Abstract
Background
Glucocorticoid receptor (GR) activity plays a role in many aspects of human physiology and may play a crucial role in chemotherapy resistance in a wide variety of solid tumors. A novel immunohistochemistry (IHC) based assay has been previously developed and validated in order to assess GR immunoreactivity in triple-negative breast cancer. The current study investigates the standardized use of this validated assay to assess GR expression in a broad range of solid tumor malignancies.
Methods
Archived formalin-fixed paraffin-embedded tumor bank samples (n=236) from 20 different solid tumor types were analyzed immunohistochemically. Nuclear staining was reported based on the H-score method using differential intensity scores (0, 1+, 2+, or 3+) with the percent stained (out of at least 100 carcinoma cells) recorded at each intensity.
Results
GR was expressed in all tumor types that had been evaluated. Renal cell carcinoma, sarcoma, cervical cancer, and melanoma were those with the highest mean H-scores, indicating high levels of GR expression. Colon, endometrial, and gastric cancers had lower GR staining percentages and intensities, resulting in the lowest mean H-scores.
Conclusion
A validated IHC assay revealed GR immunoreactivity in all solid tumor types studied and allowed for standardized comparison of reactivity among the different malignancies.
Impact
Baseline expression levels of GR may be a useful biomarker when pharmaceutically targeting GR in research or clinical setting.
Keywords: glucocorticoid receptor expression, immunohistochemistry, solid tumors
Introduction
Glucocorticoid receptor (GR), a member of the steroid nuclear receptor superfamily, is responsible for modulating many processes, including cell homeostasis, cellular development, metabolic function,1–4 immune function,5 and central nervous system and psychiatric function.6,7 In oncology, GR has been implicated in the development of cancer cell resistance by modulating the intracellular apoptosis balance point and influencing several well-documented cascade mechanisms.8 GR modulation of gene expression is complex and involves the binding of ligand and receptor in the cytoplasm, dissociation of heat shock proteins, and homodimerization and translocation of the ligand/receptor complex to the nucleus.3 The ligand/receptor complex then binds directly to the DNA and acts as a transcription factor for multiple gene products. This activity is further influenced by the complex environment of coactivator and corepressor molecules, which contribute additional effects to gene expression.2,6 Investigators have identified the role of GR in the development of chemotherapy resistance in experimental models involving tumors of epithelial origin, especially triple-negative breast cancer (TNBC), prostate cancer, and ovarian cancer.9–15
The addition of the GR agonist, dexamethasone, to cell line chemotherapy models reduces apoptosis and increases transcription of pro-cell-survival genes. These effects were reversed in the presence of the GR antagonist, mifepristone.15–17 A TNBC cell line MDA-MB-231 xenograft model demonstrated that addition of dexamethasone reduced cell death induced by paclitaxel, whereas the addition of mifepristone enhanced the efficacy of the taxanes.16 The effect of mifepristone has been replicated in in vitro and in vivo experiments across multiple cancer cell lines and in combination with multiple chemotherapeutic agents.14–17
These in vitro data indicate that glucocorticoids, whether endogenous or exogenous, may cause expression of prosurvival/antiapoptotic genes and protect tumor cells from the effects of chemotherapy.18,19 Conversely, benefits of GR antagonists have been shown preclinically in several tumor types expressing GR including TNBC, ovarian, lung, and prostate cancer.9–14,16,17,20 In addition to the preclinical evidence,16 clinical evidence21–23 suggests that GR expression plays a substantial role in TNBC. These findings have considerable clinical implications as the coadministration of glucocorticoids is common to counteract hypersensitivity to various treatments as well as nausea.
An immunohistochemistry (IHC) assay has been developed and validated for evaluating GR expression in TNBC.24 Several clinical trials are currently evaluating GR antagonists in conjunction with chemotherapy in patients with TNBC.25 These studies further measure GR tumor expression using the validated IHC assay. During development of the previously reported TNBC assay, 50 archival, formalin-fixed paraffin-embedded (FFPE) tissue samples of TNBC from individual treatment-naïve patients, collected as diagnostic tissue blocks and stored in a commercial tissue bank, were evaluated. While the earlier literature suggested rates of GR positivity of ~25%–50% in TNBC samples,26–28 rates based on the IHC assay were ≥80%24 using a minimum cutoff of 10% tumor cells staining positively.
The objective of this exploratory GR expression study was to extend this research beyond TNBC by using the previously developed IHC GR assay to survey the degree of GR expression across other tumor types. Little is known about rates of GR expression in other tumor types even though the role of GR in chemotherapy resistance has been suggested in many tumors of epithelial origin.9–13 As understanding of the role of GR in oncology progresses, the findings from this study may guide decisions about how to successfully introduce GR antagonists into clinical testing.
Methods and materials
GR IHC protocol
For each of the 20 selected tumor types, archival FFPE tumor bank tissues were randomly chosen for this study. With the exception of the pancreatic cancer samples that were purchased from Pantomics Inc. (Richmond, CA, USA), all other tissues samples were obtained from the QualTek tissue bank (QualTek Molecular Laboratories, Newtown, PA, USA). These tumor tissues were ethically acquired from various clinical sites with diverse patient populations with personal identifiers redacted.
The FFPE tumor tissues were sectioned (4–5 μm) onto slides with a positive charge (Fisher ProbeOn Plus™; Thermo Fisher Scientific, Waltham, MA, USA) and dry heated for 1 h at 65°C within 1 week of testing. Deparaffinization included a series of four 100% xylene changes followed by rehydration with a graded series of ethanol (100%, 70%, 30%) to distilled water.
Based on a previous publication, rabbit monoclonal antibody anti-GR (D8H2; Cell Signaling Technology [#3660S], Danvers, MA, USA), was chosen from three different GR antibody candidates for further assay development and validation.24
Antigen retrieval consisted of a 20-min incubation of slides in Citra Plus Target Retrieval Solution (BioGenex [catalog number HK080-9K], Fremont, CA, USA), heated to 98°C with a commercial steamer (Black and Decker HS1000 model steamer; Black and Decker, Baltimore, MD, USA). Following a 5-min cool down, slides were transferred onto an automated IHC platform (TechMate™ 500 or 1000 with WorkMate software version 3.96; Roche Diagnostics, Oro Valley, AZ, USA). All reagent changes were automated, including all detection kit steps (Rabbit Polink2+ HRP and DAB chromogen; Golden Bridge International, [catalog numbers D39-110 and C09-100], Bothell, WA, USA), a protein blocking step (QualTek proprietary reagent), primary antibody incubation (anti-GR [D8H2]; Cell Signaling Technology [catalog number 3660S]; diluted to 1:1,750 in primary antibody diluent [QualTek proprietary]), a peroxidase blocking step (3% United States Pharmacopeia H2O2, with ~0.02% v/v Tween® 20 detergent; Thermo Fisher Scientific), hematoxylin counterstaining and all intervening washes (tris-buffered saline containing 0.02% v/v Tween 20), by a capillary gap process29 at room temperature (25°C) using the previously optimized assay conditions.24 The slides were then dehydrated in a series of ethanol (95%, 100%) and 100% xylene changes, and mounted with a coverslip (Cytoseal™ XYL mounting media, Thermo Fisher Scientific).
Evaluation of GR expression in various tumors
The assay was previously validated24 in a Clinical Laboratory Investigation Amendment (CLIA)-accredited facility (QualTek Clinical Laboratories). GR expression levels, detected by the validated IHC assay, were evaluated in a panel of 20 tumor types (Table 1) using archival, treatment-naïve, FFPE tissue samples (individual or within tissue microarrays [TMAs]). Tissues included bladder cancer (n=10), breast cancer (non-TNBC) (n=10), cervical cancer (n=15), colon cancer (n=16), endometrial cancer (n=13), esophageal cancer (n=8), gall bladder cancer (n=10), gastric cancer (n=11), glioblastoma (n=8), head and neck cancer (n=10), hepatocellular carcinoma (n=10), lung cancer (n=17), melanoma (n=11), mesothelioma (n=8, including TMA [n=4; four cores each, 1.5 mm; US Biomax, Inc., catalog number T392a, Rockville, MD, USA]), neuroendocrine cancer (n=11), ovarian cancer (n=11), pancreatic cancer (n=16, including TMA [two cores each, 2 mm]; Pantomics Inc. [catalog number PAC481]), prostate cancer (n=11), renal cell carcinoma (RCC) (n=10), and sarcoma (n=14).
Table 1.
Solid tumor types evaluated using glucocorticoid receptor IHC sensitivity screening
| Bladder cancer | Hepatocellular carcinoma |
| Breast cancer (non-TNBC) | Lung cancer |
| Cervical cancer | Melanoma |
| Colon cancer | Mesothelioma |
| Endometrial cancer | Neuroendocrine cancer |
| Esophageal cancer | Ovarian cancer |
| Gall bladder cancer | Pancreatic cancer |
| Gastric cancer | Prostate cancer |
| Glioblastoma | Renal cell carcinoma |
| Head and neck cancer | Sarcoma |
Abbreviations: IHC, immunohistochemical; TNBC, triple-negative breast cancer.
GR staining is sensitive to poor tissue fixation, common among nuclear antigens, and may result in a potential false-negative IHC result. However, because GR strongly stains stromal cells (which act as an internal positive control), areas of poor fixation that would contribute to a false-negative result are easily identifiable. In such cases, those regions were not scored or the samples were rejected.
Tissue sections were scored for nuclear staining within tumor cells across the total evaluable area of the tissue. Staining within the cytoplasm of tumor cells or stroma was not scored. Necrotic regions, in situ carcinoma, and poorly fixed regions of the tissue were also excluded from scoring. GR reactivity was scored using differential intensity scores (0, null; 1+, low or weak; 2+, moderate; 3+, high or strong). A total percentage score (% of tumor cells staining ≥1+ intensity; ie, the sum of the percentage of cells at 1+, 2+, and 3+ intensities) was used to semiquantitatively evaluate tumor expression of GR. Only samples with at least 100 invasive carcinoma cells were included. The H-score, which numerates staining ratios with respect to both intensity and frequency, was used to capture the pattern of nuclear staining observed. The H-score was calculated using the following formula: H-score = [(% at 0)×0]+[(% at 1+)×1]+[(% at 2+)×2]+ [(% at 3+)×3]. The H-score produces a continuous variable that ranges from 0 to 300.
Results
GR staining was evaluated in 20 different tumor types (Table 1) with 236 total samples analyzed and 5–16 samples per tumor type (Table 2). Although GR staining is generally broad and is observed in most normal tissue, stromal staining served as an internal positive control to help identify samples to reject or regions to avoid during the scoring process. Positive and negative controls were included in all tests and were evaluated for appropriate reactivity before GR staining was evaluated.
Table 2.
Immunohistochemistry scoring for glucocorticoid receptor status in 20 tumor types
| Cancer type | N | Intensity staining ≥1+ (%)
|
H-scorea
|
||
|---|---|---|---|---|---|
| Mean (SD) | Range | Mean (SD) | Range | ||
| Melanoma | 11 | 94.1 (5.8) | 80–100 | 241.8 (33.3) | 180–290 |
| Renal cell carcinoma | 10 | 93.5 (7.1) | 80–100 | 227.5 (48.9) | 130–300 |
| Sarcoma | 14 | 92.9 (4.3) | 85–100 | 212.9 (35.8) | 155–270 |
| Cervical | 15 | 82.0 (21.5) | 40–100 | 210.0 (75.6) | 70–290 |
| Head and neck | 10 | 86.5 (7.5) | 70–95 | 191.5 (37.7) | 130–250 |
| Hepatocellular carcinoma | 10 | 82.0 (12.1) | 60–95 | 185.0 (55.5) | 90–265 |
| Neuroendocrine | 11 | 79.1 (33.2) | 10–100 | 175.5 (88.3) | 10–260 |
| Mesothelioma | 8 | 84.4 (13.7) | 65–100 | 165.9 (50.1) | 120–260 |
| Glioblastoma | 8 | 71.9 (28.0) | 20–100 | 164.4 (80.3) | 25–260 |
| Lung | 17 | 75.3 (30.8) | 0–100 | 152.9 (84.8) | 0–290 |
| Esophageal | 8 | 62.6 (42.8) | 0–100 | 151.4 (113.0) | 0–280 |
| Prostate | 11 | 85.9 (26.4) | 10–100 | 144.5 (63.2) | 10–220 |
| Pancreas | 16 | 78.3 (25.1) | 30–100 | 131.1 (78.4) | 30–280 |
| Bladder | 10 | 56.8 (46.0) | 0–98 | 117.3 (99.9) | 0–255 |
| Breast (non-TNBC) | 10 | 62.9 (44.1) | 1–100 | 111.5 (89.3) | 1–250 |
| Gall bladder | 10 | 73.5 (14.9) | 50–95 | 108.5 (43.3) | 70–205 |
| Ovarian | 11 | 57.7 (33.9) | 0–100 | 106.8 (87.8) | 0–240 |
| Endometrial | 13 | 17.5 (27.6) | 0–95 | 32.5 (60.5) | 0–215 |
| Gastric | 11 | 14.5 (26.9) | 0–90 | 22.7 (53.1) | 0–180 |
| Colon | 16 | 6.3 (22.4) | 0–90 | 12.9 (47.3) | 0–190 |
Note:
Sorted by mean H-score.
Abbreviation: SD, standard deviation; TNBC, triple-negative breast cancer.
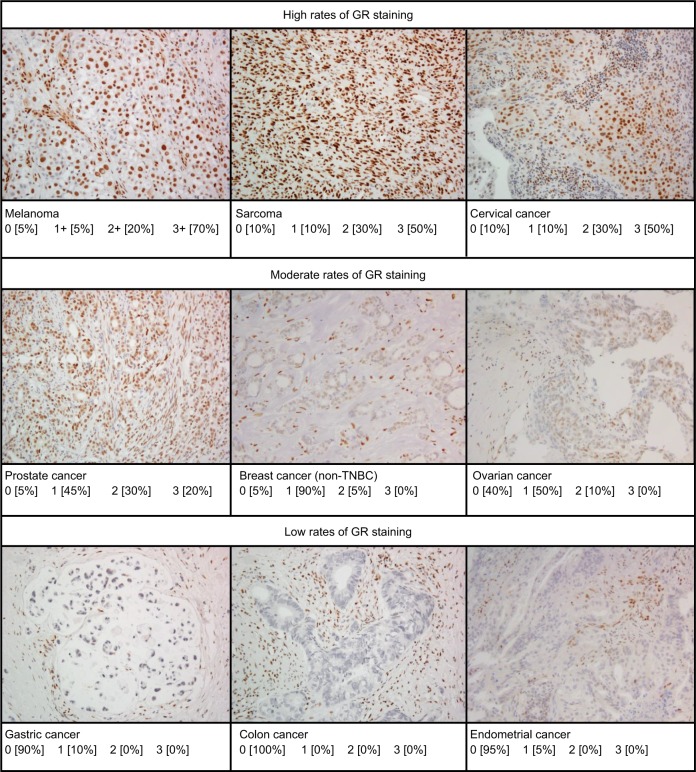
GR reactivity was observed in the nuclei of all tumor cell types examined and was highly expressed in the majority of tumors (Table 2) with little to no cytoplasmic staining. Representative micrographs showing a range of staining intensities are provided in Figure 1.
Figure 1.
Representative micrographs of immunohistochemical GR staining in various tumor types showing variations in levels of staining.
Note: Tumors with ≥10% GR staining at the following intensities: 0, 1+, 2+, and 3+.
Abbreviation: GR, glucocorticoid receptor; TNBC, triple-negative breast cancer.
With GR staining depicted as a fraction of cells with 0, 1+, 2+, and 3+ staining, melanoma, RCC, and sarcoma tumor samples show a high fraction of 2+ and 3+ staining, and few samples (<10%) show 0 staining. In contrast, >80% of colon, gastric, and endometrial cancer samples had no GR staining. For the other tumors evaluated, at least 50% of the tumors expressed 1+ to 3+ staining.
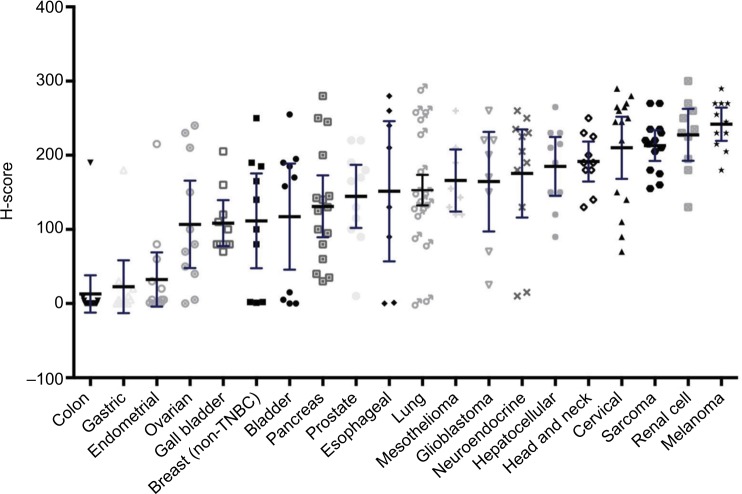
Evaluating GR expression of the tumor types by H-score (accounting for both intensity and frequency) versus frequency of GR positivity showed similar results (Figures 2 and 3). Several cancer types have high mean and median H-scores along with high variability of H-scores within tumor types. An exception is seen with melanoma, cervical cancer, RCC, and sarcoma tumor types. Their H-scores were high with little variability and the GR staining was uniformly high. Colon, gastric, and endometrial cancer were distinctly different from the other tumors by virtue of their very low GR staining intensity fraction and H-score.
Figure 2.
Glucocorticoid receptor positivity by mean H-score.
Note: The H-score was calculated as [(% at 0)×0]+[(% at 1+)×1]+[(% at 2+)×2]+[(% at 3+)×3]; thus, the H-score produces a continuous variable that ranges from 0 to 300.
Abbreviation: TNBC, triple-negative breast cancer.
Figure 3.
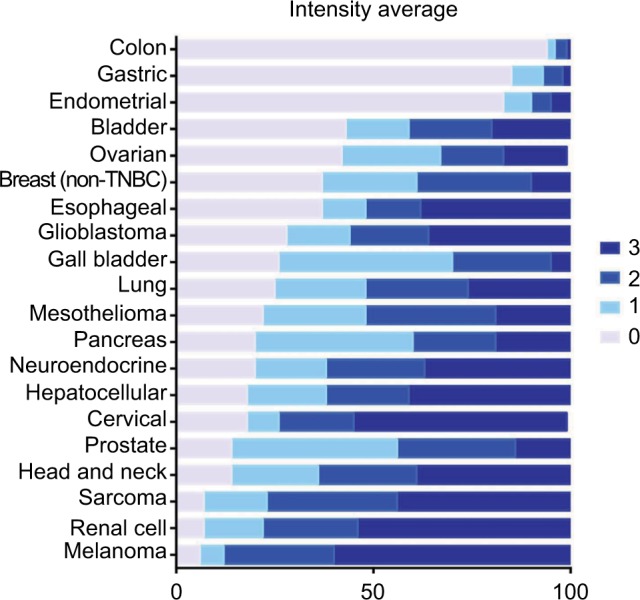
Average percent staining of glucocorticoid receptor at various intensities.
Note: Bars show the distribution of average percent staining across the 0, 1+, 2+, and 3+ categories.
Abbreviation: TNBC, triple-negative breast cancer.
The limited histologic subtypes that were analyzed (data not shown) indicated that GR expression varies by tumor subtype. This analysis showed that small-cell lung cancer (SCLC) has very low GR expression (mean H-score 81, range 0–230), whereas non-small-cell lung cancer (NSCLC) adenocarcinoma and squamous cell carcinoma have high GR expression (mean H-score 193, range 140–290, and 173, range 120–260, respectively). In the samples studied, the subtypes of the RCC were limited to clear cell type, all of high GR staining. The sarcoma tumor group included soft tissue sarcomas only, also all of high GR staining.
Discussion
GRs play an important role in tumor response to microenvironment, and GRs have been implicated in tumor cell survival and response to chemotherapy.12–15,17,18,20 To date, GR expression has been reported only within a limited number of solid tumor types (eg, breast and colon),26–28,30 and assay results have been highly variable. Hence, the clinical relevance of the results in the literature is difficult to interpret. A CLIA-validated,24 IHC-based, GR assay has been developed and assessments of 20 different tumor types have been standardized with more than 200 samples. Using surrounding tissues (ie, positively staining fibroblasts, endothelial cells, and a subset of lymphocytes) as an internal stromal control provides high confidence of antibody specificity to GR protein. The antibody used in this assay, D8H2, is both specific and sensitive to the Leu368 region of both alpha and beta isoforms of the GR protein24 without cross-reactivity to the mineralocorticoid receptor and nonspecific binding.
GR reactivity was observed in all 20 tumor types that had been evaluated, many of which had no prior published GR expression rates. GR staining was detectable at a wide range of intensities across and within the various tumor types.
Applying the validated GR IHC assay, it was observed that GR expression varies by tumor type. Among the tumors with an overall high degree of GR staining, some tumors were consistently high staining (clear cell RCC, soft tissue sarcoma, melanoma, and cervical cancer), and others showed great variability among individual samples. Colon, gastric, and endometrial cancer have very low GR staining by intensity and H-score, with the majority of samples showing no GR expression. As seen with the lung cancer samples evaluated, GR expression was different for SCLC and NSCLC. Further analyses of GR expression by other tumor subtypes is needed.
As with most established tumor receptors that have been successfully targeted, much effort has been placed on correlating clinical outcomes with target expression levels and activity. The definition of what is considered “positive” for estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 research31,32 has been modified over time. The definition of “positive” for both ER and PR decreased over time from ≥10% of tumor cells staining positive to ≥1%.31 Similar efforts are currently ongoing with programmed cell death protein 1 (PD1) and PD1-ligand 1 (PD-L1) therapies. Thus, the data were presented as intensity and frequency of staining across the 20 tumor types, without applying a threshold cutoff.
The wide range of staining intensity and H-scores seen warrants further research to understand the role of GR expression, both as a prognostic factor and as a predictive factor in the response to chemotherapy. For some tumor types (melanoma, sarcoma, and RCC), GR assessment may not be necessary due to the consistently high staining intensity and H-scores. Research is needed to determine if tumors with low staining and H-scores (colon, gastric, and endometrial cancers) are good candidates for GR-targeted therapeutics.
With the introduction of more selective GR antagonists, GR may become a relevant therapeutic target. Hence, this validated assay may provide new opportunities in developing companion diagnostic and predictive assays for GR expression. Establishing the optimal threshold for GR expression qualitatively or quantitatively that correlates with clinical benefit in patients with specific tumor types will require larger studies with clinical endpoints. An initial assessment of GR as a therapeutic target and predictive factor is ongoing in a clinical trial testing the selective GR modulator CORT125134 in combination with nab-paclitaxel in solid tumors (NCT02762981).
The current assessment of GR positivity include the limited sample size and absence of subtyping within a tumor type. Furthermore, assessment of single sections of an FFPE sample may not completely represent GR distribution within the tumor and may only reflect a portion of a tumor. Also, GR expression may evolve in response to anticancer therapy and be further influenced by the use of concomitant medication, particularly those including steroids. This validated assay provides the first steps of assessing GR as a clinical surrogate and pharmacological target. Further studies will be needed to determine the most robust technical method to prepare samples through the course of treatment and to determine which quantitative assessment provides the strongest correlation with receptor expression (eg, GR protein correlation with mRNA levels) and clinical outcomes.
Conclusion
GR expression using a validated IHC assay showed that GR is expressed in multiple different solid tumor malignancies, and may be a predictive tool to guide the clinical development of GR antagonists. Further clinical correlation will be needed to determine, within individual tumor types, what the optimal thresholds for GR expression will be for patient selection and stratification.
Acknowledgments
The authors thank Ruth Ann Gover of Corcept Therapeutics for administrative assistance in assembling the references; Grayce Fjeld of QualTek Molecular Laboratories for managing project logistics during conduct of the assays and obtaining the IHC images included in this manuscript; and Caren Rickhoff, MWC, of MedGraphica Medical Writing Services for providing medical writing and editorial assistance. This manuscript was prepared according to the International Society for Medical Publication Professionals’ “Good Publication Practice for Communicating Company-Sponsored Medical Research: the GPP3 Guidelines” and the International Committee of Medical Journal Editors’ “Uniform Requirements for Manuscripts Submitted to Biomedical Journals.” Funding for the preparation of this manuscript was provided by Corcept Therapeutics. The authors did not receive any grants in support of writing this manuscript.
Footnotes
Disclosure
TSB and DPN are employees of Corcept Therapeutics, Menlo Park, CA, USA. Corcept Therapeutics sponsored the research reported here. TIM and FJL are employees of QualTek Molecular Laboratories, contracted to perform the assays. The authors report no other conflicts of interest in this work.
References
- 1.Sapolsky RM, Romero LM, Munck AU. How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr Rev. 2000;21(1):55–89. doi: 10.1210/edrv.21.1.0389. [DOI] [PubMed] [Google Scholar]
- 2.Kadmiel M, Cidlowski JA. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol Sci. 2013;34(9):518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhen T, Cidlowski JA. Antiinflammatory aciton of glucocorticoids—New mechanisms for old drugs. N Engl J Med. 2005;353(16):1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 4.Cole TJ, Blendy JA, Monaghan AP, et al. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995;9:1608–1621. doi: 10.1101/gad.9.13.1608. [DOI] [PubMed] [Google Scholar]
- 5.Baschant U, Tuckermann J. The role of the glucocorticoid receptor in inflammation and immunity. J Steroid Biochem Mol Biol. 2010;120:69–75. doi: 10.1016/j.jsbmb.2010.03.058. [DOI] [PubMed] [Google Scholar]
- 6.Moraitis AG, Block T, Nguyen D, Belanoff JK. The role of glucocorticoid receptors in metabolic syndrome and psychiatric illness. J Steroid Biochem Mol Biol. 2017;165:114–120. doi: 10.1016/j.jsbmb.2016.03.023. [DOI] [PubMed] [Google Scholar]
- 7.Judd LL, Schettler PJ, Brown ES, et al. Adverse consequences of glucocorticoid medication: psychological, cognitive, and behavioral effects. Am J Psychiatry. 2014;171(10):1045–1052. doi: 10.1176/appi.ajp.2014.13091264. [DOI] [PubMed] [Google Scholar]
- 8.Schlossmacher G, Stevens A, White A. Glucocorticoid receptor-mediated apoptosis: mechanisms of resistance in cancer cells. J Endocrinol. 2011;211:17–25. doi: 10.1530/JOE-11-0135. [DOI] [PubMed] [Google Scholar]
- 9.Herr I, Ucur E, Herzer K, et al. Glucocorticoid cotreatment induces apoptosis resistance toward cancer therapy in carcinomas. Cancer Res. 2003;63:3112–120. [PubMed] [Google Scholar]
- 10.Schmidt S, Rainer J, Ploner C, Presul E, Riml S, Kofler R. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ. 2004;11:S45–S55. doi: 10.1038/sj.cdd.4401456. [DOI] [PubMed] [Google Scholar]
- 11.Gassler N, Zhang C, Wenger T, et al. Dexamethasone-induced cisplatin and gemcitabine resistance in lung carcinoma samples treated ex vivo. Br J Cancer. 2005;92:1084–1088. doi: 10.1038/sj.bjc.6602453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pang D, Kocherginsky M, Krausz T, Kim S-Y, Conzen SD. Dexamethasone decreases xenograft response to paclitaxel through inhibition of tumor cell apoptosis. Cancer Biol Ther. 2006;5(8):933–940. doi: 10.4161/cbt.5.8.2875. [DOI] [PubMed] [Google Scholar]
- 13.Conzen SD. Nuclear receptors and breast cancer. Mol Endocrinol. 2008;22(10):2215–2228. doi: 10.1210/me.2007-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isikbay M, Otto K, Kregel S, et al. Glucocorticoid receptor activity contributes to resistance to androgen-targeted therapy in prostate cancer. Hormone Cancer. 2014;5(2):72–89. doi: 10.1007/s12672-014-0173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Beckermann B, Kallifatidis G, et al. Corticosteroids induce chemotherapy resistance in the majority of tumour cells from bone, brain, breast, cervix, melanoma and neuroblastoma. Int J Oncol. 2006;29:1295–1301. [PubMed] [Google Scholar]
- 16.Skor MN, Wonder EL, Kocherginsky M, et al. Glucocorticoid receptor antagonism as a novel therapy for triple-negative breast cancer. Clin Cancer Res. 2013;19(22):6163–6172. doi: 10.1158/1078-0432.CCR-12-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stringer-Reasor EM, Baker GM, Skor MN, et al. Glucocorticoid receptor activation inhibits chemotherapy-induced call death in high-grade serous ovarian carcinoma. Gynecol Oncol. 2015;138:656–662. doi: 10.1016/j.ygyno.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross KL, Lu NZ, Cidlowski JA. Molecular mechanisms regulating glucocorticoid sensitivity and resistance. Mol Cell Endocrinol. 2009;300:7–16. doi: 10.1016/j.mce.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith LK, Cidlowski JA. Glucocorticoid-induced apoptosis of healthy and malignant lymphocytes. Prog Brain Res. 2010;182:1–30. doi: 10.1016/S0079-6123(10)82001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruver-Yates AL, Cidlowski JA. Tissue-specific actions of glucocorticoids on apoptosis: a double-edged sword. Cells. 2013;2:202–223. doi: 10.3390/cells2020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan D, Kocherginsky M, Conzen SD. Activation of the glucocorticoid receptor is associated with poor prognosis in estrogen receptor-negative breast cancer. Cancer Res. 2011;71(20):6360–6370. doi: 10.1158/0008-5472.CAN-11-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nanda R, Chennamaneni P, Stringer E, et al. A randomized phase I trial of nanoparticle albumin bound paclitaxel (Agraxane) with or without mifepristone for advanced breast cancer. San Antonio Breast Cancer Symposium. 2013 [Google Scholar]
- 23.Nanda R, Stringer-Reasor EM, Saha P, et al. A randomized phase I trial of nanoparticle albumin-bound paclitaxel with or without mifepristone for advanced breast cancer. Springer Plus. 2016;5:947. doi: 10.1186/s40064-016-2457-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker GM, Murphy T, Block T, Nguyen D, Lynch FJ. Development and validation of an immunohistochemistry assay to assess glucocorticoid receptor expression for clinical trials of mifepristone in breast cancer. Cancer Manag Res. 2015;7:361–368. doi: 10.2147/CMAR.S91546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. ClinicalTrials.gov [Accessed October 11, 2016]. (Study identifiers: NCT02014337, NCT01493310, NCT02788981, and NCT02046421). Available from: https://clinicaltrials.gov/
- 26.Abduljabbar R, Negm OH, Lai C-F, et al. Clinical and biological significance of glucocorticoid receptor (GR) expression in breast cancer. Breast Cancer Res Treat. 2015;150:335–346. doi: 10.1007/s10549-015-3335-1. [DOI] [PubMed] [Google Scholar]
- 27.Buxant F, Engohan-Aloghe C, Noel J-C. Estrogen receptor, progesterone receptor, and glucocorticoid receptor expression in normal breast tissue, breast in situ carcinoma, and invasive breast cancer. Appl Immunohistochem Mol Morphol. 2010;18:254–257. doi: 10.1097/PAI.0b013e3181c10180. [DOI] [PubMed] [Google Scholar]
- 28.Belova L, Delgado B, Kocherginsky M, Melhem A, Olopade OI, Conzen SD. Glucocorticoid receptor expression in breast cancer associates with older patient age. Breast Cancer Res Treat. 2009;116:444–447. doi: 10.1007/s10549-008-0136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed JA, Manahan LJ, Park C-S, Brigati DJ. Complete one-hour immunocytochemistry based on capillary action. Biotechniques. 1992;13(3):434–443. [PubMed] [Google Scholar]
- 30.Lien H-C, Lu Y-S, Shun C-T, Yao Y-T, Chang W-C, Cheng A-L. Differential expression of glucocorticoid receptor in carcinomas of the human digestive system. Histopathology. 2008;52:314–324. doi: 10.1111/j.1365-2559.2007.02953.x. [DOI] [PubMed] [Google Scholar]
- 31.Hammond MEH, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med. 2010;134:907–922. doi: 10.5858/134.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolff AC, Hammond MEH, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138(2):241–256. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]