2, 1%) or 5 (severe – 14, 8%) (Table 4) in accordance with criteria outlined by Flint et al. (2010). Examples of lesions of various severities are provided in Fig. 1. The number of spirorchiid species present appeared to increase with the presence and severity of granulomas (Table 4), ranging from 1.6 species where no granulomas were evident through to 3.5 species in the presence of severe lesions.
Table 4.
Summary table of occurrence and severity of trematode ova associated granulomas in tissues examined by histology.
| Granuloma severity | Adrenal gland | Aorta | Bladder | Brain | Cornea | Fibropapilloma | Gall bladder | GIT | Gonads | Heart | Kidney | Liver | Lung | Pancreas | Parathyroid | Salt gland | Spleen | Thyroid | Total | Average no. species |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| None observed | 7 | 7 | 4 | 15 | 0 | 0 | 0 | 9 | 8 | 13 | 17 | 17 | 12 | 6 | 1 | 0 | 5 | 14 | 135 | 1.6 (42) |
| 1 - Mild | 6 | 2 | 1 | 7 | 0 | 1 | 0 | 10 | 0 | 8 | 10 | 6 | 8 | 11 | 0 | 3 | 8 | 1 | 82 | 2.6 (22) |
| 2 - Mild/moderate | 3 | 0 | 0 | 8 | 0 | 0 | 0 | 4 | 1 | 1 | 1 | 2 | 5 | 3 | 0 | 3 | 6 | 0 | 37 | 3.5 (8) |
| 3 - Moderate | 0 | 0 | 0 | 3 | 1 | 0 | 1 | 2 | 0 | 2 | 0 | 3 | 4 | 2 | 0 | 8 | 7 | 0 | 33 | 3.5 (3) |
| 4 – Moderate/severe | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2.0 (1) |
| 5 - Severe | 0 | 10 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 14 | 3.5 (4) |
| Unclassified | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | na (0) |
| Total examined | 16 | 19 | 5 | 36 | 1 | 1 | 1 | 26 | 9 | 26 | 28 | 28 | 30 | 22 | 1 | 14 | 26 | 15 | 304 | na |
Gross lesions are not included in these figures. Numbers in brackets denote number of samples with full molecular characterisation of spirorchiid assemblages.
Fig. 1.
Lesions associated with spirorchiid blood flukes in Chelonia mydas. a) Mild (score = 1) granulomatous lesions (G) and lymphocytic inflammation within the cerebral meninges of an adult green turtle, centred on a brown-shelled fluke ovum. HE stain, scale bar = 125 μm b) Moderate (score = 3) granulomatous lesions (G) and lymphocytic inflammation within the cerebral meninges of an adult green turtle, centred on several brown-shelled fluke ova. HE stain, scale bar = 350 μm c) Severe (score = 5) granulomatous lesions (G) and lymphocytic inflammation within the meninges of a small immature green turtle, centred on multiple numerous fluke ova. HE stain, scale bar = 350 μm d) Severe (score = 5) granulomatous lesion (G) with necrotic centre protruding into the lumen of the aorta of an adult green turtle, with dense lymphocytic inflammation in surrounding tissues. Numerous adult flukes (Hapalotrema pambanensis) were recovered from the heart and major vessels. HE stain, scale bar = 1.7 mm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3.1. Brain
Moderate to severe brain lesions (Flint et al., 2010) were observed in four turtles. The most severe were observed in a small immature turtle in fair body condition; three spirorchiid species were detected in this sample, being H. postorchis, H. pambanensis and Neospirorchis Genotype 2. Another small immature turtle, this time in very poor body condition, had moderate to severe lesions associated with Neospirorchis Genotypes 1 and 2. The two final turtles had moderate lesions, with the same assemblage of spirorchiids, being L. learedi and Neospirorchis Genotypes 1 and 2. Adult Neospirorchis were recovered from the brain of one turtle (Genotype 1), however, were not directly associated with any notable abnormalities other than mild to moderate ova associated granulomatous inflammation.
3.3.2. Heart and great vessels
Aortic or heart lesions were observed grossly or histologically in twenty-nine turtles, and, in fourteen of these, adult flukes were recovered from one or both locations. Lesions (arteritis with or without thrombi) were found in all three major vessels arising from the heart, often forming inflammatory masses extending into the vessel lumen (Fig. 1D). Thrombi, found in twelve cases, were generally small (∼1–2 cm in diameter). In one particularly severe case, arteritis of the aortic trunk and associated thrombus formation expanded the vessel to a maximum of approximately 5 cm wide and extending for approximately 12 cm along its length. Adult H. postorchis were found attached to thrombi in this and five other instances, on one occasion accompanied by H. pambanensis.
Molecular testing of an aortic vessel wall containing multiple severe granulomas (up to 5 mm in diameter) indicated that associated ova were H. pambanensis and Neospirorchis Genotype 2. In another, there was transmural, chronic-active arteritis containing small aggregates of ova with associated granulomatous inflammation. Spirorchiids detected by molecular testing were H. postorchis and Neospirorchis Genotypes 1 and 2.
Lesions in the heart itself were detected in thirteen turtles. In seven cases, no adult flukes could be found and lesions were usually mild ova-associated granulomas. Large numbers of H. pambanensis were recovered in two cases where ventricles had large and densely cellular inflammatory lesions protruding from the endocardial surface into the lumen. In the remaining four cases, adult flukes present were single species infections of C. hawaiiensis, H. pambanensis and Neospirorchis Genotype 2 and one mixed infection of H. pambanensis, L. learedi and Neospirorchis Genotype 2. Pathology in these four cases was restricted to mild or moderate granulomatous reactions to the presence of ova.
3.3.3. Lungs
An adult L. learedi and fragments of an unidentified Neospirorchis sp. were recovered separately from the lungs of two animals, with no notable associated pathology observed. Multifocal granulomatous pneumonia was observed in a third turtle. While trematode ova were present, there was no clear association between these and the lesions, and the aetiology could not be conclusively resolved. T-RFLP detected five species of spirorchiid in this case; Neospirorchis Genotypes 1, 2 and 3, H. pambanensis and L. learedi. The same turtle also had moderately severe granulomatous hepatitis involving trematode ova (Neospirorchis Genotypes 1 and 2, H. pambanensis and H. postorchis).
3.4. Statistical modelling
In the univariable analysis, the presence of brain granulomas was significantly associated with infection with more than one spirorchiid species, but not with age, sex or body condition (Table 5). In the multivariable analysis, infection with more than one spirorchiid species was significantly associated with presence of brain granulomas (OR = 20.10; 95%CI: 1.62–250.14) after adjusting for the above factors (Table 5). No significant effects of age, body condition, sex or infection type on presence of granulomas in other organs was indicated using Fisher's Exact tests.
Table 5.
Results of generalised linear models analysing the effects of variables on granuloma formation in the brain.
| Univariate analysis |
Multivariate analysis |
|||||||
|---|---|---|---|---|---|---|---|---|
| OR | CI | SE | P | OR | CI | SE | P | |
| Age | 2.17 | 0.45–10.44 | 1.74 | 0.34 | 0.62 | 0.07–5.16 | 0.67 | 0.66 |
| Sex | 0.96 | 0.21–4.34 | 0.74 | 0.96 | 0.52 | 0.07–4.06 | 0.54 | 0.53 |
| Body condition | 1.04 | 0.23–4.70 | 0.80 | 0.96 | 1.96 | 0.22–17.34 | 2.12 | 0.55 |
| Multispecies infection | 16.20 | 1.57–167.74 | 19.32 | 0.02 | 20.10 | 1.62–250.14 | 25.86 | 0.02 |
Abbreviations: OR = Odds ratio, CI = 95% confidence interval, SE = standard error, P = P-value (0.05 significance limit).
4. Discussion
In this study we provide the first detailed molecular investigation into the characteristics of spirorchiidiasis in a sample of stranded green sea turtles. These were from the coastal waters of Queensland, a known hotspot for this disease (Gordon et al., 1998, Flint et al., 2010). Our approach, T-RFLP, proved efficient and effective in detecting and distinguishing fluke species, including Neospirorchis spp., which have previously been difficult to detect and identify grossly due to their fragility and small size. Whereas an adult fluke may easily be overlooked during gross necropsy, it may produce and disperse large numbers of ova, and molecular detection of low level infections can reduce the reporting of false negatives. This approach improves accuracy of data on infection rates and egg dispersal, and may assist in the detection of resistance to anthelminthics. Further, in many cases turtles are frozen prior to necropsy; in such cases molecular tools remain effective, whereas histological analysis and morphological identification of flukes becomes limited due to parasite sample degradation as a result of freeze-thaw cycles.
The overwhelming finding of the study was that eggs of all eight detected spirorchiid species occurred in multiple tissues. Six of the eight species were found in ten or more distinct tissues and even the rarest species detected, C. hawaiiensis, was detected in three. The level of spirorchiid diversity in various tissues is likely to be affected by the function and morphology of the organ, i.e. high levels of diversity may be expected in highly perfused filtration organs such as the spleen, where ova may be prone to embolization in small vessels. Patterns of tissue tropism may also assist in determining how spirorchiid eggs exit the host. The most probable route of ova elimination from the host is via the gastrointestinal tract, and spirorchiid ova can indeed be recovered from faeces (Greiner, 2013). If this were to be the primary route for all species, it would be expected that T-RFLP would commonly detect each within gastrointestinal samples, however, the ova of no species showed a detectable tropism for this tissue. A second possible site of exit is the lungs, but samples from there were also unremarkable in their spirorchiid diversity. We thus found no evidence of a characteristic pathway of the ova to the exterior; instead the results suggest that a high proportion become lost in dead-end tissues. Potentially, these ova are not released to the environment until the death of the host, which may be facilitated by associated pathology. Further investigation will be required in order to determine the definitive route of release from the host and there may well be species-specific distinctions.
The T-RFLP assay detected Neospirorchis spp. in the great majority of samples. Tissues from all but one of the 44 turtles tested positive, with Neospirorchis Genotype 2 being most common. Meanwhile, Carettacola spp. were relatively rare. The ubiquitous nature of Neospirorchis infections in green sea turtles has previously been noted in Queensland (Gordon et al., 1998) and Florida (Stacy et al., 2010). Meanwhile, Work et al. (2005) noted the low prevalence of Carettacola infections in Hawaiian green turtles, and although Stacy et al. (2010) found C. bipora in loggerhead turtles (Caretta caretta) from Florida, they did not find any Carettacola species in green turtles. Life cycle factors may contribute to the difference in frequency of Neospirorchis and Carettacola infections, e.g. relative fecundity and/or abundance of intermediate hosts. We also considered whether the lower detection rate for Carettacola found here might relate to the sensitivity of the primers targeting this genus. However, although the validation of this assay (Chapman et al., 2016b) found that the Carettacola primer pair had a higher detection limit than those for other genera, it was still sufficient to detect low numbers of eggs. Conversely, adult Carettacola were recovered more often than adult Neospirorchis, likely a result of differences in size, body morphology, and microhabitat usage.
The number of spirorchiid species detected in the brain was found to be significantly greater when granulomas were present. Also, when all granulomatous tissues were considered, the average number of species detected increased with the severity of lesions. One possible mechanism contributing to this finding is the onset of local alteration of blood flow that occurs in response to the initial embolization of ova, which in turn predisposes the accumulation of ova of further species within the lesion. General debility of the host permitting infection by a larger number of species is also likely to contribute to the number of species observed in a lesion, and may simultaneously facilitate progression to significant inflammatory responses. No individual species appeared to be disproportionately associated with granulomatous lesions. Neospirorchis Genotype 2 was detected in all brains with granulomas, sometimes as the only species present, but was also found in brains showing no observable granuloma formation. It is therefore not possible to implicate this genotype alone as a cause of lesions. The development of inflammatory lesions is likely to be influenced by multiple factors (e.g. individual response to insult, length of period of infection, environmental stressors, injury or coinfection with other pathogens). The relationship between brain lesions and clinical disease is a matter of interest because clinical neurological cases in recent times in Australia and the United States have been presumptively diagnosed as spirorchiidiasis. Neurological infection with Neospirorchis spp. was associated with a mass mortality event in loggerhead turtles (Caretta caretta) in Florida in the early 2000s (Jacobson et al., 2006) however the exact nature of the parasite's role was not resolved. Neurological impairment has also been observed alongside spirorchiid-associated brain lesions in green sea turtles from Queensland (Flint et al., 2010) and Western Australia (Raidal et al., 1998). Such neurological deficits, while injurious in themselves, may also predispose the animal to misfortunes such as boat strike or predation. The availability of molecular tools enables greater capacity to determine which Neospirorchis species are associated with neurological lesions. The methodology used here identifies the species involved, paving the way for the development of specific anti-mortem diagnostics. In this study, the species associated with the most severe neurological lesions observed are commonly present in turtles of all age and health statuses. Continued surveillance and development of quantitative ante-mortem testing for implicated species is likely to assist in unravelling the factors that lead to severe clinical effects.
In this study, overall spirorchiid diversity and the prevalence of individual species (particularly of species within the Hapalotrema/Learedius group) generally decreased with age. An exception was H. mistroides, which, in contrast to other Hapalotrema species, was detected more often in mature turtles than small immature ones, potentially due to greater accumulated exposure in large immature or mature turtles. The overall tendency for parasitic infections to decrease with age is likely to reflect progressive development of immunity after immunologically naïve small immature turtles are exposed to spirorchiids at the time of recruitment to neritic habitats (Work et al., 2005, Flint et al., 2010). Acquired immunity to blood flukes in various vertebrate hosts has been routinely observed. For example, infections of the related Schistosoma spp. in species such as humans (Cheever et al., 1977, Agnew et al., 1996, Drake and Bundy, 2001), primates (Cheever et al., 1974, He et al., 1992, Li et al., 2015), water buffalo (Li, 2014), cattle and pigs (Vercruysse and Gabriel, 2005) generally decrease in intensity as the host ages. Meanwhile, the ability of poikilotherms to develop immune responses to blood flukes is demonstrated by the progressive development of humoral responses by Pacific Bluefin tuna (Thunnus maccoyii) to aporocotylid flukes (Cardicola spp.) (Aiken et al., 2008, Kirchhoff et al., 2012, Polinski et al., 2014). Despite this, green turtle populations in the Pacific (Work et al., 2005, Flint et al., 2010) and the Atlantic (Santoro et al., 2007, Stacy et al., 2010) show different relationships between age and apparent susceptibility to spirorchiid infection. The factors causing variations in epizootiology between populations in different geographic regions remain unclear. Potential factors include different spirorchiid species assemblages, and environmental factors which may cause differential exposure to infective stages (e.g. microhabitat variations and presence of intermediate hosts), or cause stress to hosts (e.g. pollutants and poor water quality), resulting in increased susceptibility to disease (Flint et al., 2015b). Thus, spirorchiidiasis requires analysis in the context of distinct host species, parasite species and habitats.
Possible future research directions following up from our findings could include quantitative assessment of the number of spirorchiid eggs of each species or genotype in a sample. This approach would allow estimation of the relative contribution of each type to lesion development. Currently, quantitative molecular methods are largely restricted to quantitative real-time PCR (qPCR). However, the development of species/genotype specific primer/probe sets to target each spirorchiid would prove difficult given relatively low observed levels of interspecific variation in commonly examined spirorchiid genes and the high number of target genotypes. Visual methods of quantitation i.e. histology or tissue digestion and microscopic counts can produce data only to a genus level. In the case of histology, this can be difficult to achieve accurately due to sectional angles and degeneration of eggs.
As discussed, low numbers of samples were available overall and disproportionate numbers of samples came from small immature turtles. The relationship between age, the number of spirorchiid species present and presence of lesions varied between organs, which is likely due to low sample numbers as evidenced by broad confidence intervals. However, genus level infection rates and tissue tropisms were generally reflective of those reported previously. Full comparable sets of tissue samples were not available from all turtles, with infection rates calculated per organ rather than at the host level, but this in itself provides insight into tissue tropisms. Some parasite species detected here are yet to be fully identified due to a lack of suitable samples for morphological characterisation, either due to their cryptic and fragile nature or freezing of the turtle prior to collection for necropsy. No effect on molecular results due to freezing is likely, and it is anticipated that the parasites will be definitively identified and/or described in future studies. Until recently, a lack of life cycle data has constrained our understanding of disease transmission and ability to control infections, however, recent discoveries in this area (Cribb et al., 2017), paired with specific molecular diagnostic tools, will lead to improved knowledge of disease epidemiology.
This study has provided the first molecular study of the distribution of spirorchiids in the tissues of green sea turtles, and provides data at a level of specificity not previously attainable. It demonstrates that young turtles in this region are likely to be infected with the greatest diversity of spirorchiids, and that at least in the brain, there is a relationship between the presence and severity of granulomatous lesions and the number of spirorchiid species present. It is clear that variation in disease presentation between geographic regions is significant, yet not understood. The influence of spatially fluctuating environmental factors and stressors is of particular interest in understanding the triggers for the development of clinical disease, as is the elucidation of further spirorchiid life cycles and the development of ante-mortem diagnostics for investigation and monitoring of live turtle populations.
Acknowledgements
This work was supported by the Australian Research Council (Linkage Grant number LP110100569). We acknowledge staff at the wildlife rehabilitation facilities Australia Zoo (Beerwah, QLD) and Underwater World (Mooloolaba, QLD) as well as the Queensland Department of Environment and Heritage Protection (EHP) and Queensland Parks and Wildlife Service (QPWS) for facilitating access to dead turtles for examination. Chris Cazier (histology) and the Animal Genetics Laboratory (capillary electrophoresis) at The University of Queensland's School of Veterinary Science provided technical services. Thanks to Dr Paul Eden for initial work in sample collection.
References
- Agnew A., Fulford A.J.C., Mwanje M.T., Gachuhi K., Gutsmann V., Krijger F.W., Sturrock R.F., Vennervald B.J., Ouma J.H., Butterworth A.E., Deelder A.M. Age-dependent reduction of schistosome fecundity in Schistosoma haematobium but not Schistosoma mansoni infections in humans. Am. J. Trop. Med. Hyg. 1996;55:338–343. doi: 10.4269/ajtmh.1996.55.338. [DOI] [PubMed] [Google Scholar]
- Aguirre A.A., Spraker T.R., Balazs G.H., Zimmerman B. Spirorchidiasis and fibropapillomatosis in green turtles from the Hawaiian Islands. J. Wildl. Dis. 1998;34:91–98. doi: 10.7589/0090-3558-34.1.91. [DOI] [PubMed] [Google Scholar]
- Aiken H.M., Hayward C.J., Crosbie P., Watts M., Nowak B.F. Serological evidence of an antibody response in farmed southern bluefin tuna naturally infected with the blood fluke. Cardicola Forsteri. Fish. Shellfish Immunol. 2008;25:66–75. doi: 10.1016/j.fsi.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Chapman P.A., Cribb T.H., Blair D., Traub R.J., Kyaw-Tanner M.T., Flint M., Mills P.C. Molecular analysis of the genera Hapalotrema Looss, 1899 and Learedius Price, 1934 (Digenea: Spirorchiidae) reveals potential cryptic species, with comments on the validity of the genus Learedius. Syst. Parasitol. 2015;90:67–79. doi: 10.1007/s11230-014-9535-y. [DOI] [PubMed] [Google Scholar]
- Chapman P.A., Owen H., Flint M., Traub R.J., Cribb T.H., Mills P.C. Molecular characterization of coccidia associated with an epizootic in green sea turtles (Chelonia mydas) in south east Queensland, Australia. PLoS One. 2016;11:e0149962. doi: 10.1371/journal.pone.0149962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman P.A., Traub R.J., Kyaw-Tanner M.T., Owen H., Flint M., Cribb T.H., Mills P.C. Terminal restriction fragment length polymorphism for the identification of spirorchiid ova in tissues from the green sea turtle, Chelonia mydas. PLoS One. 2016;11:e0162114. doi: 10.1371/journal.pone.0162114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever A.W., Erickson D.G., Sadun E.H., von Lichtenberg F. Schistosoma japonicum infection in monkeys and baboons: parasitological and pathological findings. Am. J. Trop. Med. Hyg. 1974;23:51–64. doi: 10.4269/ajtmh.1974.23.51. [DOI] [PubMed] [Google Scholar]
- Cheever A.W., Kamel I.A., Elwi A.M., Mosimann J.E., Danner R. Schistosoma mansoni and S. haematobium infections in Egypt: II. Quantitative parasitological findings at necropsy. Am. J. Trop. Med. Hyg. 1977;26:702–716. doi: 10.4269/ajtmh.1977.26.702. [DOI] [PubMed] [Google Scholar]
- Cribb T.H., Crespo-Picazo J.L., Cutmore S.C., Stacy B.A., Chapman P.A., García-Párraga D. Elucidation of the first definitively identified life cycle for a marine turtle blood fluke (Trematoda: Spirorchiidae) enables informed control. Int. J. Parasitol. 2017;47:61–67. doi: 10.1016/j.ijpara.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Drake L.J., Bundy D.A.P. Multiple helminth infections in children: impact and control. Parasitology. 2001;122:S73–S81. doi: 10.1017/s0031182000017662. [DOI] [PubMed] [Google Scholar]
- Fajardo V., Gonzalez I., Martin I., Rojas M. Real-time PCR for detection and quantification of red deer and roe deer (Capreolus capreolus) in meat mixtures. Meat Sci. 2008;79:289–298. doi: 10.1016/j.meatsci.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Flint J., Flint M., Limpus C.J., Mills P.C. Trends in marine turtle strandings along the east Queensland, Australia coast, between 1996 and 2013. J. Mar. Biol. 2015;2015:7. [Google Scholar]
- Flint M., Eden P.A., Limpus C.J., Owen H., Gaus C., Mills P. Clinical and pathological findings in green turtles (Chelonia mydas) from Gladstone, Queensland: investigations of a stranding epidemic. EcoHealth. 2015;12:298–309. doi: 10.1007/s10393-014-0972-5. [DOI] [PubMed] [Google Scholar]
- Flint M., Patterson-Kane J.C., Limpus C.J., Mills P.C. Health surveillance of stranded green turtles in Southern Queensland, Australia (2006-2009): an epidemiological analysis of causes of disease and mortality. EcoHealth. 2010;7:135–145. doi: 10.1007/s10393-010-0300-7. [DOI] [PubMed] [Google Scholar]
- Flint M., Patterson-Kane J.C., Limpus C.J., Work T.M., Blair D., Mills P.C. Postmortem diagnostic investigation of disease in free-ranging marine turtle populations: a review of common pathologic findings and protocols. J. Vet. Diagn Investig. 2009;21:733–759. doi: 10.1177/104063870902100601. [DOI] [PubMed] [Google Scholar]
- Flint M., Patterson-Kane J.C., Mills P.C., Limpus C.J. School of Veterinary Science, The University of Queensland; Gatton, Queensland: 2009. A Veterinarian's Guide to Sea Turtle Post Mortem Examination and Histological Investigation. [Google Scholar]
- Glazebrook J.S., Campbell R.S.F. A survey of the diseases of marine turtles in northern Australia. II. Oceanarium-reared and wild turtles. Dis. Aquat. Org. 1990;9:97–104. [Google Scholar]
- Glazebrook J.S., Campbell R.S.F., Blair D. Studies on cardiovascular fluke (Digenea: Spirorchiidae) infections in sea turtles from the great barrier reef, Queensland, Australia. J. Comp. Pathol. 1989;101:231–250. doi: 10.1016/0021-9975(89)90033-9. [DOI] [PubMed] [Google Scholar]
- Gordon A.N., Kelly W.R., Cribb T.H. Lesions caused by cardiovascular flukes (Digenea: Spirorchidae) in stranded green turtles (Chelonia mydas) Vet. Pathol. 1998;35:21–30. doi: 10.1177/030098589803500102. [DOI] [PubMed] [Google Scholar]
- Gordon A.N., Kelly W.R., Lester R.J.G. Epizootic mortality of free-living green turtles, Chelonia mydas, due to coccidiosis. J. Wildl. Dis. 1993;29:490–494. doi: 10.7589/0090-3558-29.3.490. [DOI] [PubMed] [Google Scholar]
- Graczyk T.K., Aguirre A.A., Balazs G.H. Detection by ELISA of circulating anti-blood fluke (Carettacola, Hapalotrema, and Learedius) immunoglobulins in Hawaiian green turtles (Chelonia mydas) J. Parasitol. 1995;81:416–421. [PubMed] [Google Scholar]
- Greiner E.C. Parasites of marine turtles. In: Wyneken J.L.K.J., Musick J.A., editors. The Biology of Sea Turtles, Volume III. Marine Biology. CRC Press; 2013. pp. 427–446. [Google Scholar]
- He Y.X., Yu Q.F., Hu Y.Q. Parasitological and histopathological studies on rhesus monkeys infected with Chinese mainland strain of Schistosoma japonicum. Southeast Asian J. Trop. Med. Public Health. 1992;23:254–260. [PubMed] [Google Scholar]
- Jacobson E.R., Homer B.L., Stacy B.A., Greiner E.C., Szabo N.J., Chrisman C.L., Origgi F., Coberley S., Foley A.M., Landsberg J.H., Flewelling L., Ewing R.Y., Moretti R., Schaf S., Rose C., Mader D.R., Harman G.R., Manire C.A., Mettee N.S., Mizisin A.P., Shelton G.D. Neurological disease in wild loggerhead sea turtles Caretta caretta. Dis. Aquat. Org. 2006;70:139–154. doi: 10.3354/dao070139. [DOI] [PubMed] [Google Scholar]
- Kirchhoff N.T., Leef M.J., Valdenegro V., Hayward C.J., Nowak B.F. Correlation of humoral immune response in southern bluefin tuna, T. maccoyii, with infection stage of the blood fluke, Cardicola forsteri. PLoS One. 2012;7:e45742. doi: 10.1371/journal.pone.0045742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-H., Xu Y.-X., Vance G., Wang Y., Lv L.-B., van Dam G.J., Cao J.-P., Wilson R.A. Evidence that rhesus macaques self-cure from a Schistosoma japonicum infection by disrupting worm esophageal function: a new route to an effective vaccine? PLoS Negl. Trop. Dis. 2015;9:e0003925. doi: 10.1371/journal.pntd.0003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-S. The Schistosoma japonicum self-cure phenomenon in water buffaloes: potential impact on the control and elimination of schistosomiasis in China. Int. J. Parasitol. 2014;44:167–171. doi: 10.1016/j.ijpara.2013.10.007. [DOI] [PubMed] [Google Scholar]
- McManus D.P., Bowles J. Molecular genetic approaches to parasite identification: their value in diagnostic parasitology and systematics. Int. J. Parasitol. 1996;26:687–704. doi: 10.1016/0020-7519(96)82612-9. [DOI] [PubMed] [Google Scholar]
- Meager J.J., Limpus C.J. vol. 3. Department of Environment and Heritage Protection; Queensland: 2012. (Marine wildlife Stranding and Mortality Database Annual Report 2011 III. Marine Turtle). [Google Scholar]
- Ndao M. 2009. Diagnosis of Parasitic Diseases: Old and New Approaches. Interdiscip Perspect Infect Dis 2009, pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polinski M., Shirakashi S., Bridle A., Nowak B. Transcriptional immune response of cage-cultured Pacific bluefin tuna during infection by two Cardicola blood fluke species. Fish. Shellfish Immunol. 2014;36:61–67. doi: 10.1016/j.fsi.2013.10.008. [DOI] [PubMed] [Google Scholar]
- Queensland Department of Environment and Heritage Protection . 2013. Marine wildlife Strandings Data.http://www.ehp.qld.gov.au/wildlife/caring-for-wildlife/marine-strandings-data.html (Accessed 24 August 2016) [Google Scholar]
- Queensland Department of Environment and Heritage Protection . 2015. Marine strandings Update 2015.https://www.ehp.qld.gov.au/wildlife/caring-for-wildlife/marine-strandings-update.html (Accessed 5 May 2016) [Google Scholar]
- Raidal S.R., Ohara M., Hobbs R.P., Prince R.I.T. Gram-negative bacterial infections and cardiovascular parasitism in green sea turtles (Chelonia mydas) Aust. Vet. J. 1998;76:415–417. doi: 10.1111/j.1751-0813.1998.tb12392.x. [DOI] [PubMed] [Google Scholar]
- Santoro M., Morales J.A., Rodriguez-Ortiz B. Spirorchiidiosis (Digenea: Spirorchiidae) and lesions associated with parasites in Caribbean green turtles (Chelonia mydas) Vet. Rec. 2007;161:482–486. doi: 10.1136/vr.161.14.482. [DOI] [PubMed] [Google Scholar]
- Stacy B.A., Foley A.M., Greiner E., Herbst L.H., Bolten A., Klein P., Manire C.A., Jacobson E.R. Spirorchiidiasis in stranded loggerhead Caretta caretta and green turtles Chelonia mydas in Florida (USA): host pathology and significance. Dis. Aquat. Org. 2010;89:237–259. doi: 10.3354/dao02195. [DOI] [PubMed] [Google Scholar]
- Vercruysse J., Gabriel S. Immunity to schistosomiasis in animals: an update. Parasite Immunol. 2005;27:289–295. doi: 10.1111/j.1365-3024.2005.00766.x. [DOI] [PubMed] [Google Scholar]
- Work T.M., Balazs G.H., Rameyer R.A., Morris R.A. Retrospective pathology survey of green turtles Chelonia mydas with fibropapillomatosis in the Hawaiian Islands, 1993-2003. Dis. Aquat. Org. 2004;62:163–176. doi: 10.3354/dao062163. [DOI] [PubMed] [Google Scholar]
- Work T.M., Balazs G.H., Schumacher J.L., Marie A. Epizootiology of spirorchiid infection in green turtles (Chelonia mydas) in Hawaii. J. Parasitol. 2005;91:871–876. doi: 10.1645/GE-454R.1. [DOI] [PubMed] [Google Scholar]
- Work T.M., Balazs G.H., Summers T.M., Hapdei J.R., Tagarino A.P. Causes of mortality in green turtles from Hawaii and the insular Pacific exclusive of fibropapillomatosis. Dis. Aquat. Org. 2015;115:103–110. doi: 10.3354/dao02890. [DOI] [PubMed] [Google Scholar]