Abstract
OBJECTIVES
Nicotine metabolism rates differ greatly among individuals, even after controlling for variation in the major nicotine metabolizing enzyme, CYP2A6. In this study, the impact of genetic variation in alternative metabolic enzymes and transporters on nicotine and cotinine pharmacokinetics and smoking was investigated.
METHODS
We examined the impact of UGT2B10, UGT2B17, FMO3, NAT1, and OCT2 variation on pharmacokinetics and smoking (total nicotine equivalents and topography), before and after stratifying by CYP2A6 genotype in 60 African American smokers who received a simultaneous intravenous infusion of deuterium-labeled nicotine and cotinine.
RESULTS
Variants in UGT2B10 and UGT2B17 were associated with urinary glucuronidation ratios (glucuronide/free substrate). UGT2B10 rs116294140 was associated with significant alterations in cotinine and modest alterations in nicotine pharmacokinetics. These alterations, however, were not sufficient to change nicotine intake or topography. Neither UGT2B10 rs61750900, UGT2B17*2, FMO3 rs2266782, nor NAT1 rs13253389 altered nicotine or cotinine pharmacokinetics among all subjects (n=60); or among individuals with reduced CYP2A6 activity (n=23). The organic cation transporter OCT2 rs316019 significantly increased nicotine and cotinine Cmax (p=0.005, p=0.02, respectively) and decreased nicotine clearance (p=0.05). UGT2B10 rs116294140 had no significant impact on the plasma or urinary trans-3’-hydroxycotinine/cotinine ratio, commonly used as a biomarker of CYP2A6 activity.
CONCLUSIONS
We demonstrated that polymorphisms in genes other than CYP2A6 represent minor sources of variation in nicotine pharmacokinetics, insufficient to alter smoking in African Americans. The change in cotinine pharmacokinetics with UGT2B10 rs116294140 highlights the UGT2B10 gene as a source of variability in cotinine as a biomarker of tobacco exposure among African American smokers.
Keywords: Nicotine, Cotinine, Pharmacokinetics, African American, Glucuronidation, UGT2B10, UGT2B17, FMO3, OCT2
Introduction
African American (AA) smokers display higher tobacco-related disease risk despite smoking on average fewer cigarettes per day compared to European Americans (EA) [1–3]. They also differ in the average rate of nicotine metabolism [4] which is an important determinant of smoking behavior [5], and smoking-induced disease risk [6].
Nicotine, the main psychoactive compound in cigarettes [7], is primarily inactivated by the liver CYP2A6 enzyme to cotinine (COT), which undergoes further CYP2A6-mediated metabolism to trans-3’-hydroxycotinine (3HC) [8]. The ratio of plasma 3HC/COT, known as the nicotine metabolite ratio (NMR), is a validated biomarker of CYP2A6 enzymatic activity [9, 10]. Large ethnic differences in the rate of nicotine and cotinine metabolism, as well as the frequency of CYP2A6 gene variants exist between African and European Americans [10–13]. CYP2A6 reduced or null-activity variants such as CYP2A6*17, *20, *23-*28, and *35 are found at a higher frequency in African compared to European Americans leading to lower nicotine clearance in African Americans [14–16]. Less is known about the contribution of nicotine metabolizing enzymes other than CYP2A6 to nicotine clearance, smoking behavior, and tobacco-induced disease risk among African Americans.
UDP-glucuronosyl transferase 2B10 (UGT2B10) catalyzes nicotine N’-glucuronidation, a pathway responsible for 3–5% of total nicotine urinary recovery [17–19], which can be as high as 40% in smokers who have deleted CYP2A6 [20]. The same is true for flavin-containing monoxygenase (FMO)-3 enzyme which catalyzes nicotine N’-oxidation, a pathway responsible for 4–7% of total nicotine urinary recovery [8], which increases in individuals with deleted CYP2A6 [21]. This suggests a potentially greater reliance on alternative nicotine metabolizing pathways mediated by UGT2B10 or FMO3 when CYP2A6 activity is reduced.
A missense mutation in UGT2B10 (rs61750900) reduces nicotine and cotinine glucuronidation metabolic ratios (most often expressed as glucuronidated metabolite/free substrate or glucuronidated metabolite/total nicotine equivalents) in European American smokers during ad libitum smoking [22, 23], or at a single time-point following oral administration of labeled nicotine-d2 [23]. Among African Americans, nicotine glucuronidation ratios are lower than in European Americans [24, 25]. Two previously identified UGT2B10 gene variants, the missense mutation (rs61750900) and a splice variant (rs116294140), lead to reductions in nicotine and cotinine glucuronidation ratios [24–27]. More recently, in a genome-wide association study (GWAS) of nicotine and cotinine glucuronidation ratios (glucuronidated metabolite/total substrate), the most significant hit on 4q13 was strongly correlated with the same UGT2B10 splice variant, while another chromosome 4q13 hit was correlated with rs61750900 [27]. Here, we aimed to assess the impact of these two variants on nicotine and cotinine pharmacokinetic parameters, including clearance. Studies have not, as yet, examined the role of these two UGT2B10 variants, or variants in FMO3, on the pharmacokinetics of intravenously administered nicotine and its primary metabolite, cotinine, among African American smokers.
We also examined the impact of variation in UGT2B17, an enzyme responsible for O’-glucuronidation of 3HC [28], and OCT2, a basolateral-type organic cation transporter which mediates tubular secretion of nicotine [29], on nicotine and cotinine pharmacokinetics among African Americans. Recently, LeMasters and colleagues [30] demonstrated a potential role for N-acetyltransferase 1 (NAT1) gene in the metabolic pathway of nicotine. Therefore, we aimed to study the impact of NAT1 polymorphisms on nicotine and cotinine pharmacokinetics. In addition, we investigated whether variation in glucuronidation, N’-oxidation, or transport pathways, which could differentially influence 3HC and COT levels, altered the ratio of 3HC/COT leading to inaccurate phenotyping of CYP2A6 activity among African Americans. Lastly, as changes in nicotine pharmacokinetics alter smoking behavior such as consumption and topography, we tested whether variation in alternative nicotine metabolism genes was sufficient to alter measures of smoking.
Methods
Sixty African American smokers completed a nicotine and cotinine pharmacokinetic study as previously described [31]. Participants’ demographic and pharmacokinetic characteristics are presented in supplementary Table S1. Briefly, subjects smoked their usual brand of cigarettes supplied by the study for 7 days prior to the start of the four-day pharmacokinetic study. They abstained from cigarettes starting at 10PM on the night prior to infusion. In a fasting condition, at about 9AM, subjects received a simultaneous thirty-minute infusion of deuterium-labeled nicotine-d2 and cotinine-d4 (2.0 µg/kg/min). Urine and blood samples were collected and plasma nicotine-d2 was assessed in samples collected at 0, 10, 20, 30, 45, 60, 90, 120, 180, 240, 360, and 480 minutes, and at 12, 16, 24, 48, and 72 hours after the infusion. Cotinine and 3HC levels (unlabeled, d2 and d4) from the 360 minute plasma sample were used for the determination of the 3HC/COT ratio [9]. This study was approved by the Institutional Review Board at the University of Toronto and the University of California San Francisco.
Participants were previously genotyped for CYP2A6 reduced or null activity variants found widely among different ethnic groups, CYP2A6*2, *4, *9, and *12, as well as those found predominantly in African Americans, CYP2A6*17, *20, *23-*28, *31, and *35 [31–33]. Individuals with no identified CYP2A6 gene variants were grouped as wild type (*1/*1). To examine the contribution of pathways other than CYP2A6-mediated C-oxidation to nicotine metabolism, those having at least one CYP2A6 genetic variant with reduced or null activity were considered to have reduced CYP2A6 activity (n=23) which included those with CYP2A6*1/*2 (n=1), *1/*4H (n=2), *1/*9 (n=2), *1/*17 (n=8), *1/*24 (n=1), *1/*26 (n=1), *1/*27 (n=1), *1/*35 (n=3), *4/*17 (n=1), *9/*26 (n=1), *17/*31 (n=1), *17/*35 (n=1) genotypes, as previously described [31, 34, 35].
Genotyping for UGT2B10 splice variant (rs116294140) and non-synonymous SNP (rs61750900), UGT2B17 deletion variant (*2), FMO3 non-synonymous SNP (rs2266782), NAT1 intronic SNP (rs13253389), and OCT2 non-synonymous SNP (rs316019) were performed using TaqMan SNP genotyping assays from Applied Biosystems, Foster City, USA.
Plasma nicotine and cotinine concentrations were measured by gas chromatography-mass spectrometry [36]. 3HC concentration in plasma and nicotine, cotinine, 3HC, and nicotine N-oxide concentrations in urine were determined by liquid chromatography-tandem mass spectrometry [9]. Concentrations of nicotine, cotinine, and 3HC glucuronides in urine were measured as the difference in analyte concentrations before (free) and after (total) enzymatic hydrolysis, as described previously by Benowitz et al. [37]. The limits of quantification for nicotine, cotinine, and 3HC in plasma were 0.1 ng/ml. The limits of quantification for the urine analytes were 10 ng/ml for nicotine, cotinine, 3HC, and nicotine N-oxide and 1 ng/ml for minor species (nornicotine, norcotinine, cotinine N-oxide).
Model independent methods were used to estimate pharmacokinetic parameters using blood concentration and urinary nicotine and metabolite data [38]. Nicotine clearance was computed as: CLNIC= [DoseNIC-d2]/ [AUCNIC-d2], where Dose is the dose of nicotine-d2 infused and AUC is the area under the plasma nicotine-d2 concentration time curve extrapolated to infinity. Cotinine clearance was computed the same way using cotinine-d4 dose and AUC [39]. Glucuronidation and N’-oxidation ratios were calculated using the ratio of urinary metabolites to the free parent compound excreted.
Pharmacokinetic means and urinary glucuronidation and N’-oxidation ratios were compared using non-parametric Mann-Whitney test and Kruskal-Wallis test between genotype groups. The Bonferroni method was used to correct for multiple testing across five gene variants using a significance threshold of 0.01. Standard regression modeling, assuming a dominant effect of the minor allele, was used to determine the percentage of variance in the pharmacokinetic parameters explained by each gene variant. Considering that d2- and d4-lableled cotinine and 3HC were not in steady state, the unlabeled plasma 3HC/COT ratio measured at 360 minute time point was used as a biomarker of CYP2A6 activity in the regression models. Total nicotine equivalents (TNE) was calculated based on the creatinine-corrected molar sum of all unlabeled nicotine metabolites excreted in a 24-hour urine collection during ad libitum smoking. Urinary nicotine metabolite ratios were also calculated based on unlabeled metabolites excreted during this 24-hr urine collection. Percent variance in the pharmacokinetic parameter explained by the genotype or phenotype is calculated based on the square value of the part correlation coefficients.
Results
UGT2B10 rs116294140 and rs61750900 allele frequencies were 38.3% and 3.3%, respectively. UGT2B17*2, FMO3 rs2266782, NAT1 rs13253389, and OCT2 rs316019 allele frequencies were 31.7%, 39.2%, 32.5%, and 7.5%, respectively. All variants examined were reduced or null-function variants as described in supplementary Table S2. All frequencies were in Hardy Weinberg equilibrium and consistent with previous reports in African Americans [40–42].
Effect of UGT2B10 and UGT2B17 genotype on nicotine and cotinine glucuronidation ratios
We examined the effect of UGT2B10 and UGT2B17 variation on urinary glucuronidation phenotypes using the ratio of nicotine glucuronide to free nicotine or cotinine glucuronide to free cotinine as phenotypic measures of UGT2B10 activity, and the ratio of 3HC glucuronide to free 3HC as a phenotypic measure of UGT2B17 activity. The impact of functional polymorphisms in UGT2B10 (rs61750900) and UGT2B17 (*2) on nicotine, cotinine, and 3HC glucuronidation ratios has previously been demonstrated in human liver microsomes or urine of smokers [19, 22–25, 27, 43], and are replicated here as positive controls for the impact of UGT2B10 and UGT2B17 variation on nicotine and cotinine pharmacokinetics which has not been tested previously.
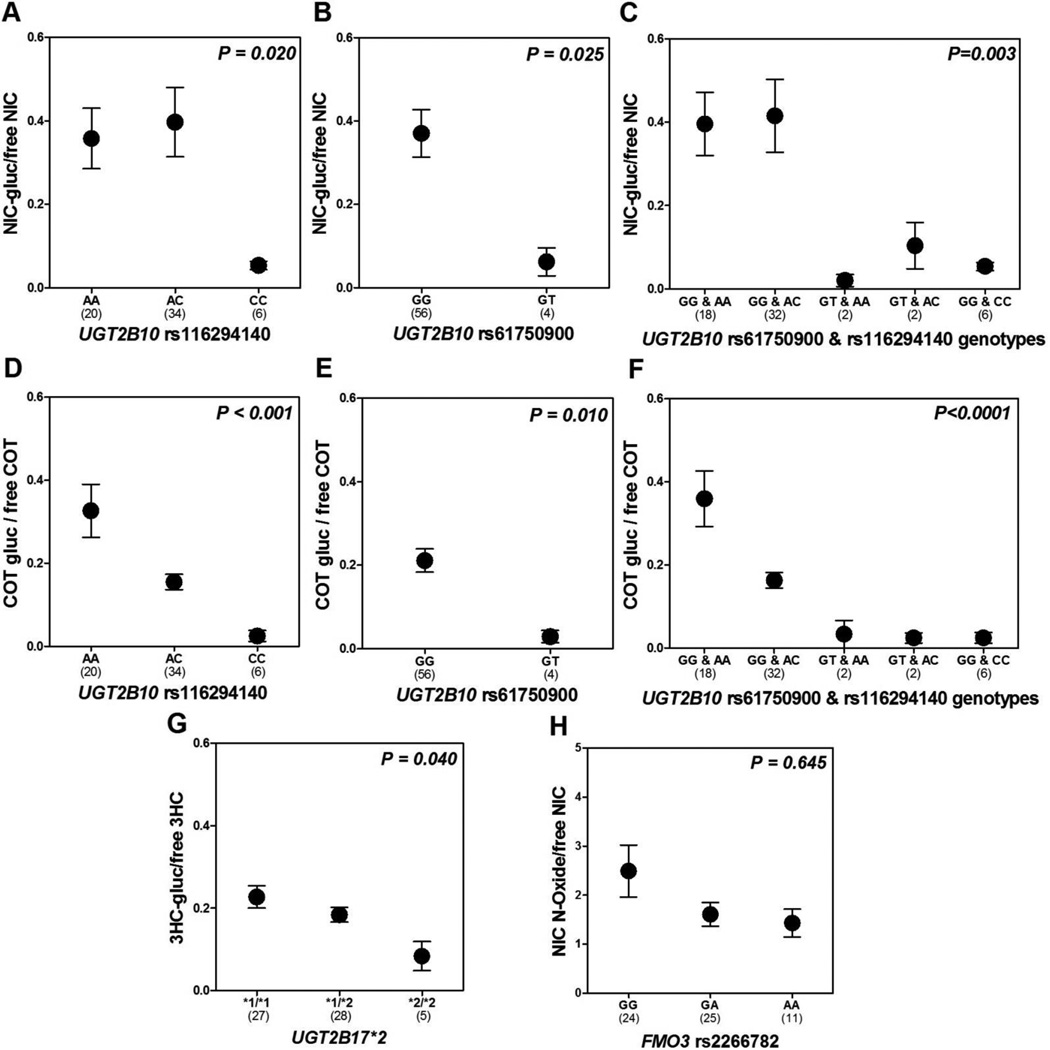
The primary role of UGT2B10 as the catalyst of nicotine and cotinine N’-glucuronidation was confirmed by the effect of UGT2B10 rs116294140 and rs61750900 on both nicotine and cotinine glucuronide ratios (Figure 1). Individuals homozygous for rs116294140 and heterozygous for rs61750900 demonstrated significantly lower nicotine and cotinine glucuronide ratios compared to wild types (Figure 1). Similarly, the 3HC glucuronide ratio was significantly lower in individuals with the variant genotypes (Figure 1).
Figure 1.
Mean ± SEM nicotine-glucuronide/free nicotine and cotinine-glucuronide/free cotinine ratios according to UGT2B10 rs116294140 genotype (A&D) and according to UGT2B10 61750900 genotype (B&E) is shown; Mean ± SEM 3HC-glucuronide/free 3HC ratio according to UGT2B17*2 genotype is shown in (G); Mean ± SEM nicotine N-oxide/free nicotine ratio according to FMO3 rs2266782 genotype is shown in (H). UGT2B10 rs116294140 and rs61750900 genotypes were combined in (C&F). Group 1 includes individuals who are wild type for both variants (n=18); group 2 includes individuals who are wild type for rs61750900 and heterozygous variant for rs116294140 (n=32); group 3 includes individuals who are heterozygous variant for rs61750900 and wild type for rs116294140 (n=2); group 4 includes individuals who are heterozygous for both variants (n=2); group 5 includes individuals who are wild type for rs61750900 and homozygous variant for rs116294140 (n=6). All P-values indicated were derived from Mann-Whitney U tests comparing UGT2B10 rs61750900 GG and GT genotypes, or Kruskal-Wallis tests comparing UGT2B10 rs116294140, UGT2B17*2, FMO3 rs2266782, and UGT2B10 rs116294140 and rs61750900 combined genotypes. Abbreviation: SEM, standard error of the mean.
Effect of UGT2B10 and UGT2B17 genotype on nicotine and cotinine disposition kinetics
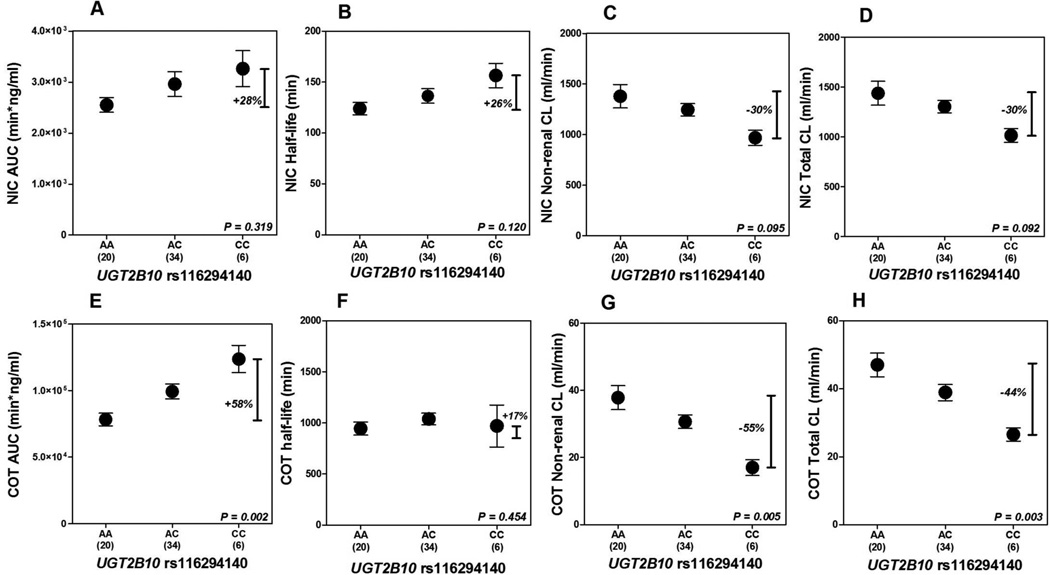
We examined the association between two functional polymorphisms in UGT2B10 and nicotine and cotinine pharmacokinetics. Although not statistically significant, among all subjects (n=60), we observed a small increase in nicotine half-life and a corresponding decrease in nicotine non-renal and total clearance in homozygous carriers of UGT2B10 rs116294140 (CC genotype) compared to wild types (AA genotype) (Table 1, Figure 2). Among individuals with reduced CYP2A6 activity (n=23), there was a non-significant increase in nicotine half-life and a decrease in nicotine non-renal and total clearance in homozygous carriers of UGT2B10 rs116294140 compared to wild types (Table 2). We found no association between UGT2B10 rs61750900 and nicotine pharmacokinetics among all subjects, or among individuals with reduced CYP2A6 activity (Table 1, Table 2).
Table 1.
Nicotine pharmacokinetic parameters by UGT2B10, FMO3 and OCT2 genotypes
| Genotype | Half-lifec | Cmaxc | AUCc | Non-renal CLc | Total CLc | |
|---|---|---|---|---|---|---|
| Meanb (SD) | Meanb (SD) | Meanb (SD) | Meanb (SD) | Meanb (SD) | ||
| UGT2B10 | AA (20)d | 124 (27) | 21.1 (7.4) | 2.55 (0.64) | 1.38 (0.52) | 1.44 (0.54) |
| (rs116294140) | AC (34) | 136 (42) | 24.0 (22.4) | 2.96 (1.41) | 1.24 (0.35) | 1.30 (0.37) |
| CC (6) | 156 (29) | 32.2 (19.7) | 3.26 (0.87) | 0.97 (0.18) | 1.01 (0.17) | |
| P-value | 0.12a | 0.29a | 0.32a | 0.09a | 0.09a | |
| UGT2B10 | GG (56) | 133 (38) | 24.0 (18.9) | 2.86 (1.20) | 1.26 (0.42) | 1.32 (0.43) |
| (rs61750900) | GT (4) | 149 (8) | 21.8 (9.8) | 2.81 (0.45) | 1.22 (0.45) | 1.26 (0.42) |
| TT (0) | NA | NA | NA | NA | NA | |
| P-value | 0.34a | 0.98a | 0.65a | 0.99a | 0.82a | |
| FMO3 | GG (24) | 143 (38) | 22.9 (12.3) | 2.93 (0.90) | 1.21 (0.41) | 1.26 (0.41) |
| (rs2266782) | GA (25) | 125 (33) | 24.2 (25.5) | 2.74 (1.40) | 1.39 (0.46) | 1.46 (0.49) |
| AA (11) | 137 (42) | 25.2 (9.6) | 2.94 (1.18) | 1.08 (0.19) | 1.13 (0.19) | |
| P-value | 0.34a | 0.19a | 0.27a | 0.05a | 0.05a | |
| OCT2 | CC (51) | 135 (37) | 20.3 (7.5) | 2.74 (0.87) | 58.2 (57.8)e | 1.36 (0.43) |
| (rs316019) | CA (9) | 127 (40) | 44.0 (40.2) | 3.50 (2.17) | 43.8 (20.7)e | 1.07 (0.31) |
| AA (0) | NA | NA | NA | NA | NA | |
| P-value | 0.53a | p<0.01a | 0.49a | 0.77a | 0.05a | |
P-values are based on Mann-Whitney test or Kruskal-Wallis test with Bonferroni correction for multiple testing.
Arithmetic means are presented.
Nicotine (d2) half-life is measured in min, Cmax in ng/ml, AUC in min*µg/ml, and non-renal and total clearance in L/min.
Genotype frequencies are presented in the brackets.
OCT2 variation was examined with respect to nicotine renal as opposed to non-renal clearance (ml/min).
Abbreviation: SD, standard deviation; AUC, area under the curve; CL, clearance.
Figure 2.
Mean ± SEM nicotine (A–D) and cotinine (E–H) AUC, half-life, non-renal, and total clearance according to UGT2B10 rs116294140 genotype is shown; All P-values indicated were derived from Kruskal-Wallis tests comparing UGT2B10 rs116294140 genotypes. Δ (% change) is calculated based on the value of (CC genotype – AA genotype)/AA genotype. Abbreviation: NIC, nicotine; AUC, area under the curve; CL, clearance; COT, cotinine; SEM, standard error of the mean.
Table 2.
Nicotine pharmacokinetic parameters by UGT2B10 FMO3 and OCT2 genotypes among individuals with reduced CYP2A6 activity (n=23)
| Genotype | Half-lifec | Cmaxc | AUCc | Non-renal CLc | Total CLc | |
|---|---|---|---|---|---|---|
| Meanb (SD) | Meanb (SD) | Meanb (SD) | Meanb (SD) | Meanb (SD) | ||
| UGT2B10 | AA (6)d | 130 (29) | 21.4 (5.6) | 2.86 (0.57) | 1.09 (0.32) | 1.16 (0.33) |
| (rs116294140) | AC (12) | 157 (47) | 18.8 (7.5) | 3.19 (1.29) | 1.27 (0.43) | 1.31 (0.44) |
| CC (5) | 156 (32) | 34.3 (21.3) | 3.36 (0.93) | 0.99 (0.20) | 1.04 (0.18) | |
| P-value | 0.49a | 0.14a | 0.76a | 0.22a | 0.32a | |
| UGT2B10 | GG (20) | 150 (43) | 23.6 (13.1) | 3.21 (1.10) | 1.14 (0.36) | 1.19 (0.37) |
| (rs61750900) | GT (3) | 149 (9) | 17.7 (6.7) | 2.70 (0.48) | 1.33 (0.48) | 1.36 (0.44) |
| TT (0) | NA | NA | NA | NA | NA | |
| P-value | 0.93a | 0.39a | 0.41a | 0.41a | 0.52a | |
| FMO3 | GG (4) | 157 (50) | 27.0 (8.5) | 3.73 (1.50) | 0.98 (0.23) | 1.01 (0.24) |
| (rs2266782) | GA (9) | 139 (32) | 19.2 (5.5) | 2.62 (0.43) | 1.33 (0.27) | 1.38 (0.29) |
| AA (10) | 156 (45) | 24.5 (17.5) | 3.38 (1.12) | 1.09 (0.45) | 1.13 (0.44) | |
| P-value | 0.59a | 0.34a | 0.10a | 0.13a | 0.11a | |
| OCT2 | CC (19) | 147 (43) | 20.6 (8.3) | 3.13 (1.10) | 43.0 (38.4)e | 1.24 (0.39) |
| (rs316019) | CA (4) | 162 (20) | 33.7 (23.3) | 3.23 (0.84) | 60.0 (15.1)e | 1.07 (0.30) |
| AA (0) | NA | NA | NA | NA | NA | |
| P-value | 0.29a | 0.12a | 0.57a | 0.11a | 0.33a | |
P-values are based on Mann-Whitney test or Kruskal-Wallis test with Bonferroni correction for multiple testing.
Arithmetic means are presented.
Nicotine (d2) half-life is measured in min, Cmax in ng/ml, AUC in min*µg/ml, and non-renal and total clearance in L/min.
Genotype frequencies are presented in the brackets.
OCT2 variation was examined with respect to nicotine renal as opposed to non-renal clearance (ml/min).
Abbreviation: SD, standard deviation; AUC, area under the curve; CL, clearance.
Cotinine pharmacokinetics were substantially altered by UGT2B10 rs116294140: individuals who were homozygous for rs116294140 had 58% higher cotinine AUC, 55% lower cotinine non-renal clearance and 44% lower cotinine total clearance (Table 3, Figure 2). No other associations between UGT2B10 rs61750900 and UGT2B17*2 and cotinine pharmacokinetics were observed (Table 3).
Table 3.
Cotinine (d4) pharmacokinetic parameters by UGT2B10, UGT2B17, FMO3 and OCT2 genotypes
| Genotype | Half-lifec | Cmaxc | AUCc | Non-renal CLc | Total CLc | |
|---|---|---|---|---|---|---|
| Meanb (SD) | Meanb (SD) | Meanb (SD) | Meanb (SD) | Meanb (SD) | ||
| UGT2B10 | AA (20)d | 994 (181) | 81.3 (23.1) | 78.3 (20.7) | 37.8 (15.6) | 47.0 (15.6) |
| (rs116294140) | AC (34) | 1070 (285) | 85.1 (21.4) | 99.3 (32.1) | 30.6 (11.5) | 38.9 (13.8) |
| CC (6) | 1165 (185) | 99.5 (27.7) | 124.0 (22.8) | 17.0 (5.4) | 26.5 (4.7) | |
| P-value | 0.24a | 0.26a | p<0.01a | p<0.01a | p<0.01a | |
| UGT2B10 | GG (56) | 1051 (255) | 85.1 (23.1) | 93.5 (31.5) | 32.2 (14.1) | 41.0 (15.2) |
| (rs61750900) | GT (4) | 1075 (164) | 84.9 (18.2) | 107.0 (11.9) | 26.6 (5.6) | 31.7 (5.0) |
| TT (0) | NA | NA | NA | NA | NA | |
| P-value | 0.61a | 0.96a | 0.22a | 0.52a | 0.16a | |
| UGT2B17 | *1/*1 (27) | 1050 (251) | 89.1 (24.1) | 95.8 (29.9) | 30.5 (13.5) | 40.6 (15.4) |
| (*2) | *1/*2 (28) | 1036 (223) | 79.6 (19.6) | 90.0 (26.4) | 33.5 (14.5) | 41.5 (15.0) |
| *2/*2 (5) | 1153 (389) | 95.1 (27.6) | 111.0 (52.8) | 28.1 (11.4) | 32.5 (11.8) | |
| P-value | 0.81a | 0.31a | 0.65a | 0.80a | 0.53a | |
| FMO3 | GG (24) | 1067 (263) | 87.6 (24.3) | 105.0 (33.2) | 28.8 (12.0) | 36.9 (15.3) |
| (rs2266782) | GA (25) | 1047 (192) | 83.1 (21.1) | 88.1 (25.7) | 35.0 (14.1) | 44.0 (13.4) |
| AA (11) | 1031 (347) | 84.2 (23.9) | 85.2 (31.4) | 31.1 (16.2) | 39.7 (16.7) | |
| P-value | 0.89a | 0.51a | 0.11a | 0.30a | 0.09a | |
| OCT2 | CC (51) | 1073 (242) | 82.5 (22.4) | 95.3 (30.6) | 8.16 (3.72)e | 40.3 (14.5) |
| (rs316019) | CA (9) | 943 (272) | 99.1 (19.1) | 90.0 (32.7) | 6.46 (2.99)e | 40.8 (18.1) |
| AA (0) | NA | NA | NA | NA | NA | |
| P-value | 0.25a | 0.02a | 0.99a | 0.21a | 0.82a | |
P-values are based on Mann-Whitney test or Kruskal-Wallis test with Bonferroni correction for multiple testing.
Arithmetic means are presented.
Cotinine (d4) half-life is measured in min, Cmax in ng/ml, AUC in min*µg/ml, and non-renal and total clearance in ml/min.
Genotype frequencies are presented in the brackets.
OCT2 variation was examined with respect to cotinine renal as opposed to non-renal clearance (ml/min).
Abbreviation: SD, standard deviation; AUC, area under the curve; CL, clearance.
Using standard multiple regression models, with the number of minor allele copies entered as the predictor, we found that UGT2B10 rs116294140 explained ~4% of variance in nicotine pharmacokinetics after controlling for variation in CYP2A6, FMO3, OCT2, and UGT2B10 rs61750900. UGT2B10 rs116294140 and rs61750900 together accounted for 5–6% of variation in nicotine pharmacokinetics (Table 4). The contribution of UGT2B10 rs116294140 to cotinine pharmacokinetics, however, was more substantial, explaining up to 14% of variance in cotinine AUC, non-renal, and total clearance after taking into account variation in CYP2A6, FMO3, OCT2, and UGT2B10 rs61750900 (Table 5). As expected (Tables 1–3), neither UGT2B10 rs61750900 nor UGT2B17*2 made significant contributions to nicotine or cotinine pharmacokinetics, respectively; less than 3% of the variance in nicotine or cotinine pharmacokinetics was explained by UGT2B10 rs61750900 or UGT2B17*2 (Table 4, Table 5). Of note, findings from standard multiple regression models including or excluding the unlabeled plasma 3HC/COT ratio were similar.
Table 4.
Determinants of nicotine disposition kinetics
| Gene SNP/Variant | Alleles | MAF | P-Value% | Variance explained |
|---|---|---|---|---|
| Nicotine half-life | ||||
| 3HC/COT (d0) 360 min | p<0.0001 | 37.2% | ||
| UGT2B10 rs116294140 | A:C | 0.38 | 0.09 | 2.9% |
| UGT2B10 rs61750900 | G:T | 0.03 | 0.38 | 0.8% |
| FMO3 rs2266782 | G:A | 0.39 | 0.29 | 1.1% |
| OCT2 rs316019 | C:A | 0.07 | 0.54 | 0.4% |
| Nicotine Cmax | ||||
| 3HC/COT (d0) 360 min | 0.84 | 0.1% | ||
| UGT2B10 rs116294140 | A:C | 0.38 | 0.35 | 1.3% |
| UGT2B10 rs61750900 | G:T | 0.03 | 0.68 | 0.3% |
| FMO3 rs2266782 | G:A | 0.39 | 0.97 | 0.0% |
| OCT2 rs316019 | C:A | 0.07 | 0.001 | 18.0% |
| Nicotine AUC | ||||
| 3HC/COT (d0) 360 min | 0.005 | 12.5% | ||
| UGT2B10 rs116294140 | A:C | 0.38 | 0.34 | 1.4% |
| UGT2B10 rs61750900 | G:T | 0.03 | 0.74 | 0.2% |
| FMO3 rs2266782 | G:A | 0.39 | 0.70 | 0.2% |
| OCT2 rs316019 | C:A | 0.07 | 0.01 | 9.4% |
| Nicotine non-renal CL | ||||
| 3HC/COT (d0) 360 min | 0.003 | 13.5% | ||
| UGT2B10 rs116294140 | A:C | 0.38 | 0.09 | 4.2% |
| UGT2B10 rs61750900 | G:T | 0.03 | 0.99 | 0.0% |
| FMO3 rs2266782 | G:A | 0.39 | 0.97 | 0.0% |
| OCT2 rs316019 | C:A | 0.07 | 0.01 | 8.8% |
| Nicotine total CL | ||||
| 3HC/COT (d0) 360 min | 0.002 | 14.1% | ||
| UGT2B10 rs116294140 | A:C | 0.38 | 0.09 | 4.0% |
| UGT2B10 rs61750900 | G:T | 0.03 | 0.93 | 0.0% |
| FMO3 rs2266782 | G:A | 0.39 | 0.98 | 0.0% |
| OCT2 rs316019 | C:A | 0.07 | 0.01 | 9.1% |
Alleles indicated as major:minor with impaired allele in bolded italics. P-value and percentage of variance in the pharmacokinetic parameters explained by the variables are from standard multiple regression modelling assuming a dominant effect of the minor. % Variance explained is calculated based on the square value of the part correlation coefficients. Abbreviation: MAF, minor allele frequency; CL, clearance.
Table 5.
Determinants of cotinine disposition kinetics
| Gene SNP/Variant | Alleles | MAF | P-Value | % Variance explained |
|---|---|---|---|---|
| Cotinine half-life | ||||
| 3HC/COT (d0) 360 min | p<0.0001 | 30.9% | ||
| UGT2B10 rs116294140 | A:C | 0.38 | 0.78 | 0.1% |
| UGT2B10 rs61750900 | G:T | 0.03 | 0.76 | 0.1% |
| UGT2B17*2 | 1:0 | 0.32 | 0.33 | 1.2% |
| FMO3 rs2266782 | G:A | 0.39 | 0.42 | 0.8% |
| OCT2 rs316019 | C:A | 0.07 | 0.48 | 0.6% |
| Cotinine Cmax | ||||
| 3HC/COT (d0) 360 min | 0.22 | 2.6% | ||
| UGT2B10 rs116294140 | A:C | 0.38 | 0.80 | 0.1% |
| UGT2B10 rs61750900 | G:T | 0.03 | 0.95 | 0.0% |
| UGT2B17*2 | 1:0 | 0.32 | 0.72 | 0.2% |
| FMO3 rs2266782 | G:A | 0.39 | 0.30 | 1.8% |
| OCT2 rs316019 | C:A | 0.07 | 0.02 | 9.4% |
| Cotinine AUC | ||||
| 3HC/COT (d0) 360 min | p<0.0001 | 24.0% | ||
| UGT2B10 rs116294140 | A:C | 0.38 | p<0.0001 | 13.8% |
| UGT2B10 rs61750900 | G:T | 0.03 | 0.13 | 2.2% |
| UGT2B17*2 | 1:0 | 0.32 | 0.06 | 3.5% |
| FMO3 rs2266782 | G:A | 0.39 | 0.06 | 3.5% |
| OCT2 rs316019 | C:A | 0.07 | 0.45 | 0.5% |
| Cotinine non-renal CL | ||||
| 3HC/COT (d0) 360 min | p<0.0001 | 35.9% | ||
| UGT2B10 rs116294140 | A:C | 0.38 | 0.001 | 10.7% |
| UGT2B10 rs61750900 | G:T | 0.03 | 0.17 | 1.6% |
| UGT2B17*2 | 1:0 | 0.32 | 0.24 | 1.2% |
| FMO3 rs2266782 | G:A | 0.39 | 0.74 | 0.1% |
| OCT2 rs316019 | C:A | 0.07 | 0.45 | 0.5% |
| Cotinine total CL | ||||
| 3HC/COT (d0) 360 min | p<0.0001 | 35.6% | ||
| UGT2B10 rs116294140 | A:C | 0.38 | p<0.0001 | 12.7% |
| UGT2B10 rs61750900 | G:T | 0.03 | 0.05 | 3.0% |
| UGT2B17*2 | 1:0 | 0.32 | 0.02 | 4.3% |
| FMO3 rs2266782 | G:A | 0.39 | 0.36 | 0.7% |
| OCT2 rs316019 | C:A | 0.07 | 0.18 | 1.3% |
UGT2B17 0 allele: deletion allele. Alleles indicated as major:minor with impaired allele in bolded italics. P-value and percentage of variance in the pharmacokinetic parameters explained by the variables are from standard multiple regression modelling assuming a dominant effect of the minor allele. % Variance explained is calculated based on the square value of the part correlation coefficients. Abbreviation: MAF, minor allele frequency; CL, clearance.
Effect of FMO3 and NAT1 genotype on nicotine and cotinine pharmacokinetics
To examine the effect of FMO3 variation on the N’-oxidation phenotype, we used the urinary ratio of nicotine N-oxide to free nicotine as a measure of FMO3 activity. The FMO3 rs2266782 AA and GA genotypes were associated, albeit non-significantly, with lower nicotine N-oxide to free nicotine ratio (36–44% lower versus wild type GG genotype) (Figure 1). There was a 10% reduction in nicotine non-renal and total clearance in homozygous variant individuals (AA genotypes) compared to wild types (GG genotypes). The impact on other nicotine (Table 1) and cotinine (Table 3) pharmacokinetic parameters was negligible in all subjects; or in individuals with reduced CYP2A6 activity (Table 2). Regression analyses confirmed that the contribution of FMO3 rs2266782 to variance in nicotine (Table 4) or cotinine (Table 5) pharmacokinetics is negligible among African Americans, with <3.0% of variance in any pharmacokinetic parameter explained by the FMO3 rs2266782 genotype.
Furthermore, in contrast to LeMasters et al. findings [30], we found no impact of NAT1 rs13253389 polymorphism on total cotinine levels, or any of cotinine pharmacokinetic parameters (Supplementary Table S3).
Effect of OCT2 genotype on nicotine and cotinine pharmacokinetics
Next, we examined the association between the organic cation transporter, OCT2, genotype and nicotine and cotinine pharmacokinetics. The OCT2 CA genotype was associated with a doubling in nicotine Cmax (Table 1) and a 20% increase in cotinine Cmax compared to CC genotype (Table 3). There was also a trend towards lower nicotine total clearance, but not renal clearance, among individuals with the CA genotype compared to wild types. OCT2 genotype explained 18% of variance in nicotine Cmax, 9% of variance in nicotine total clearance, and an additional 9% of variance in cotinine Cmax (Table 4, Table 5).
Effect of variation in UGT2B10, UGT2B17, and FMO3 on the plasma and urinary 3HC/COT ratio
Given that African Americans have lower 3HC/COT and glucuronidation metabolic ratios compared to European Americans [24, 25], we examined whether the presence of UGT and FMO3 gene variants alters 3HC/COT in this population. We calculated plasma 3HC/COT ratio at 360 minutes based on the metabolites generated from tobacco use (unlabeled or d0) as well as d2-, and d4-labeled compounds. The unlabeled, d2-, and d4-labeled ratios were highly correlated with each other (p<0.001) with a Pearson’s partial coefficient, r, of 0.92 (d2- vs. d4-), 0.88 (d0- vs. d2-), and 0.90 (d0- vs. d4-). Albeit non-significantly, UGT2B10 rs116294140 was associated with a reduction in unlabeled plasma 3HC/COT ratio and in d2- and d4- labeled ratios, but no impact on the urinary ratios (Table 6). UGT2B17 and FMO3 did not significantly alter the unlabeled, d2-, d4-lableled, or urinary 3HC/COT ratios (Table 6).
Table 6.
Plasma and urinary nicotine metabolite ratio by UGT2B10, UGT2B17 and FMO3 genotypes
| Genotype | Plasma 3HC/COT (d0)d |
Plasma 3HC/COT (d2)d |
Plasma 3HC/COT (d4)d |
Urinary total 3HC/free COT |
Urinary total 3HC/total COT |
|
|---|---|---|---|---|---|---|
| Meanb (SD) | Meanb (SD) | Meanb (SD) | Meanb (SD) | Meanb (SD) | ||
| UGT2B10 | AA (20)c | 0.31 (0.13) | 0.18 (0.09) | 0.18 (0.08) | 0.98 (0.88) | 0.68 (0.43) |
| (rs116294140) | AC (34) | 0.31 (0.20) | 0.17 (0.12) | 0.16 (0.11) | 0.87 (0.10) | 0.75 (0.50) |
| CC (6) | 0.20 (0.08) | 0.11 (0.05) | 0.10 (0.03) | 0.82 (0.21) | 0.80 (0.50) | |
| P-value | 0.22a | 0.23a | 0.06a | 0.99 | 0.77 | |
| UGT2B10 | GG (56) | 0.30 (0.17) | 0.17 (0.11) | 0.17 (0.10) | 0.91 (0.70) | 0.73 (0.47) |
| (rs61750900) | GT (4) | 0.27 (0.09) | 0.11 (0.03) | 0.12 (0.04) | 0.85 (0.56) | 0.82 (0.54) |
| TT (0) | NA | NA | NA | NA | NA | |
| P-value | 0.78a | 0.12a | 0.40a | 0.91 | 0.73 | |
| UGT2B17 | *1/*1 (27) | 0.31 (0.21) | 0.17 (0.14) | 0.17 (0.12) | 0.82 (0.59) | 0.73 (0.53) |
| (*2) | *1/*2 (28) | 0.29 (0.12) | 0.16 (0.78) | 0.16 (0.08) | 0.97 (0.74) | 0.75 (0.44) |
| *2/*2 (5) | 0.32 (0.16) | 0.15 (0.07) | 0.15 (0.06) | 0.92 (0.97) | 0.61 (0.43) | |
| P-value | 0.80a | 0.96a | 0.97a | 0.62 | 0.71 | |
| FMO3 | GG (24) | 0.29 (0.23) | 0.18 (0.15) | 0.16 (0.14) | 0.78 (0.69) | 0.73 (0.43) |
| (rs2266782) | GA (25) | 0.31 (0.15) | 0.17 (0.11) | 0.18 (0.08) | 0.94 (0.75) | 0.73 (0.43) |
| AA (11) | 0.28 (0.15) | 0.15 (0.08) | 0.14 (0.07) | 0.92 (0.64) | 0.74 (0.67) | |
| P-value | 0.58a | 0.77a | 0.17a | 0.64 | 0.85 | |
P-values are based on Mann-Whitney test or Kruskal-Wallis test with Bonferroni correction for multiple testing.
Arithmetic means are presented.
Genotype frequencies are presented in the brackets.
Plasma 3HC/COT ratio at 360 minutes based on the metabolites generated from tobacco use i.e. unlabeled (d0) as well as d2-, and d4-labeled compounds are presented.
Abbreviation: 3HC, 3-hydroxycotinine; COT, cotinine; SD, standard deviation.
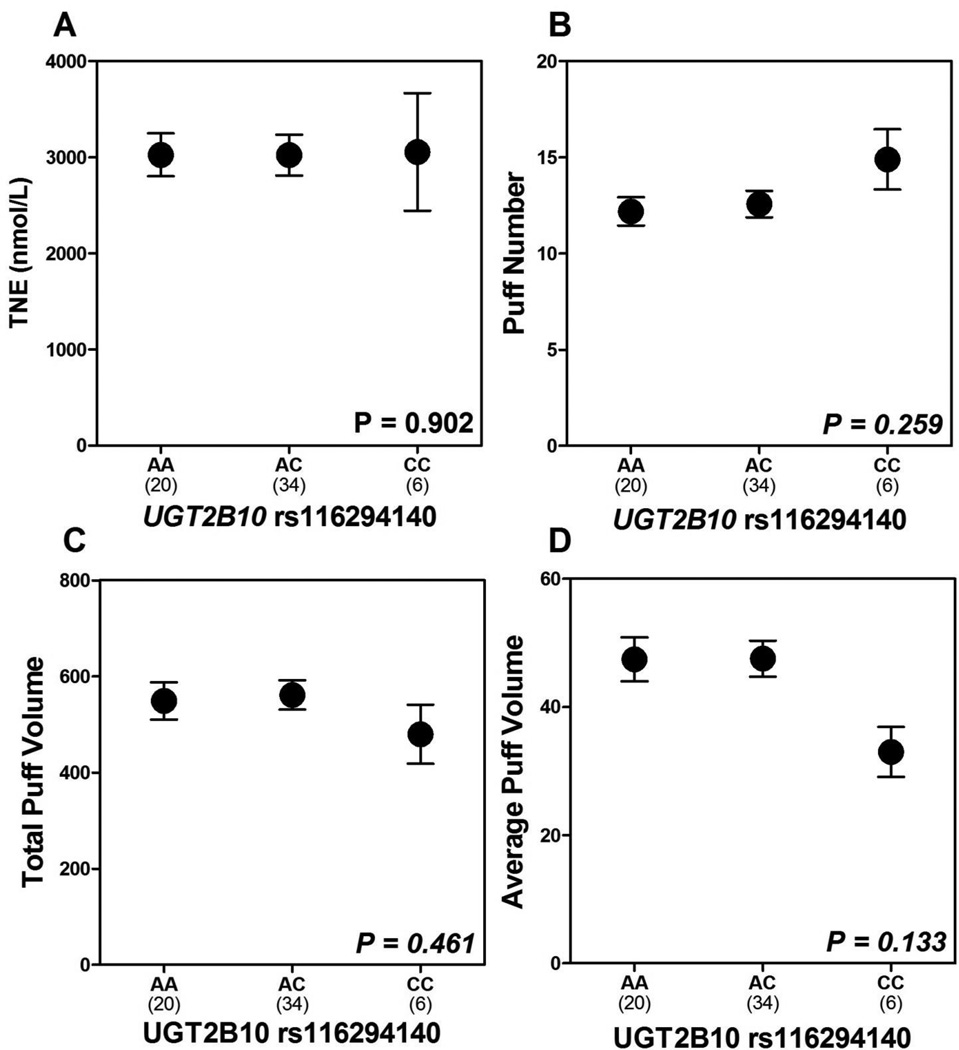
Effect of UGT2B10 rs116294140 on biomarkers of smoking behavior
Based on the effect of UGT2B10 rs116294140 genotype on nicotine clearance, albeit modest, which has been previously shown to affect smoking, we investigated its impact on biomarkers of tobacco consumption and smoking topography (i.e. urine total nicotine equivalents (TNE), number of puffs, average puff volume, and total puff volume). TNE is an excellent biomarker of daily nicotine uptake and total tobacco smoke exposure [8, 44] and is calculated as the molar sum of urinary concentration of nicotine and all of its metabolites. Consistent with the modest, non-significant reduction in nicotine clearance, we found no significant impact of UGT2B10 rs116294140 on TNE or on smoking topography variables (Figure 3) suggesting the changes in nicotine clearance with UGT2B10 rs116294140 are not sufficient to result in altered smoking behavior among African American smokers. This is consistent with findings from a GWAS study where genetic variation in nicotine and cotinine glucuronidation ratios was not significantly associated with TNE [27].
Figure 3.
Mean ± SEM of TNE (nmol/L), puff number, total puff volume (ml), and average puff volume (ml) according to UGT2B10 rs116294140 genotype (A–D) is shown. Mean TNE (3026 nmol/L AA, 3023 nmol/L AC, 3057 nmol/L CC), mean puff number (12 AA, 13 AC, 15 CC), mean total puff volume (549 ml AA, 562 ml AC, 479 CC), mean average puff volume (47.4 ml AA, 47.5 AC, 33.0 CC). All P-values indicated were derived from Kruskal-Wallis tests comparing UGT2B10 rs116294140 genotypes. Abbreviation: TNE, total nicotine equivalents; SEM, standard error of the mean.
Discussion
We present data on the functional significance of genetic variation in nicotine metabolism pathways other than C-oxidation, such as glucuronidation and N’-oxidation, on nicotine and cotinine pharmacokinetics in African Americans where these variants are prevalent. Variants in the main nicotine metabolizing gene, CYP2A6, alter nicotine pharmacokinetics in European Americans [34], and African Americans [39]. African Americans have a higher prevalence of low frequency reduced and loss of function CYP2A6 alleles, suggesting variation in alternative nicotine metabolism pathways might play a larger role in altering nicotine pharmacokinetics among African Americans. Overall, we found a potential influence of variation in UGT2B10-mediated glucuronidation, and no influence of variation in FMO-3-mediated N-oxidation pathways on nicotine pharmacokinetics among African Americans, and a substantial influence of variation in UGT2B10-mediated glucuronidation on cotinine pharmacokinetics.
The most common UGT2B10 SNP in African Americans is a splice variant (rs116294140) which is rare among European Americans (allele frequency <1%) [45]. The frequency of this splice variant was 38% in this study, similar to the 37% previously reported (NHLBI Exome Sequencing project, accessed June 23 2016). The rs116294140 has been described as a splice acceptor site between intron 2 and exon 3 which likely results in a UGT2B10 mRNA without exon 3 [46]. Such an mRNA has not been previously described in publicly available databases, suggesting that perhaps this SNP results in undetectable mRNA. Among 364 African Americans, UGT2B10 rs116294140 was shown to account for the majority of the reduced nicotine glucuronide levels compared to smokers of other ethnicities [24]. Similarly, in our study, individuals who were homozygous for UGT2B10 rs116294140 excreted little to no nicotine or cotinine glucuronide. We found a small, non-significant gene-dose effect on increasing nicotine half-life and a resulting decrease in nicotine clearance with UGT2B10 rs116294140. The modest decrease in nicotine clearance among UGT2B10 rs116294140 homozygous variants compared to wild types, however, was not sufficient to change smoking behavior such as consumption or smoking topography as observed previously [27].
The UGT2B10 rs116294140 substantially altered cotinine pharmacokinetics. This is consistent with a GWAS study where the UGT2B10 locus was highly associated with cotinine levels among smokers [47]. Cotinine has a low hepatic extraction ratio suggesting its clearance is primarily dependent on the amount and activity of metabolic enzymes such as CYP2A6 and UGT2B10. The reason for the more modest effect of the UGT2B10 rs116294140 variant on nicotine pharmacokinetics may be due to nicotine’s high hepatic extraction resulting in its clearance being more dependent on other factors such as hepatic blood flow [48]. The effect of UGT2B10 rs116294140 on cotinine pharmacokinetics may explain why African Americans have particularly low cotinine clearance compared to Asians, both of whom have similarly decreased nicotine clearance compared to European Americans [7]. Cotinine is the most widely used biomarker of tobacco, nicotine, and carcinogen intake among regular smokers [48]; lower cotinine glucuronidation ratios in African American smokers compared to other ethnicities has been previously noted [24, 25]. Any given cotinine level may not represent the same nicotine intake between races with differing frequencies of CYP2A6 gene variants [49]. Using pharmacokinetic data we also show here that cotinine levels may be misleading when compared among those with differing UGT2B10 rs116294140 genotypes within a race i.e. cotinine levels may overestimate tobacco exposure in African Americans with UGT2B10 rs116294140 variants.
UGT2B17 catalyzes the O-glucuronidation of 3HC, the main nicotine metabolite detected in smokers’ urine, but individuals who are homozygous null for UGT2B17 still excreted 3HC glucuronide, albeit significantly less than individuals with two active copies of this gene. The small amount of 3HC glucuronide excreted in homozygous null individuals could be the product of UGT2B7 which is a minor enzyme in the 3HC O’-glucuronidation pathway [43].
FMO3 rs2266782 is associated with reduced activity, thus a significant reduction in nicotine N’-oxide/free nicotine ratio with increasing copies of the FMO3 rs2266782 variant allele was expected, but not observed. It is possible that enzymes other than FMO3 are involved in nicotine N’-oxidation and compensate for the lack of activity among those with FMO3 variants. Consistent with the lack of effect on N’-oxidation ratios, we observed no impact of FMO3 rs2266782 on nicotine or cotinine pharmacokinetics among all subjects, or among those with reduced CYP2A6 activity. It is likely that FMO3 rs2266782 is a very minor source of variation in nicotine metabolism, insufficient to significantly alter nicotine or cotinine pharmacokinetics in African American smokers.
The OCT2 rs316019 CA genotype was associated with significantly higher nicotine and cotinine Cmax which was unexpected given that labeled nicotine-d2 and cotinine-d4 were administered intravenously. The functional impact of the OCT2 rs316019 variant is unclear. Those with the OCT2 rs316019 minor allele demonstrated reduced metformin clearance (in a laboratory study of 15 Chinese [50]) and reduced cisplatin-related nephrotoxicity (among 78 Dutch and 53 Japanese cancer patients [51, 52]), but was also associated with reduced metformin plasma levels (among 23 individuals [53]). Here, among 60 African American smokers, OCT2 rs316019 was associated with a non-significant, modest decrease in nicotine clearance. The increase in nicotine Cmax in people with this variant, without a substantial effect on nicotine renal clearance, suggests a depot effect where those with the variant have potentially reduced tissue uptake of nicotine, for example into skeletal muscle or brain.
The ratio of 3HC/COT has been widely used as a biomarker of CYP2A6 activity, and of nicotine and cotinine clearance. Considering that variation in UGT2B10 and UGT2B17 altered cotinine and 3HC glucuronidation ratios, respectively, we investigated the potential effect of variation in these genes on the 3HC/COT ratio. Genetic variation in UGT2B10 was associated with a small, non-significant reduction in plasma, but not urinary, 3HC/COT ratio. Moreover, the 3HC/COT ratio was essentially unaffected by variation in FMO3 or UGT2B17 genes.
Despite being the largest study of nicotine and cotinine pharmacokinetics in African Americans, our analysis is limited by small sample sizes in some genotype groups. Moreover, due to low power, we could not investigate the effects of combined glucuronidation and N-oxidation variation (UGT/FMO3 genotypes) on nicotine and cotinine pharmacokinetics.
In summary, among African Americans, variation in UGT2B10 was associated with a small, non-significant impact on nicotine pharmacokinetics, no impact on smoking (TNE and topography), but a substantial impact on cotinine pharmacokinetics. Therefore, variation in glucuronidation contributes to changes in the widely used tobacco exposure biomarker, cotinine, among African Americans and may lead to overestimation of tobacco exposure in African Americans compared to smokers of other races.
Supplementary Material
Acknowledgments
We would like to thank Sandra Tinetti for research coordination, Ewa Hoffmann for CYP2A6 genotyping, Olivia Yturralda, Trisha Mao and Lita Ramos for analytical chemistry and Faith Allen for data management. We appreciate the assistance of the nurses on the Clinical Research Center-Clinical Translational Science Institute research ward at San Francisco General Hospital.
Sources of Funding:NLB serves as a paid consultant to pharmaceutical companies that are developing or that market smoking cessation medications. He also has been a paid expert witness in litigation against tobacco companies, including on issues related to light cigarettes. RFT has served as paid consultant to Apotex. The research was supported by US Public Health Service grants DA02277, DA 020830, DA 12393 from the National Institute on Drug Abuse. We acknowledge the support of the Endowed Chair in Addictions for the Department of Psychiatry (R.F. Tyndale), CIHR grant TMH-109787 (R.F. Tyndale), the Campbell Family Mental Health Research Institute of CAMH, the CAMH Foundation, the Canadian Foundation for Innovation (#20289 and #16014 to R.F. Tyndale) and the Ontario Ministry of Research and Innovation. Clinical studies were performed at the General Clinical Research Center at Zuckerberg San Francisco General Hospital with support of grant UL1 RO24131 from the National Institute of Health/National Center for Research Resources.
Footnotes
Conflicts of InterestNone of the other authors have any competing interests to declare.
References
- 1.Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE, et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med. 2006;354(4):333–342. doi: 10.1056/NEJMoa033250. [DOI] [PubMed] [Google Scholar]
- 2.Siahpush M, Singh GK, Jones PR, Timsina LR. Racial/ethnic and socioeconomic variations in duration of smoking: results from 2003, 2006 and 2007 Tobacco Use Supplement of the Current Population Survey. J Public Health. 2010;32(2):210–218. doi: 10.1093/pubmed/fdp104. [DOI] [PubMed] [Google Scholar]
- 3.Trinidad DR, Perez-Stable EJ, Emery SL, White MM, Grana RA, Messer KS. Intermittent and light daily smoking across racial/ethnic groups in the United States. Nicotine Tob Res. 2009;11(2):203–210. doi: 10.1093/ntr/ntn018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez-Stable EJ, Herrera B, Jacob P, 3rd, Benowitz NL. Nicotine metabolism and intake in black and white smokers. Jama. 1998;280(2):152–156. doi: 10.1001/jama.280.2.152. [DOI] [PubMed] [Google Scholar]
- 5.Malaiyandi V, Sellers EM, Tyndale RF. Implications of CYP2A6 genetic variation for smoking behaviors and nicotine dependence. Clin Pharmacol Ther. 2005;77(3):145–158. doi: 10.1016/j.clpt.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship Between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 Variation and Smoking Behaviors and Lung Cancer Risk. Journal of the National Cancer Institute. 2011;103(17):1342–1346. doi: 10.1093/jnci/djr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benowitz NL. Clinical pharmacology of nicotine: implications for understanding, preventing, and treating tobacco addiction. Clin Pharmacol Ther. 2008;83(4):531–541. doi: 10.1038/clpt.2008.3. [DOI] [PubMed] [Google Scholar]
- 8.Hukkanen J, Jacob P, 3rd, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev. 2005;57(1):79–115. doi: 10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- 9.Dempsey D, Tutka P, Jacob P, 3rd, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76(1):64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 10.St Helen G, Novalen M, Heitjan DF, Dempsey D, Jacob P, 3rd, Aziziyeh A, et al. Reproducibility of the nicotine metabolite ratio in cigarette smokers. Cancer Epidemiol Biomarkers Prev. 2012;21(7):1105–1114. doi: 10.1158/1055-9965.EPI-12-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chenoweth MJ, O’Loughlin J, Sylvestre MP, Tyndale RF. CYP2A6 slow nicotine metabolism is associated with increased quitting by adolescent smokers. Pharmacogenet Genomics. 2013;23(4):232–235. doi: 10.1097/FPC.0b013e32835f834d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu AZ, Zhou Q, Cox LS, David SP, Ahluwalia JS, Benowitz NL, et al. Association of CHRNA5-A3-B4 SNP rs2036527 with smoking cessation therapy response in African-American smokers. Clin Pharmacol Ther. 2014;96(2):256–265. doi: 10.1038/clpt.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonagh EM, Wassenaar C, David SP, Tyndale RF, Altman RB, Whirl-Carrillo M, et al. PharmGKB summary: very important pharmacogene information for cytochrome P-450, family 2, subfamily A, polypeptide 6. Pharmacogenet Genomics. 2012;22(9):695–708. doi: 10.1097/FPC.0b013e3283540217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al Koudsi N, Ahluwalia JS, Lin SK, Sellers EM, Tyndale RF. A novel CYP2A6 allele (CYP2A6*35) resulting in an amino-acid substitution (Asn438Tyr) is associated with lower CYP2A6 activity in vivo. Pharmacogenomics J. 2009;9(4):274–282. doi: 10.1038/tpj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mwenifumbo JC, Al Koudsi N, Ho MK, Zhou Q, Hoffmann EB, Sellers EM, et al. Novel and established CYP2A6 alleles impair in vivo nicotine metabolism in a population of Black African descent. Hum Mutat. 2008;29(5):679–688. doi: 10.1002/humu.20698. [DOI] [PubMed] [Google Scholar]
- 16.Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14(9):615–626. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Benowitz NL, Jacob P., 3rd Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin Pharmacol Ther. 1994;56(5):483–493. doi: 10.1038/clpt.1994.169. [DOI] [PubMed] [Google Scholar]
- 18.Byrd GD, Chang KM, Greene JM, deBethizy JD. Evidence for urinary excretion of glucuronide conjugates of nicotine, cotinine, and trans-3’-hydroxycotinine in smokers. Drug Metab Dispos. 1992;20(2):192–197. [PubMed] [Google Scholar]
- 19.Chen G, Blevins-Primeau AS, Dellinger RW, Muscat JE, Lazarus P. Glucuronidation of nicotine and cotinine by UGT2B10: loss of function by the UGT2B10 Codon 67 (Asp>Tyr) polymorphism. Cancer Res. 2007;67(19):9024–9029. doi: 10.1158/0008-5472.CAN-07-2245. [DOI] [PubMed] [Google Scholar]
- 20.Yamanaka H, Nakajima M, Nishimura K, Yoshida R, Fukami T, Katoh M, et al. Metabolic profile of nicotine in subjects whose CYP2A6 gene is deleted. Eur J Pharm Sci. 2004;22(5):419–425. doi: 10.1016/j.ejps.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Bloom AJ, Murphy SE, Martinez M, von Weymarn LB, Bierut LJ, Goate A. Effects upon in-vivo nicotine metabolism reveal functional variation in FMO3 associated with cigarette consumption. Pharmacogenet Genomics. 2013;23(2):62–68. doi: 10.1097/FPC.0b013e32835c3b48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen G, Giambrone NE, Jr, Dluzen DF, Muscat JE, Berg A, Gallagher CJ, et al. Glucuronidation genotypes and nicotine metabolic phenotypes: importance of functional UGT2B10 and UGT2B17 polymorphisms. Cancer Res. 2010;70(19):7543–7552. doi: 10.1158/0008-5472.CAN-09-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloom AJ, von Weymarn LB, Martinez M, Bierut LJ, Goate A, Murphy SE. The contribution of common UGT2B10 and CYP2A6 alleles to variation in nicotine glucuronidation among European Americans. Pharmacogenet Genomics. 2013;23(12):706–716. doi: 10.1097/FPC.0000000000000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy SE, Park SS, Thompson EF, Wilkens LR, Patel Y, Stram DO, et al. Nicotine N-glucuronidation relative to N-oxidation and C-oxidation and UGT2B10 genotype in five ethnic/racial groups. Carcinogenesis. 2014;35(11):2526–2533. doi: 10.1093/carcin/bgu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg JZ, Mason J, Boettcher AJ, Hatsukami DK, Murphy SE. Nicotine metabolism in African Americans and European Americans: variation in glucuronidation by ethnicity and UGT2B10 haplotype. J Pharmacol Exp Ther. 2010;332(1):202–209. doi: 10.1124/jpet.109.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg JZ, von Weymarn LB, Thompson EA, Wickham KM, Weisensel NA, Hatsukami DK, et al. UGT2B10 genotype influences nicotine glucuronidation, oxidation, and consumption. Cancer Epidemiol Biomarkers Prev. 2010;19(6):1423–1431. doi: 10.1158/1055-9965.EPI-09-0959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel YM, Stram DO, Wilkens LR, Park SS, Henderson BE, Le Marchand L, et al. The contribution of common genetic variation to nicotine and cotinine glucuronidation in multiple ethnic/racial populations. Cancer Epidemiol Biomarkers Prev. 2015;24(1):119–127. doi: 10.1158/1055-9965.EPI-14-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuehl GE, Murphy SE. N-glucuronidation of nicotine and cotinine by human liver microsomes and heterologously expressed UDP-glucuronosyltransferases. Drug Metab Dispos. 2003;31(11):1361–1368. doi: 10.1124/dmd.31.11.1361. [DOI] [PubMed] [Google Scholar]
- 29.Urakami Y, Okuda M, Masuda S, Saito H, Inui KI. Functional characteristics and membrane localization of rat multispecific organic cation transporters, OCT1 and OCT2, mediating tubular secretion of cationic drugs. J Pharmacol Exp Ther. 1998;287(2):800–805. [PubMed] [Google Scholar]
- 30.LeMasters GK, Khurana Hershey GK, Sivaprasad U, Martin LJ, Pilipenko V, Ericksen MB, et al. N-acetyltransferase 1 polymorphism increases cotinine levels in Caucasian children exposed to secondhand smoke: the CCAAPS birth cohort. Pharmacogenomics J. 2015;15(2):189–195. doi: 10.1038/tpj.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benowitz NL, St Helen G, Dempsey DA, Jacob P, 3rd, Tyndale RF. Disposition kinetics and metabolism of nicotine and cotinine in African American smokers: impact of CYP2A6 genetic variation and enzymatic activity. Pharmacogenet Genomics. 2016;31:31. doi: 10.1097/FPC.0000000000000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho MK, Mwenifumbo JC, Al Koudsi N, Okuyemi KS, Ahluwalia JS, Benowitz NL, et al. Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clin Pharmacol Ther. 2009;85(6):635–643. doi: 10.1038/clpt.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piliguian M, Zhu AZ, Zhou Q, Benowitz NL, Ahluwalia JS, Sanderson Cox L, et al. Novel CYP2A6 variants identified in African Americans are associated with slow nicotine metabolism in vitro and in vivo. Pharmacogenet Genomics. 2014;24(2):118–128. doi: 10.1097/FPC.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benowitz NL, Swan GE, Jacob P, Lessov-Schlaggar CN, Tyndale RF. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clinical Pharmacology & Therapeutics. 2006;80(5):457–467. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 35.Malaiyandi V, Lerman C, Benowitz NL, Jepson C, Patterson F, Tyndale RF. Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Mol Psychiatry. 2006;11(4):400–409. doi: 10.1038/sj.mp.4001794. [DOI] [PubMed] [Google Scholar]
- 36.Jacob P, 3rd, Yu L, Wilson M, Benowitz NL. Selected ion monitoring method for determination of nicotine, cotinine and deuterium-labeled analogs: absence of an isotope effect in the clearance of (S)-nicotine-3’,3’-d2 in humans. Biological mass spectrometry. 1991;20(5):247–252. doi: 10.1002/bms.1200200503. [DOI] [PubMed] [Google Scholar]
- 37.Benowitz NL, Jacob P, 3rd, Fong I, Gupta S. Nicotine metabolic profile in man: comparison of cigarette smoking and transdermal nicotine. The Journal of pharmacology and experimental therapeutics. 1994;268(1):296–303. [PubMed] [Google Scholar]
- 38.Benowitz NL, Swan GE, Jacob P, 3rd, Lessov-Schlaggar CN, Tyndale RF. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clinical pharmacology and therapeutics. 2006;80(5):457–467. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Benowitz NL, St Helen G, Dempsey DA, Jacob P, 3rd, Tyndale RF. Disposition kinetics and metabolism of nicotine and cotinine in African American smokers: impact of CYP2A6 genetic variation and enzymatic activity. Pharmacogenet Genomics. 2016;26(7):340–350. doi: 10.1097/FPC.0000000000000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergen AW, Javitz HS, Krasnow R, Michel M, Nishita D, Conti DV, et al. Organic cation transporter variation and response to smoking cessation therapies. Nicotine Tob Res. 2014;16(12):1638–1646. doi: 10.1093/ntr/ntu161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wassenaar CA, Conti DV, Das S, Chen P, Cook EH, Ratain MJ, et al. UGT1A and UGT2B genetic variation alters nicotine and nitrosamine glucuronidation in european and african american smokers. Cancer Epidemiol Biomarkers Prev. 2015;24(1):94–104. doi: 10.1158/1055-9965.EPI-14-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hisamuddin IM, Yang VW. Genetic polymorphisms of human flavin-containing monooxygenase 3: implications for drug metabolism and clinical perspectives. PharmacoGenomics. 2007;8(6):635–643. doi: 10.2217/14622416.8.6.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen G, Giambrone NE, Lazarus P. Glucuronidation of trans-3’-hydroxycotinine by UGT2B17 and UGT2B10. Pharmacogenet Genomics. 2012;22(3):183–190. doi: 10.1097/FPC.0b013e32834ff3a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Liang Q, Mendes P, Sarkar M. Is 24h nicotine equivalents a surrogate for smoke exposure based on its relationship with other biomarkers of exposure? Biomarkers. 2011;16(2):144–154. doi: 10.3109/1354750X.2010.536257. [DOI] [PubMed] [Google Scholar]
- 45.Fowler S, Kletzl H, Finel M, Manevski N, Schmid P, Tuerck D, et al. A UGT2B10 splicing polymorphism common in african populations may greatly increase drug exposure. J Pharmacol Exp Ther. 2015;352(2):358–367. doi: 10.1124/jpet.114.220194. [DOI] [PubMed] [Google Scholar]
- 46.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ware JJ, Chen X, Vink J, Loukola A, Minica C, Pool R, et al. Genome-Wide Meta-Analysis of Cotinine Levels in Cigarette Smokers Identifies Locus at 4q13.2. Sci Rep. 2016;6(20092) doi: 10.1038/srep20092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benowitz NL, Hukkanen J, Jacob P., 3rd Nicotine chemistry, metabolism, kinetics and biomarkers. Handb Exp Pharmacol. 2009;192:29–60. doi: 10.1007/978-3-540-69248-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu AZX, Renner CC, Hatsukami DK, Swan GE, Lerman C, Benowitz NL, et al. The ability of plasma cotinine to predict nicotine and carcinogen exposure is altered by differences in CYP2A6: the influence of genetics, race and sex: Cancer Epidemiol Biomarkers Prev. 2013 Apr;22(4):708–718. doi: 10.1158/1055-9965.EPI-12-1234-T. Epub 2013 Jan 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang ZJ, Yin OQ, Tomlinson B, Chow MS. OCT2 polymorphisms and in-vivo renal functional consequence: studies with metformin and cimetidine. Pharmacogenet Genomics. 2008;18(7):637–645. doi: 10.1097/FPC.0b013e328302cd41. [DOI] [PubMed] [Google Scholar]
- 51.Filipski KK, Mathijssen RH, Mikkelsen TS, Schinkel AH, Sparreboom A. Contribution of organic cation transporter 2 (OCT2) to cisplatin-induced nephrotoxicity. Clin Pharmacol Ther. 2009;86(4):396–402. doi: 10.1038/clpt.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwata K, Aizawa K, Kamitsu S, Jingami S, Fukunaga E, Yoshida M, et al. Effects of genetic variants in SLC22A2 organic cation transporter 2 and SLC47A1 multidrug and toxin extrusion 1 transporter on cisplatin-induced adverse events. Clin Exp Nephrol. 2012;16(6):843–851. doi: 10.1007/s10157-012-0638-y. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y, Li S, Brown C, Cheatham S, Castro RA, Leabman MK, et al. Effect of genetic variation in the organic cation transporter 2 on the renal elimination of metformin. Pharmacogenet Genomics. 2009;19(7):497–504. doi: 10.1097/FPC.0b013e32832cc7e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.