Abstract
Clopidogrel is one of the most commonly used therapeutics for secondary prevention of cardiovascular events in patients with acute coronary syndromes. However, substantial inter-individual variation in clopidogrel response has been documented, resulting in suboptimal therapy and an increased risk of recurrent events for some patients. In this investigation, we conducted the first genome-wide association study of circulating clopidogrel active metabolite levels in 513 healthy participants in order to directly measure clopidogrel pharmacokinetics. We observed that the CYP2C19 locus was the strongest genetic determinant of active metabolite formation (P=9.5×10−15). In addition, we identified novel genome-wide significant variants on chromosomes 3p25 (rs187941554, P=3.3×10−11) and 17q11 (rs80343429, P=1.3×10−8), as well as 6 additional loci that showed suggestive evidence of association (P≤1.0×10−6). Four of these loci demonstrated nominal associations with on-clopidogrel ADP-stimulated platelet aggregation (P≤0.05). Evaluation of clopidogrel active metabolite concentration may help identify novel genetic determinants of clopidogrel response, which has implications for the development of novel therapeutics and improved anti-platelet treatment for at-risk patients in the future.
Keywords: pharmacogenomics, precision medicine, clopidogrel, pharmacokinetics, platelet aggregation, genome-wide association study, CYP2C19
Clopidogrel is a thienopyridine prodrug frequently prescribed to reduce atherothrombotic events in patients with acute coronary syndromes. However, up to 40% of patients do not receive adequate benefit from clopidogrel therapy [1, 2]. Furthermore, up to 73% of the inter-individual variation in clopidogrel response is heritable [3]; yet, aside from well-described variants such as CYP2C19*2, few polymorphisms have been linked with clopidogrel response, suggesting that most of the genetic variation that leads to variability in drug efficacy remains unidentified [3–7].
Most investigations to date have measured platelet reactivity to assess clopidogrel response. While evaluation of circulating levels of the clopidogrel active metabolite is a more direct measure of clopidogrel metabolism, it has been underutilized due to the instability of the clopidogrel active metabolite, which requires rapid derivatization for accurate assessment [8]. Herein, we performed a genome-wide association study (GWAS) of circulating clopidogrel active metabolite levels in 513 healthy subjects of the Pharmacogenomics of Anti-Platelet Intervention (PAPI) Study after 8 days of clopidogrel (300 mg loading dose followed by 75 mg/day for 7 days). Furthermore, we assessed the influence of identified variants on adenosine diphosphate (ADP)-stimulated platelet aggregation post-clopidogrel administration.
Participants
Amish individuals from Lancaster, Pennsylvania were recruited into the PAPI Study between 2006 and 2012 [3, 9]. Participants were over age 20, generally healthy, and agreed to discontinue the use of medications, supplements, and vitamins at least one week prior to study initiation. Baseline platelet aggregation measurements were recorded after overnight fast, after which participants received a 300 mg oral loading dose of clopidogrel followed by 75 mg/day for 7 days (Supplemental Digital Content, Figure S1). Follow-up platelet aggregation and clopidogrel metabolite measurements were obtained 1 hour following the last clopidogrel dose. Exclusion criteria and additional information regarding recruitment and design of the PAPI Study have been previously described [3, 9]. The protocol was approved by the University of Maryland, Baltimore Institutional Review Board. All subjects provided written informed consent.
Genotyping
Genotyping was performed with Affymetrix 500k and 6.0 genome-wide arrays (Affymetrix Inc., Santa Clara, California). Variants were called using Birdseed V2 and BRLMM, and imputed using the 1000 Genomes Project reference panel (Phase I version 3, March 2012) with Impute2 [10] and ShapeIt [11]. Polymorphisms with an imputation quality score < 0.5 and a minor allele frequency < 1% were excluded from analysis.
Clopidogrel metabolite quantification
Blood samples were collected within 1 hour after the last clopidogrel dose into EDTA tubes containing 2mmol/l (E)-2-bromo-3′-methoxyacetophenone (MPB; Sigma Aldrich, St Louis, Missouri) to derivatize the clopidogrel active metabolite. Plasma levels of clopidogrel MPB-derivatized active metabolite and parent drug were assessed using an ultra-high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) assay as previously described [5, 12].
Platelet aggregation
Platelet-rich plasma was isolated from blood samples drawn into tubes containing 3.2% sodium citrate (Becton-Dickinson, Franklin Lakes, New Jersey), and platelet counts were adjusted to 200,000 platelets/μl with platelet-poor plasma (PPP). Platelet function was assessed using a PAP8E aggregometer (Bio/Data Corporation, Horsham, Pennsylvania) after stimulation with ADP (20 μmol/L, Chronolog Corp., Havertown, Pennsylvania) and was expressed as the maximal percentage change in light transmittance using PPP as a referent.
Statistical analysis
Association analyses were conducted with a variance component method using Mixed Model Analysis for Pedigrees (MMAP) (http://edn.som.umaryland.edu/mmap/), [3, 13–15]. Analyses of clopidogrel active metabolite were performed under an additive model adjusted for age, sex, and relatedness. Analyses of on-clopidogrel ADP-mediated platelet aggregation were performed under an additive model and adjusted for age, sex, relatedness, and baseline platelet aggregation. Clopidogrel prodrug concentration was assessed for the top GWAS signals and conditional analyses were conducted with adjustment for the index SNP at each locus. A genome-wide significance threshold of P = 5 × 10−8 was used for GWAS analysis and P < 0.05 was used to establish significance of ADP-stimulated platelet aggregation analyses.
We tested 7,884,700 SNPs for association with clopidogrel active metabolite concentration in PAPI Study subjects (Figure 1, Supplemental Digital Content, Figure S2). Characteristics of these individuals are listed in Table 1. A locus on chromosome 10 near the CYP2C9-CYP2C18-CYP2C19 gene cluster was significantly associated with active metabolite concentration (rs137891020, P = 9.5 × 10−15, Supplemental Digital Content, Figure S3). After adjustment for the CYP2C19*2 variant, association of rs137891020 was markedly attenuated (P = 9.1 × 10−4), and no other variant in this region showed suggestive evidence of association (P ≥ 1.0 × 10−6). In addition to CYP2C19*2, novel variants on chromosomes 3p25 (rs187941554, P = 3.3 × 10−11, Supplemental Digital Content, Figure S4) and 17q11 (rs80343429, P = 1.3 × 10−8, Supplemental Digital Content, Figure S5) were significantly associated with clopidogrel active metabolite concentration. Another 6 independent loci exhibited suggestive evidence of association (P ≤ 1.0 × 10−6, Table 2). The full list of SNPs with significant (P ≤ 5.0 × 10−8) or suggestive (P ≤ 1.0 × 10−6) evidence of association with active metabolite concentration can be found in Supplemental Digital Content, Table S1. Conditional analyses adjusted for the most significantly associated SNP at each locus fully accounted for the associations observed at all loci (Supplemental Digital Content, Table S2). We extended our findings by examining whether polymorphisms that influence active metabolite levels also impacted on-clopidogrel ADP-stimulated platelet aggregation. Indeed, nominal associations (P < 0.05) were observed between on-clopidogrel platelet aggregation and 4 regions identified in the active metabolite GWAS (Table 2). No associated SNPs were significantly associated with clopidogrel prodrug levels and adjustment for prodrug levels did not appreciably change our results (data not shown).
Figure 1.
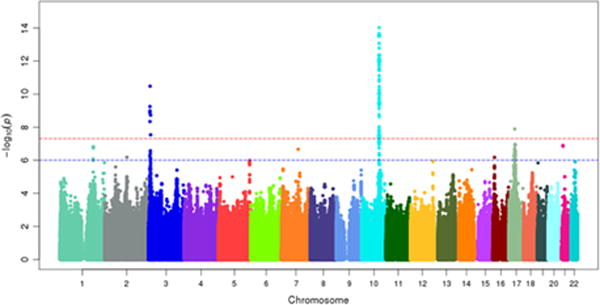
Manhattan plot of circulating clopidogrel active metabolite concentration as measured by HPLC MS/MS after clopidogrel administration (300 mg loading dose followed by 75 mg/day for 7 days). N=513. Red line indicates P = 5.0 × 10−8. Blue line indicates P = 1.0 × 10−6.
Table 1.
Characteristics of PAPI Study Participants
| Characteristics (units) | Total (N = 513) | Females (N = 256) | Males (N = 257) |
|---|---|---|---|
| Age (years) | 44.9 ± 13.2 | 45.6 ± 13.6 | 44.2 ± 12.7 |
| BMI (kg/m2) | 27.1 ± 4.7 | 28.2 ± 5.4 | 26.0 ± 3.6 |
| Platelet Count (thousands) | 239.7 ± 48.2 | 245.7 ± 51.3 | 234.1 ± 44.3 |
| SBP (mm Hg) | 116.8 ± 12.3 | 117.0 ± 13.1 | 116.6 ± 11.4 |
| DBP (mm Hg) | 70.2 ± 7.1 | 69.6 ± 7.0 | 70.8 ± 7.2 |
| Cholesterol (mg/dl) | 208.9 ± 46.8 | 210.0 ± 49.4 | 207.6 ± 43.5 |
| LDL (mg/dl) | 135.9 ± 42.7 | 132.6 ± 44.7 | 138.6 ± 40.2 |
| HDL (mg/dl) | 58.7 ± 15.2 | 62.4 ± 15.5 | 55.6 ± 14.5 |
| Triglycerides (mg/dl) | 71.4 ± 40.2 | 74.9 ± 42.1 | 67.3 ± 37.3 |
| Diabetes (%) | 0.4% | 0.4% | 0.4% |
Values are listed as mean ± standard deviation or percent. Abbreviations: BMI, body mass index; DBP, diastolic blood pressure; HDL; high-density lipoprotein; LDL, low-density lipoprotein; PAPI, Pharmacogenomics of Anti-Platelet Intervention; SBP, systolic blood pressure.
Table 2.
Novel genomic regions with significant (P ≤ 5.0 × 10−8) or suggestive (P ≤ 1.0 × 10−6) associations with clopidogrel active metabolite concentration
| Active Metabolite | On-Clopidogrel Platelet Aggregation | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Index SNP | Chr:Pos | Minor Allele | MAF | Gene | β | P-value | β | P-value |
| rs187941554 | 3:8987233 | A | 0.01 | RAD18 | 25.2 ± 3.7 | 3.3 × 10−11 | −3.8 ± 4.8 | 0.42 |
| rs80343429 | 17:31556681 | A | 0.05 | ASIC2 | 8.1 ± 1.4 | 1.3 × 10−8 | −3.5 ± 1.7 | 0.04 |
| rs79172967 | 21:18571112 | T | 0.02 | N/A | 15.5 ± 2.9 | 1.3 × 10−7 | −2.2 ± 3.7 | 0.55 |
| rs72392086 | 1:185033816 | D | 0.04 | RNF2 | 7.6 ± 1.4 | 1.6 × 10−7 | −5.0 ± 1.8 | 5.3 × 10−3 |
| rs73407739 | 7:92491283 | T | 0.01 | LOC101927497 | 16.8 ± 3.2 | 2.2 × 10−7 | −11.9 ± 3.9 | 2.2 × 10−3 |
| rs181524103 | 2:123209400 | G | 0.03 | N/A | 12.0 ± 2.4 | 6.6 × 10−7 | −3.1 ± 3.0 | 0.31 |
| rs138852022 | 16:9155981 | G | 0.01 | N/A | 17.0 ± 3.4 | 6.7 × 10−7 | −6.9 ± 4.1 | 0.09 |
| rs6892003 | 5:174191465 | A | 0.29 | N/A | 3.3 ± 0.7 | 1.0 × 10−6 | −2.2 ± 0.9 | 9. 7 × 10−3 |
Betas are listed as mean ± standard deviation or percent. Abbreviations: MAF, minor allele frequency; Chr:Pos, chromosome and position; P, p-value; N/A, not applicable. D, deletion
We report the first GWAS of clopidogrel active metabolite concentration, a direct measure of clopidogrel pharmacokinetics. Clopidogrel is among the most widely used antiplatelet agents available; however, genetic background results in substantial variation in drug response [3]. To date, most studies have utilized ADP-based platelet function tests, a pharmacodynamic measure, in order to examine clopidogrel response. However, apart from CYP2C19*2, few actionable variants have been identified. We hypothesized that the genome-wide analysis of clopidogrel active metabolite concentration would facilitate the identification of novel pharmacokinetic variants that may remain unidentified in genetic investigations focused solely on pharmacodynamic measures.
The active metabolite of clopidogrel is highly unstable [16] and must be derivatized to be measured accurately [12]. Through genome-wide genotyping and assessment of clopidogrel active metabolite in a large cohort, we successfully identified novel loci on chromosomes 3p25 and 17q11 that are significantly correlated with formation of the clopidogrel active metabolite, as well as an additional 6 loci that may warrant further investigation. Consistent with our initial results, 4 of the 8 loci that influence formation of the clopidogrel active metabolite also impact on-clopidogrel ADP-stimulated platelet aggregation. Importantly, none of these variants were associated with ADP-mediated platelet aggregation at a genome-wide level and would not have been identified in traditional genome-wide investigations that solely utilize platelet function testing (Supplemental Digital Content, Figures S6 and S7). In addition, while we observed strong evidence of association between CYP2C19*2 and formation of the clopidogrel active metabolite, no other variant in this or other cytochrome P450 genes was correlated with active metabolite levels after adjustment for CYP2C19*2 and correction for multiple testing. It should be noted that our investigation was not properly powered to identify previously reported loss-of-function variants that are exceedingly rare in European-derived populations (e.g. CYP2C19*3); however, these data do provide initial evidence of little to no impact of other more common variants in CYP2C19 (e.g. CYP2C19*17) or other cytochrome P450 genes on active metabolite formation.
Several variants identified in this study are in close proximity to genes with potential roles in clopidogrel metabolism. For instance, a cluster of SNPs on chromosome 3p25 are near ATP2B2, which codes for a plasma membrane calcium transporter [17] that is expressed in the liver [18] and was previously implicated in clopidogrel response [19]. On chromosome 1p25, the most strongly associated variant is located within Ring Finger Nuclease 2 (RNF2), which encodes a polycomb group protein that interacts with P-glycoprotein, a key enzyme for the intestinal absorption of clopidogrel [20]. Moreover, prior investigations have shown that RNF2 protein levels are negatively correlated with P-glycoprotein expression [20].
We acknowledge some limitations of this investigation. Clopidogrel active metabolite is not routinely measured, and no suitable replication cohort was available. This study is also potentially limited by the methodology used to quantify the clopidogrel active metabolite. The derivatization process with MPB does not differentiate between the racemic H4 (active) and H3 (inactive) stereoisomer of clopidogrel metabolites, potentially leading to some measurement error. While high concordance between H3 and H4 has been noted previously [21], which may minimize the import of this potential limitation, the use of novel methodologies to isolate the H4 isomer, such as the method developed by Hua and colleagues [22], may improve variant detection in future studies. Additionally, all work presented herein was conducted in the Amish of Lancaster, PA, which could influence translatability of findings to non-Amish populations. However, this has not been the case in the past [3, 13–15].
While the identification of polymorphisms such as CYP2C19*2 has informed clinicians in prescribing the most effective antiplatelet regimen and has improved outcomes in a percentage of cardiovascular disease patients, there is still significant ‘missing’ heritability that leaves many patients at an unnecessarily high risk of recurrent ischemic events. In this study, we aimed to identify novel genetic variants that influence clopidogrel active metabolite formation. Indeed, a number of novel variants with potential effects on clopidogrel efficacy were identified. While verification in an independent population is needed, this study demonstrates the utility of directly assessing clopidogrel metabolites. Furthermore, such investigations have implications for improved next-generation antiplatelet drug discovery through identification of variants that negatively influence drug efficacy thereby facilitating the development of novel drugs whose metabolism/efficacy are not dependent on proteins that harbor these mutations.
Supplementary Material
Acknowledgments
We gratefully acknowledge the Amish community, our Amish liaisons, and field workers for their extraordinary cooperation, without which this investigation would not have been possible.
Sources of Funding: Drs. Lewis and Shuldiner receive grant support from NIH to study the pharmacogenomics of anti-platelet therapy. Dr. Shuldiner is now an employee of Regeneron Pharmaceuticals. Dr. Yerges-Armstrong is now an employee of GlaxoSmithKline.
This investigation was supported by National Institutes of Health grants U01 GM074518, U01 GM074518-05S1, U01 HL105198, and K23 GM102678, the University of Maryland General Clinical Research Center, Grant M01 RR 16500, General Clinical Research Centers Program, National Center for Research Resources (NCRR), and the Baltimore Veterans Administration Geriatric Research and Education Clinical Center (GRECC).
Footnotes
Conflicts of Interest
All other authors have no conflict of interest to declare.
References
- 1.Nguyen TA, Diodati JG, Pharand C. Resistance to clopidogrel: a review of the evidence. J Am Coll Cardiol. 2005;45:1157–64. doi: 10.1016/j.jacc.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 2.Kolandaivelu K, Bhatt DL. Overcoming ‘resistance’ to antiplatelet therapy: targeting the issue of nonadherence. Nature Reviews Cardiology. 2010;7:461–467. doi: 10.1038/nrcardio.2010.71. [DOI] [PubMed] [Google Scholar]
- 3.Shuldiner AR, O’Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–57. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su J, Xu J, Li X, Zhang H, Hu J, Fang R, et al. ABCB1 C3435T Polymorphism and Response to Clopidogrel Treatment in Coronary Artery Disease (CAD) Patients: A Meta-Analysis. PLoS One. 2012;7:e46366. doi: 10.1371/journal.pone.0046366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis JP, Horenstein RB, Ryan K, O’Connell JR, Gibson Q, Mitchell BD, et al. The functional G143E variant of carboxylesterase 1 is associated with increased clopidogrel active metabolite levels and greater clopidogrel response. Pharmacogenet Genomics. 2013;23:1–8. doi: 10.1097/FPC.0b013e32835aa8a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calderon-Cruz B, Rodriguez-Galvan K, Manzo-Francisco LA, Vargas-Alarcon G, Fragoso JM, Pena-Duque MA, et al. C3435T polymorphism of the ABCB1 gene is associated with poor clopidogrel responsiveness in a Mexican population undergoing percutaneous coronary intervention. Thromb Res. 2015;136:894–898. doi: 10.1016/j.thromres.2015.08.025. [DOI] [PubMed] [Google Scholar]
- 7.Johnson JA, Cavallari LH. Pharmacogenetics and Cardiovascular Disease—Implications for Personalized Medicine. Pharmacol Rev. 2013;65:987–1009. doi: 10.1124/pr.112.007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi M, Pang H, Kawabata K, Farid NA, Kurihara A. Quantitative determination of clopidogrel active metabolite in human plasma by LC–MS/MS. J Pharm Biomed Anal. 2008;48:1219–1224. doi: 10.1016/j.jpba.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 9.Bozzi LM, Mitchell BD, Lewis JP, Ryan KA, Herzog WR, O’Connell JR, et al. The Pharmacogenomics of Anti-Platelet Intervention (PAPI) Study: Variation in Platelet Response to Clopidogrel and Aspirin. Curr Vasc Pharmacol. 2015 doi: 10.2174/1570161113666150916094829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delaneau O, Marchini J, Genomes Project, C, and Genomes Project, C Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peer CJ, Spencer SD, VanDenBerg DA, Pacanowski MA, Horenstein RB, Figg WD. A sensitive and rapid ultra HPLC-MS/MS method for the simultaneous detection of clopidogrel and its derivatized active thiol metabolite in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;880:132–9. doi: 10.1016/j.jchromb.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollin TI, Damcott CM, Shen H, Ott SH, Shelton J, Horenstein RB, et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322:1702–5. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albert JS, Yerges-Armstrong L, Horenstein RB, Pollin TI, Sreenivasan UT, Chai S, et al. Null Mutation in Hormone-Sensitive Lipase Gene and Risk of Type 2 Diabetes. N Engl J Med. 2014;370:2307–2315. doi: 10.1056/NEJMoa1315496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis JP, Ryan K, O’Connell JR, Horenstein RB, Damcott CM, Gibson Q, et al. Genetic variation in PEAR1 is associated with platelet aggregation and cardiovascular outcomes. Circ Cardiovasc Genet. 2013;6:184–92. doi: 10.1161/CIRCGENETICS.111.964627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plavix® prescribing information. 2015 Jul; [cited 2016 May 17]; Available from: http://products.sanofi-aventis.us/PLAVIX/PLAVIX.html.
- 17.Tempel BL, Shilling DJ. The plasma membrane calcium ATPase and disease. Subcell Biochem. 2007;45:365–83. doi: 10.1007/978-1-4020-6191-2_13. [DOI] [PubMed] [Google Scholar]
- 18.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price MJ, Carson AR, Murray SS, Phillips T, Janel L, Tisch R, et al. First Pharmacogenomic Analysis Using Whole Exome Sequencing to Identify Novel Genetic Determinants of Clopidogrel Response Variability: Results of the Genotype Information and Functional Testing (Gift) Exome Study. J Am Coll Cardiol. 2012;59:E9–E9. [Google Scholar]
- 20.Rao PS, Mallya KB, Srivenugopal KS, Balaji KC, Rao US. RNF2 interacts with the linker region of the human P-glycoprotein. Int J Oncol. 2006;29:1413–9. [PubMed] [Google Scholar]
- 21.Karaźniewicz-Łada M, Danielak D, Burchardt P, Kruszyna Ł, Komosa A, Lesiak M, et al. Clinical Pharmacokinetics of Clopidogrel and Its Metabolites in Patients with Cardiovascular Diseases. Clin Pharmacokinet. 2014;53:155–164. doi: 10.1007/s40262-013-0105-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua W, Lesslie M, Hoffman BT, Binns C, Mulvana D. Development of a sensitive and fast UHPLC-MS/MS method for determination of clopidogrel, clopidogrel acid and clopidogrel active metabolite H4 in human plasma. Bioanalysis. 2015;7:1471–82. doi: 10.4155/bio.15.82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


