Abstract
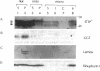
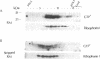
Subfractionation of a canine pancreatic homogenate was performed by several differential centrifugation steps, which gave rise to fractions with distinct marker profiles. Specific binding of guanosine 5'-[gamma-[35S]thio]triphosphate (GTP[gamma-35S]) was assayed in each fraction. Enrichment of GTP[gamma-35S] binding was greatest in the interfacial "smooth" microsomal fraction, expected to contain Golgi and other smooth vesicles. There was also marked enrichment in the rough microsomal fraction. Electron microscopy and marker protein analysis revealed the rough microsomes (RMs) to be highly purified rough endoplasmic reticulum (RER). The distribution of small (low molecular weight) GTP binding proteins was examined by a [alpha-32P]GTP blot-overlay assay. Several apparent GTP binding proteins of molecular masses 22-25 kDa were detected in various subcellular fractions. In particular, at least two such proteins were found in the Golgi-enriched and RM fractions, suggesting that these small GTP binding proteins were localized to the Golgi and RER. To more precisely localize these proteins to the RER, native RMs and RMs stripped of ribosomes by puromycin/high salt were subjected to isopycnic centrifugation. The total GTP[gamma-35S] binding, as well as the small GTP binding proteins detected by the [alpha-32P]GTP blot overlay, distributed into fractions of high sucrose density, as did the RER marker ribophorin I. Consistent with a RER localization, when the RMs were stripped of ribosomes and subjected to isopycnic centrifugation, the total GTP[gamma-35S] binding and the small GTP binding proteins detected in the blot-overlay assay shifted to fractions of lighter sucrose density along with the RER marker.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelman M. R., Sabatini D. D., Blobel G. Ribosome-membrane interaction. Nondestructive disassembly of rat liver rough microsomes into ribosomal and membranous components. J Cell Biol. 1973 Jan;56(1):206–229. doi: 10.1083/jcb.56.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Potter V. R. Studies on free and membrane-bound ribosomes in rat liver. I. Distribution as related to total cellular RNA. J Mol Biol. 1967 Jun 14;26(2):279–292. doi: 10.1016/0022-2836(67)90297-5. [DOI] [PubMed] [Google Scholar]
- Bolender R. P. Stereological analysis of the guinea pig pancreas. I. Analytical model and quantitative description of nonstimulated pancreatic exocrine cells. J Cell Biol. 1974 May;61(2):269–287. doi: 10.1083/jcb.61.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne H. R. Do GTPases direct membrane traffic in secretion? Cell. 1988 Jun 3;53(5):669–671. doi: 10.1016/0092-8674(88)90081-5. [DOI] [PubMed] [Google Scholar]
- Gill D. L., Ueda T., Chueh S. H., Noel M. W. Ca2+ release from endoplasmic reticulum is mediated by a guanine nucleotide regulatory mechanism. Nature. 1986 Apr 3;320(6061):461–464. doi: 10.1038/320461a0. [DOI] [PubMed] [Google Scholar]
- Lapetina E. G., Reep B. R. Specific binding of [alpha-32P]GTP to cytosolic and membrane-bound proteins of human platelets correlates with the activation of phospholipase C. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2261–2265. doi: 10.1073/pnas.84.8.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melançon P., Glick B. S., Malhotra V., Weidman P. J., Serafini T., Gleason M. L., Orci L., Rothman J. E. Involvement of GTP-binding "G" proteins in transport through the Golgi stack. Cell. 1987 Dec 24;51(6):1053–1062. doi: 10.1016/0092-8674(87)90591-5. [DOI] [PubMed] [Google Scholar]
- Mignery G. A., Südhof T. C., Takei K., De Camilli P. Putative receptor for inositol 1,4,5-trisphosphate similar to ryanodine receptor. Nature. 1989 Nov 9;342(6246):192–195. doi: 10.1038/342192a0. [DOI] [PubMed] [Google Scholar]
- Mullaney J. M., Chueh S. H., Ghosh T. K., Gill D. L. Intracellular calcium uptake activated by GTP. Evidence for a possible guanine nucleotide-induced transmembrane conveyance of intracellular calcium. J Biol Chem. 1987 Oct 5;262(28):13865–13872. [PubMed] [Google Scholar]
- Neer E. J., Clapham D. E. Roles of G protein subunits in transmembrane signalling. Nature. 1988 May 12;333(6169):129–134. doi: 10.1038/333129a0. [DOI] [PubMed] [Google Scholar]
- Nigam S. K., Blobel G. Cyclic AMP-dependent protein kinase in canine pancreatic rough endoplasmic reticulum. J Biol Chem. 1989 Oct 5;264(28):16927–16932. [PubMed] [Google Scholar]
- Northup J. K., Smigel M. D., Gilman A. G. The guanine nucleotide activating site of the regulatory component of adenylate cyclase. Identification by ligand binding. J Biol Chem. 1982 Oct 10;257(19):11416–11423. [PubMed] [Google Scholar]
- Paiement J., Bergeron J. J. Localization of GTP-stimulated core glycosylation to fused microsomes. J Cell Biol. 1983 Jun;96(6):1791–1796. doi: 10.1083/jcb.96.6.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz I., Thévenod F., Dehlinger-Kremer M. Modulation of intracellular free Ca2+ concentration by IP3-sensitive and IP3-insensitive nonmitochondrial Ca2+ pools. Cell Calcium. 1989 Jul;10(5):325–336. doi: 10.1016/0143-4160(89)90058-4. [DOI] [PubMed] [Google Scholar]
- Supattapone S., Danoff S. K., Theibert A., Joseph S. K., Steiner J., Snyder S. H. Cyclic AMP-dependent phosphorylation of a brain inositol trisphosphate receptor decreases its release of calcium. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8747–8750. doi: 10.1073/pnas.85.22.8747. [DOI] [PMC free article] [PubMed] [Google Scholar]