Abstract
Mitochondrial transcription factor A (TFAM) stimulates transcription from mitochondrial DNA (mtDNA) promoters in vitro and in organello. To investigate whether changes of TFAM levels also modulate transcription and replication in situ, the protein was transiently overexpressed in cultured cells. Mitochondrial mRNAs were significantly elevated at early time points, when no expansion of the TFAM pool was yet observed, but were decreased when TFAM levels had doubled, resemb-ling in vitro results. HEK cells contain about 35 molecules of TFAM per mtDNA. High levels of TFAM were not associated with increases of full-length mtDNA, but nucleic acid species sensitive to RNAse H increased. Stimulation of transcription was more evident when TFAM was transiently overexpressed in cells pre-treated with ethidium bromide (EBr) having lowered mtDNA, TFAM and mitochondrial transcript levels. EBr rapidly inhibited mtDNA transcription, while decay of mtDNA was delayed and preferentially slowly migrating, relaxed mtDNA species were depleted. In conclusion, we show that transcription of mtDNA is submaximal in cultured cells and that a subtle increase of TFAM within the matrix is sufficient to stimulate mitochondrial transcription. Thus, this protein meets all criteria for being a key factor regulating mitochondrial transcription in vivo, but other factors are necessary for increasing mtDNA copy number, at least in cultured cells.
INTRODUCTION
Transcription and replication of mitochondrial DNA (mtDNA) are important steps in mitochondrial biogenesis; however, the regulation of these processes in vivo is unclear, especially during developmental or physiological adaptation of the mitochondrial content of cells to different energy demands. In vitro transcription systems were reconstituted initially from a partially purified mitochondrial RNA polymerase (POLRMT) preparation and a single additional protein, mitochondrial transcription factor A (TFAM) (1), and more recently from pure recombinant proteins (POLRMT, TFAM, TFB1M or TFB2M) (2). In these systems, it was consistently shown that elevating the concentration of the DNA-binding protein TFAM increases the yield of run-off transcripts from mtDNA promoters, and that an optimal stoichiometry between this protein and template DNA exists, since high concentrations of TFAM have an inhibitory effect on transcription (3,4). In contrast, the TFBM proteins bind directly to POLRMT with high affinity, and at a 1:1 stoichiometry a maximal transcription rate is obtained (2). Thus, in vitro TFAM regulates the rate of mtDNA transcription, and therefore meets the criteria of a bona fide transcription factor. However, it remains to be shown whether this is also true in vivo.
Also, replication of mammalian mtDNA was thought to be transcription primed from the light strand promoter (LSP), giving rise to a leading DNA strand starting at OH and a lagging strand starting at OL in the asymmetric replication model (5). Recently, this model has been questioned and mammalian mtDNA may be replicated in a conventional, strand symmetric mode (6). Moreover, initiation of synthesis may even occur at multiple sites on both strands within a broad initiation zone (7), and OH may rather play a role in fork arrest, but in any case, TFAM will probably still participate in the initiation of replication by RNA primer formation.
In mice, where TFAM was ablated by homologous recombination techniques, lethality in homozygous embryos clearly demonstrated that it is absolutely required for the maintenance of mtDNA. In heterozygous and tissue-specific knockout animals, its levels influenced copy number of mtDNA, but only moderately affected transcript levels (8–11). Therefore, its actual role in the process of transcription regulation still remained unclear, since copy number control and transcription regulation could not be dissected from each other in these animals. Very recently, however, constitutive overexpression of human TFAM in transgenic mice showed that the elevated total TFAM levels correlate well with increased mtDNA copy number, and also with 7S DNA (12), which is thought to be transcription primed from LSP (13). ND6 mRNA derived from LSP was also increased, but transcripts derived from the heavy strand promoter (HSP) were unaffected. Finally, evidence was provided that TFAM may be a high abundance structural protein (14), so its main function could be to form a scaffold for mitochondrial nucleoid formation (12).
Using an alternative approach, we have previously shown that import of TFAM into rat liver mitochondria in amounts stoichiometric to endogenous mtDNA stimulates synthesis rate of 7S DNA derived from LSP (15) as well as mRNAs and rRNAs derived from HSP (16), strongly indicating a causal link between intramitochondrial TFAM levels and transcription rate. However, isolated mitochondria are deprived from a constant supply of nuclear-encoded proteins, and indeed, while they faithfully transcribe mRNAs for several hours, the processing of the rRNAs is rapidly impaired (17). Therefore, binding sites may have been generated during the isolation procedure and may not represent free binding sites accessible for regulation by TFAM in vivo.
Thus, in order to probe the acute effect of increased intramitochondrial levels of TFAM on mitochondrial transcription, and also replication and mtDNA topology in situ, the protein was overexpressed by transient transfection in cultured cells. As appropriate controls, a C-terminal truncated version of TFAM was used, which had been demonstrated to bind DNA in vitro, but has little transcriptional activity (4).
MATERIALS AND METHODS
Cloning and subcloning of human TFAM and truncated versions of TFAM
The complete human TFAM coding sequence (1) was amplified by RT–PCR using total human RNA as template. The PCR product was cloned into the XbaI–ClaI sites downstream of the HCMV-IE promoter in pKEX-2-XR (18), yielding pKEX-TFAM; restriction sites had been introduced by the primers (forward-CCTCTAGAGCGATGGCGTTTCTCCGAA, reverse-CCATCGATCCATTGTGAACACATCTC). The sequence of the TFAM insert was confirmed by dideoxy-sequencing of both strands. The insert was then subcloned into the BamHI–HindIII sites of pQE9 (Qiagen) for the expression of recombinant TFAM in Escherichia coli employing PCR, pBS-TFAM and the following primers (forward-CCGGGGATCCATGGCGTTTCTCCGAAGCAT, reverse-GGCCAAGCTTTTAAGATCTACACTCCTCAGCACCATATT), thus also introducing a poly-Histidin-tail at the N-terminus. Truncated versions of TFAM (ΔC-8-TFAM, ΔC-15-TFAM and ΔC-25-TFAM) were also produced lacking 8, 15 or 25 amino acids at the C-terminal end, respectively. The PCR products were cloned into the XbaI–XhoI sites of pKEX-2-XR using pBS-TFAM as template and appropriate primers containing XbaI and XhoI restrictions sites. (forward-CCTCTAGAGCGATGGCGTTTCTCCGAA; ΔC-8-reverse-CCGGCTCGAGTTATTGTTTCTTTATTGTGCGACG; ΔC-15-reverse-CCGGCTCGAGTTATAGAAGATCCTTTCGTCCAA; ΔC-25-reverse-CCGGCTCGAGTTATTGTTCTTCCCAAGACTTCA).
Transient overexpression of TFAM in HeLa or HEK293 cells
HeLa cells of the HtTa strain (19) were grown in DMEM supplemented with 10% fetal calf serum (FCS) on 21 cm2 culture dishes with gas-permeable bottoms (Bachofer) to ensure optimal oxygenation of the cells. HEK cells were cultivated in large regular 150 cm2 dishes in DMEM supplemented with 10% FCS. In some instances, the cells were pre-cultivated in medium including ethidium bromide (EBr) (100 ng/ml), uridine (50 μg/ml) and sodium pyruvate (110 μg/ml) for 4 days in order to reduce the level of mtDNA and cultivated further after the removal of EBr for the indicated times. Prior to transfection experiments, such cells were washed three times with incomplete medium in order to remove EBr completely. For each time point, a total of 9 to 18 dishes were either transfected with the empty vector pKEX, pKEX-TFAM or versions of pKEX-ΔC-TFAM, respectively, by the calcium phosphate precipitate method. Transfection efficiency was monitored by co-transfection with pCMV-lacZ. Staining for β-galactosidase activity showed that routinely 40–60% of cells had been successfully transfected.
For rapid and high-level expression of TFAM proteins, HEK cells were transfected by nucleoporation using the electroporation device, the appropriate buffer as well as the electric pulse parameters recommended for this cell line by the manufacturer (Amaxa, Köln).
The cells were harvested at various time points after transfection and analysed by western, northern and Southern blot assays.
Analysis of cellular TFAM levels
Cellular protein was routinely dissolved in Laemmli buffer (2 μg), quantitated by the Bradford assay, separated by SDS–PAGE and electroblotted to nitrocellulose (Sartorius). Equal loading of the lanes was routinely checked by staining blots with the Ponceau S reagent (Sigma) prior to further analysis. Blots were subsequently blocked in 20 mM Tris, pH 7.5, 150 mM NaCl, 0.5% Tween-20, 2% BSA for 2 h, and incubated overnight in the same buffer containing a rabbit anti-human TFAM antiserum (1:4000), prepared as described previously (20). To obtain quantitative data, the blot was cut horizontally and the lower half was incubated with TFAM antiserum, the upper half with a monoclonal antibody against β-tubulin (Sigma), respectively. After washing, proteins were visualized by incubation with goat anti-rabbit immunoglobulin G (IgG) or goat anti-mouse IgG antiserum, horseradish–peroxidase conjugated (Dianova) and chemoluminescence detection (ECL, NEN).
In some cases, the rabbit antiserum was used in a 1:1000 dilution, protein A labelled with 125I was used instead of the secondary antiserum (Amersham; 0.4 μCi/ml), and TFAM signals were quantitated using a phosphoImager (Raytest).
In order to show that TFAM was indeed imported into mitochondria, 18 h after transfection cells were disrupted in hypertonic isolation buffer [0.6 M mannitol, 10 mM Tris–HCl, pH 7.4, 1 mM EDTA, 0.1% BSA, phenylmethylsulfonyl fluoride (PMSF)], using a loosely fitting glass–teflon potter and pestle, unbroken cells and nuclei were sedimented by centrifugation at 500 g, mitochondria were pelleted by centrifugation at 5000 g, and incubated in the same buffer (without BSA) with proteinase K at concentrations and for incubation times as indicated.
Analysis of mitochondrial gene expression
For analysis of mitochondrial gene expression, RNA was extracted from pooled HeLa cells of three equally treated dishes (equivalent to n = 1) using a commercially available RNA-extraction kit (RNeasy, Qiagen) or from one large dish of HEK cells by TRIZOL (Invitrogen). RNA (5 μg) was separated on formaldehyde agarose gels and blotted onto nitrocellulose membranes by the capillary transfer method. Northern blots were subsequently hybridized with cDNAs or PCR products encoding human cytochrome-c-oxidase subunit I (CO I), NADH dehydrogenase subunit I (ND1), ATP synthase subunit 6/8 (ATPase 6/8) and 12S rRNA, labelled with [α-32P]dCTP by random priming under conditions previously described in detail (21). Hybridization with a 28S rRNA probe was used to normalize for unequal loading. Intensity of bands on autoradiograms were quantitated with a CCD-video-camera based analysis system (AIDA software; Raytest) and arbitrary densitometric units obtained for CO I mRNA, ND1 mRNA, ATPase 6/8 mRNA and their precursors as well as 12S rRNA were normalized to the amount of 28S rRNA present in each lane. Care was taken that the signal was in the linear range of the relation: densitometric value/blotted RNA. The densitometric value obtained for the control sample loaded in lane 1 of each gel was arbitrarily set to 100 in order to obtain mean values for control cells and cells transfected with pKEX-TFAM or pKEX-ΔC-TFAMs.
Analysis of mtDNA levels and conformations
DNA was isolated from cells lysed in SDS containing Tris–EDTA–NaCl (STE) buffer by proteinase K digestion, followed by RNAse treatment, phenol/chloroform extraction and salt/ethanol precipitation (22). DNA was run on large 0.8% agarose gels overnight at 4°C, either undigested or after linearization with PvuII, and blotted onto nitrocellulose membranes by the capillary transfer method. Where indicated, DNA was treated with RNAse H for 1 h in the PvuII digestion buffer or heat denatured by boiling for 15 min, followed by chilling on ice before loading. Southern blots were subsequently hybridized to DNAs encoding human CO I, ND6, a D-Loop probe and to a human 18S probe (kindly provided by Dr Bob Lightowlers, Newcastle) and analysed as described for northern blots.
Analysis of TFAM/mtDNA stoichiometry in HEK cells
HEK or HeLa cells were counted using a CASY1 system (Schärfe System GmbH, Reutlingen, Germany) and values were confirmed by direct analysis in a counting chamber. For this experiment, whole-cell extracts were routinely prepared from known numbers of cells in Laemmli buffer, and also in TOTEX lysis buffer (20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid [HEPES pH 7.9], 250 mM NaCl, 0.25% Nonidet P-40, 0.5 mM PMSF, 0.5 mM sodium orthovanadate, 10 mM NaF, 1 mM DTT). The cells were extracted for 10 min at 4°C and centrifuged at 15 000 g for 10 min to remove cell debris. The protein content was determined by the Bradford assay using BSA as standard.
Recombinant, full-length His-tagged TFAM protein was purified from E.coli using an appropriate isolation kit employing Ni++ chemistry (Qiagen). The TFAM concentration of the resulting, diluted solution was quantitated in comparison with a serial dilution of known amounts of BSA on Coomassie stained gels. Known amounts of recombinant TFAM protein as well as cell lysate from a known number of cells were run on SDS gels and analysed by western blotting as described above.
Accordingly, DNA was extracted from known cell numbers as described above, digested with XbaI and analysed on Southern blots together with known amounts of pBS-KS-ND1 linearized with PstI. Blots were probed with an ND1 probe derived from the same plasmid. In both cases, films were analysed as described above.
Statistical analysis
When appropriate, data are given as mean ± SD and were compared by a two-tailed Student's t-test. A confidence level of P < 0.05 was considered indicative of a statistically significant difference between groups.
RESULTS
Transient overexpression of TFAM in HeLa cells
We failed to obtain an inducible cell line overexpressing TFAM under the control of doxycycline, probably due to the inherent leakiness of these systems (0 inducible out of 472 hygromycin resistant clones; H. L. Garstka, PhD Thesis, University of Heidelberg). Thus, transient transfection using a high expression level plasmid was employed in all further experiments, which routinely yielded 40–60% of transfected cells shown by β-galactosidase activity staining. Also, labelling the protein by a His-tag, Flag-tag or green fluorescent protein added to the C-terminus in order to be able to distinguish between endogenous TFAM and the plasmid-derived protein, respectively, was not feasible, since only the precursors, but never the mature forms of these tagged proteins, accumulated after transfection (data not shown).
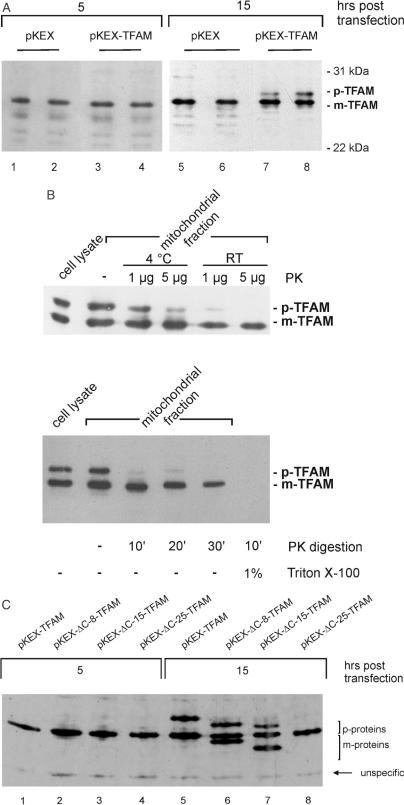
After transfection with pKEX-TFAM, the levels of the 24 kDa mature TFAM protein were not significantly elevated at 5 h, but increased 2-fold at 15 h (Figure 1A; at 15 h: 100 ± 14 versus 200 ± 16, pKEX versus pKEX-TFAM; n = 3; P < 0.05). Quantitation of TFAM was performed using 125I-protein A and phosphoimaging for detection; some unspecific bands show that the lanes were equally loaded. When transfected cells were cultivated up to 48 h, no more than such a 2-fold increase was ever observed (see below). Interestingly, a 29 kDa protein consistent with the predicted molecular mass of the unprocessed TFAM precursor had accumulated at 15 h (Figure 1A). This protein was never observed in control cells, even after loading large amounts of cellular protein and overexposure of chemoluminescent blots. The 29 kDa TFAM precursor was sedimented together with the mitochondrial fraction, but was obviously located outside the matrix, since it was accessible to protease digestion (Figure 1B, upper panel), while the mature protein was protected and thus proven to be located in the matrix. It became digestable only after lysis of mitochondria with Triton X-100 (Figure 1B, lower panel).
Figure 1.
Transient overexpression of TFAM in HeLa cells. (A) TFAM levels analysed by western blotting at 5 h (lanes 1–4) and 15 h (lanes 5–8) after transfection with pKEX and pKEX-TFAM. Two representative lanes for each group are shown. Vectors used are indicated above the lanes. (B) Localization of the TFAM precursor and mature proteins, respectively, comparing total cell lysate with a mitochondrial fraction treated with proteinase K (μg/ml) under various conditions. (C) Expression of several TFAM isoforms after transfection with pKEX-TFAM (lanes 1 and 5), pKEX-ΔC-8-TFAM (lanes 2 and 6), pKEX-ΔC-15-TFAM (lanes 3 and 7), pKEX-ΔC-25-TFAM (lanes 4 and 8). Cells were harvested at 5 and 15 h as indicated. p, precursor; m, mature protein; PK, proteinase K; RT, room temperature.
In search of an appropriate control for later experiments, several C-terminal truncated isoforms of TFAM were constructed, which have considerable DNA-binding activity, but show reduced or even no transcriptional activity in vitro (4). From pulse-chase experiments with [35S]methionine, followed by immunoprecipitation, subsequent autoradiography and densitometry, we could estimate that the half-life of wt-TFAM is >12 h in HeLa cells (data not shown). Thus, replacement of pre-existing TFAM molecules by exchange for new, vector-derived molecules is a slow process. Again, only after 15 h, the ΔC-8-TFAM and the ΔC-15-TFAM species accumulated (Figure 1C, lanes 6 and 7), both the mature form as well as the precursors, and levels of wt-TFAM again moderately increased (Figure 1C, lane 5). An unspecific band showed that lanes were equally loaded. The ΔC-25-TFAM protein could not be expressed (Figure 1C, lane 8), and remained undetectable even upon overexposure of the blots, although transcripts derived from pKEX-ΔC-25-TFAM were present in large amounts (northern blots, data not shown). In the presence of the truncated isoforms (lanes 6 and 7), the level of the endogenous TFAM remained unchanged (compare with lanes 1–4). This indicates that there is a limited number of unoccupied TFAM-binding sites available in these cells, which are equally accessible to all three isoforms.
In conclusion, transient transfection increased the level of mature wt-TFAM in the matrix of HeLa cell mitochondria only after extended times. Two C-terminal truncated versions could also be expressed, while the ΔC-25-TFAM lacking the complete C-terminus distal to the second high-mobility-group-domain seemed to be unstable, in contrast to the in organello system (16).
Effect of transient TFAM overexpression on mtDNA transcription in HeLa cells
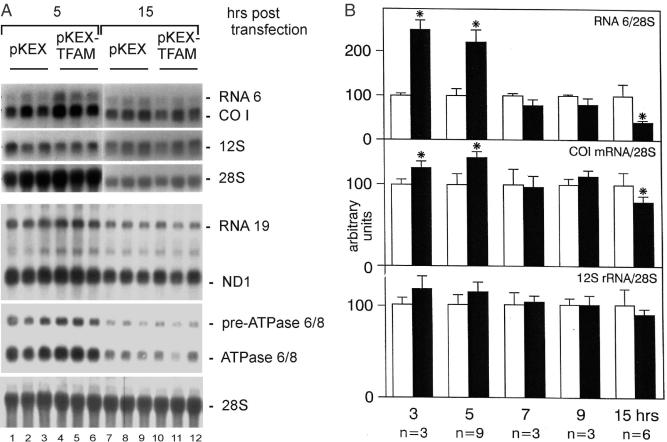
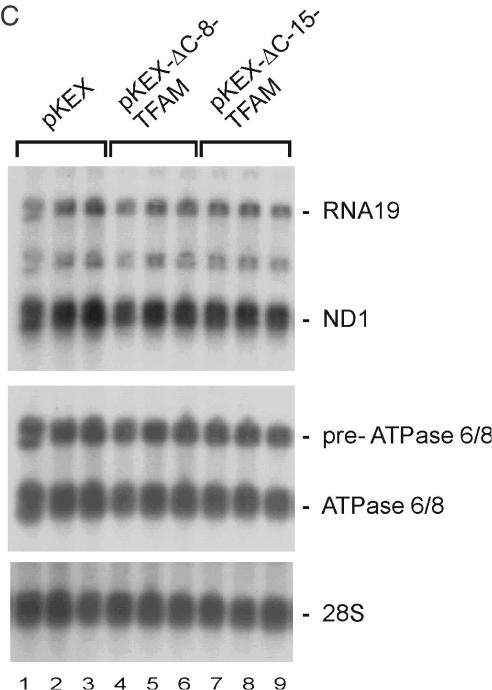
Five hours after transfection with pKEX-TFAM, levels of CO I mRNA as well as its precursor RNA 6 (23) were slightly, but clearly increased, when compared with controls (Figure 2A, upper panels). At 15 h post-transfection, transcript levels were decreased. To ensure that this is reproducible, a blot from a second, independent experiment was probed for ND1 and ATPase 6/8, obtaining similar results (Figure 2A, lower panels). Changes of CO I mRNA, RNA 6 and 12S rRNA were then analysed extensively, and compiled data of all experiments are shown in Figure 2B. Levels of CO I mRNA and more obvious its precursor RNA 6 were significantly increased after 3 and 5 h (P < 0.05), returned to control values at 7 and 9 h and were significantly decreased 15 h after transfection (P < 0.05). Small changes of the 12S rRNA levels indicated a similar time course; however, differences were not significant, probably due to the larger pool size and the slower turnover of rRNAs compared with mRNAs. It should be noted that the changes induced by overexpressed TFAM were actually 2-fold higher in the successfully transfected cells, since they represented only half of the population. Upon transfection with vectors encoding truncated TFAMs (pKEX-ΔC-8-TFAM and pKEX-ΔC-15-TFAM), no differences in mitochondrial transcript levels were found compared with controls (Figure 2C).
Figure 2.
Effect of transient overexpression of TFAM on mitochondrial transcript levels in HeLa cells. (A) Northern blot showing levels of CO I mRNA, RNA 6 and 12S rRNA (upper panel), ND1 mRNA, RNA 19, ATPase 6/8 mRNA and its precursor (lower panel) at 5 h and 15 h after transfection with pKEX (lanes 1–3 and 7–9), and pKEX-TFAM (lanes 4–6 and 10–12). Also shown are the signals obtained for cytosolic 28S rRNA used for normalization. Each lane contains RNA from pooled cells of three equally treated culture dishes. Vectors used are indicated above the lanes. (B) Levels of CO I mRNA and its precursor RNA 6, as well as 12S rRNA (arbitrary densitometric units/28S rRNA signal) at the indicated time points after transfection with pKEX-TFAM (black bars) or controls (pKEX, open bars). Numbers of RNA preparations analysed are given for each time point; data are mean ± SD. (C) Northern blot showing levels of ND1 mRNA, ATPase 6/8 mRNA and their respective precursors as well as 28S rRNA used for normalization after transfection with pKEX (lanes 1–3), pKEX-ΔC-8-TFAM (lanes 4–6) and pKEX-ΔC-15-TFAM (lanes 7–9). The cells were harvested after 5 h. Each lane contains RNA from pooled cells of three equally treated culture dishes. Asterisk indicates a statistically significant difference between cells transfected with pKEX-TFAM and controls (pKEX), P < 0.05.
In conclusion, small elevations of TFAM, but not its C-terminal truncated forms, which however do not significantly expand the total TFAM pool within the mitochondrial matrix, cause an increase, while a 4-fold elevation of the protein in the successfully transfected cells causes a decrease in mitochondrial transcript levels, thus resembling closely the situation in reconstituted systems in vitro.
Effect of enhanced TFAM levels on mtDNA replication and copy number
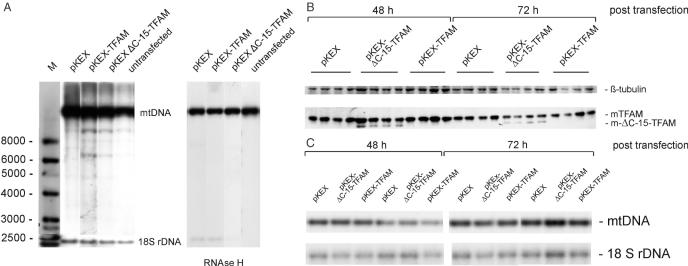
Replication may be stimulated by increased transcription of RNA primers, or TFAM may stabilize mtDNA molecules in the mitochondrial nucleoid. Therefore, mtDNA levels were measured on Southern blots (Figure 3A). After 20 h following transfection of HEK cells, the abundance of full-length mtDNA was 4.35 ± 1.48 (pKEX), 4.14 ± 2.45 (pKEX-TFAM), 5.22 ± 2.44 (pKEX-ΔC-15-TFAM) and 4.36 ± 1.75 (untransfected; mtDNA/18S rDNA; mean ± SD; two independent measurements per sample, n = 3 per group). Therefore, high levels of TFAM were not associated with a high copy number of mtDNA (see also Figure 7A and C). On the other hand, after overexpression of TFAM, several additional nucleic acid species showed up (Figure 3A and Supplementary Figure 3X). Upon close inspection of the blots, these molecules are present in all samples, though to a lesser extent in untransfected, empty vector transfected cells or in cells expressing the C-terminal truncated isoform. These species completely disappeared after digestion of the DNA preparation with RNAse H, indicating a high content of DNA/RNA hybrids, while RNAse H did not affect copy number of full-length mtDNA. Since TFAM has been reported to destabilize 7S DNA in vitro (14) and to stimulate its synthesis in organello (15), we also looked for this species but found no obvious change in the ratio of 7S DNA/mtDNA in cells overexpressing the wt-protein (see Supplementary Figure 3X).
Figure 3.
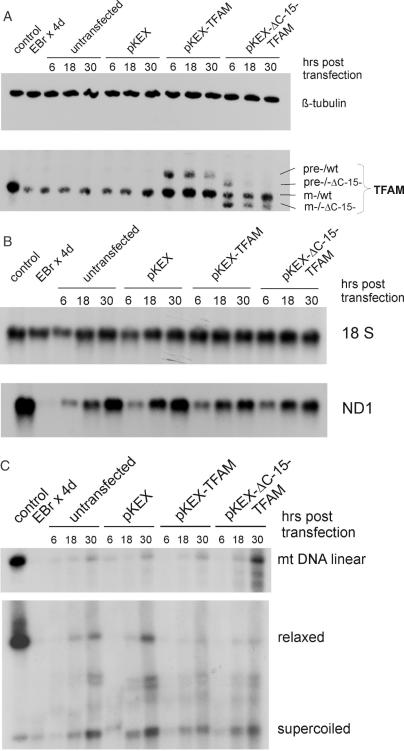
Effect of transient overexpression of TFAM on mtDNA copy number in HEK cells. (A) Southern blot showing linearized mtDNA at 20 h after transfection as well as new nucleic acid species hybridizing to a D-Loop probe (left panel), which are sensitive to RNAse H (right panel). Also shown is the signal for 18S rDNA used for normalization of mtDNA. M, double-stranded DNA size marker. (B) Western blot showing levels of TFAM and ΔC-15-TFAM at the indicated time points after transfection (four samples per group). The same blot was incubated with a β-tubulin antibody for normalization. (C) Southern blot showing levels of linearized mtDNA at the indicated time points after transfection with empty vector pKEX, pKEX-ΔC-15-TFAM and pKEX-TFAM, respectively (two samples per group). The blot was re-hybridized with an 18S rDNA probe for normalization.
Figure 7.
Transient overexpression of TFAM and its C-terminal truncated isoform by nucleoporation in HEK cells with lowered mtDNA content. (A) Western blot showing levels of TFAM and precursor proteins in untreated control cells, after 4 days of EBr treatment and 6, 18 and 30 h after nucleoporation of EBr treated cells with various plasmids or in untransfected cells. Also shown is the level of β-tubulin to demonstrate equal loading. (B) Northern blot showing levels of ND1 mRNA in the samples described in (A). (C) Southern blots showing levels of linearized mtDNA (upper panel) or conformations of mtDNA (lower panel) in the samples described in A and B.
mtDNA is synthesized rather slowly, and only a small sub-fraction of all molecules are replicated at any given time point in cultured cells (5), thus mtDNA copy number was analysed in an independent experiment 48 and 72 h after transfection of HEK cells. TFAM and the truncated isoform were still moderately elevated 48 h after transfection, while the protein precursors had disappeared (Figure 3B); however, no changes in the levels of mtDNA were observed even after this long time period (Figure 3C).
In conclusion, forced elevation of TFAM in HEK cells stimulates synthesis of RNAse H sensitive nucleic acid species or, alternatively, stabilizes preformed molecules, which may be replication intermediates, but it is not sufficient to increase copy number of full-length molecules of mtDNA.
Analysis of TFAM to mtDNA stoichiometry in HEK cells
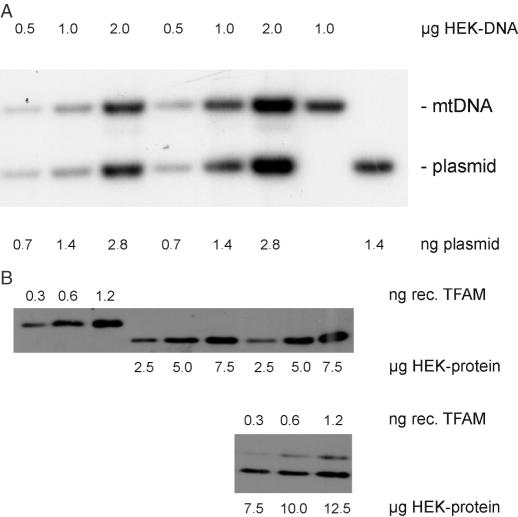
Since there is considerable uncertainty about the actual TFAM to mtDNA stoichiometry within cells, we estimated mtDNA as well as TFAM copies in HEK cells. DNA isolated from a known number of cells as well as plasmid containing the ND1 gene were digested with appropriate enzymes, so that the products were of about the same size, and were run in the same lanes, both measures were taken to ensure equal transfer of the DNA fragments during blotting. HEK cells contain 21 ± 5 pg of DNA per cell and from blots like the one shown in Figure 4A, we calculate that they harbour 7566 ± 2091 molecules of mtDNA (n = 6), which is in the lower range of values reported for a large number of cell lines analysed previously [6600 to 13900 molecules per cell, (24)].
Figure 4.
Determination of the stoichiometry of TFAM to mtDNA in HEK cells. (A) Indicated amounts of HEK DNA as well as plasmid containing the ND1 gene were run in the same (left) or separate lanes (two right lanes), the Southern blot was hybridized to an ND1 probe and data were analysed densitometrically. (B) Indicated amounts of HEK protein as well as recombinant full-length TFAM protein were run in the same lanes (lower panel) or separate lanes (upper panel), the western blot was incubated with a TFAM antiserum and data were analysed densitometrically.
Protein was isolated accordingly from a known number of cells and was run together with known amounts of full-length recombinant TFAM protein, blotted and probed with TFAM antiserum (Figure 4B); only data in the linear range of densitometric value versus loaded sample were used for quantitation. We found that HEK cells contain 117 ± 32 pg or 132 ± 14 pg of protein per cell when isolated with TOTEX or Laemmli buffer, respectively (mean value: 125 pg of protein per cell). Since the signal of 1 ng of recombinant TFAM (MW 31 kDa) is equivalent to 9.4 μg of HEK protein (value derived from seven independent dilution series like those shown in Figure 4B), these cells contain ∼260 000 molecules per cell and, consequently, 35 molecules of TFAM per mtDNA.
Effect of EBr on mtDNA, TFAM and mitochondrial transcript levels
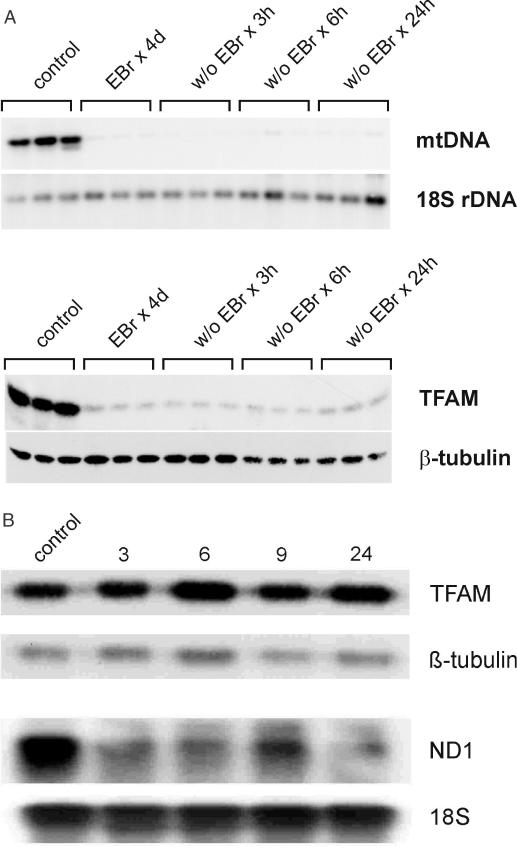
Depletion of mtDNA and mitochondrial transcripts by treatment of cells with EBr or dideoxycytidine as well as recovery from this treatment has been used to study interactions between mtDNA, TFAM and mitochondrial RNA and DNA synthesis, respectively (25,26). Since we could not unequivocally correlate an increase in TFAM with stimulation of transcription in untreated cells, we used this approach in all further experiments. In a preliminary series, the time course of changes of mtDNA, TFAM and mitochondrial transcripts as well as mtDNA conformations following EBr treatment or removal was established. In our hands, after 4 days of treatment, mtDNA was depleted to levels below 10% of control (Figure 5A, upper panel) and concomitantly, TFAM was also severely reduced (Figure 5A, lower panel); however, to a lesser extent, as reported previously (26). Upon withdrawal of the chemical, TFAM recovered somewhat faster than mtDNA in the initial phase (Figure 5A); however, contents were still severely depleted after 24 h. Southern blots of undigested native DNA showed that EBr preferentially reduced the levels of slowly migrating molecules, while the fast migrating molecules were almost unaffected (Supplementary Figure 5X, see also Figure 7C). We also noted that transcription of mtDNA is extremely sensitive to EBr: already 3 h after the addition of the drug, the ND1 mRNA was severely depleted (Figure 5B, lower panels), a time point at which TFAM levels were still unchanged (Figure 5B, upper panels). Upon EBr removal, mitochondrial transcripts remained at low levels for at least 18 h (see Supplementary Figure 5Y and Figure 6B).
Figure 5.
Effect of EBr treatment and removal in HEK cells. (A) Effect of EBr treatment for 4 days and drug removal (w/o) for 3, 6 and 24 h on levels of mtDNA (upper panel) and TFAM (lower panel). Also shown are the signals for 18S rDNA and β-tubulin used for normalization, respectively. (B) Kinetics of changes of TFAM and mitochondrial ND1 mRNA upon EBr treatment for up to 24 h.
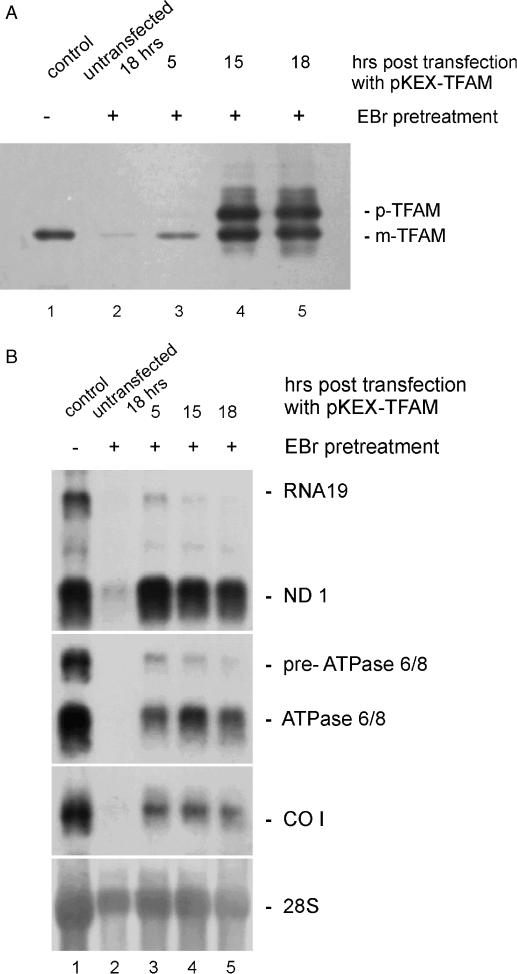
Figure 6.
Transient overexpression of TFAM in HEK cells with lowered mtDNA and TFAM content. (A) Levels of TFAM protein after transfection of cells pre-treated with EBr for 4 days. Control sample of untreated cells shows normal level of TFAM (lane 1); untransfected cells harvested after 18 h (lane 2) and cells which were transfected with pKEX-TFAM for 5 h (lane 3), 12 h (lane 4) and 18 h (lane 5) are shown. p, precursor; m, mature protein. (B) Northern blot showing levels of ND1 mRNA, RNA 19, ATPase 6/8 mRNA, pre-ATPase 6/8 and CO I mRNA as well as the signal of the 28S rRNA used for normalization. Each lane contains RNA from pooled cells of three equally treated culture dishes. Control sample of untreated cells shows normal wild-type level of various RNA species (lane 1), and lanes 2–5 show RNA levels in cells pre-treated with EBr for 4 days. Untransfected cells harvested after 18 h (lane 2) and cells that were transfected with pKEX-TFAM for 5 h (lane 3), 12 h (lane 4) and 18 h (lane 5) are shown. These samples were derived from the same plates as the samples shown on the western blot in Figure 5A.
In conclusion, inhibition of mtDNA transcription is a very rapid effect of EBr, while depletion of mtDNA and TFAM takes considerably longer, and preferentially affects the slowly migrating, relaxed circles, but not the fast migrating, probably supercoiled molecules. For a time window of at least 18 h after EBr removal, mtDNA, TFAM and mitochondrial transcripts remained at low levels, allowing us to explore the effect of overexpressed TFAM on transcription, replication and mtDNA topology under these conditions.
Transient overexpression of TFAM in HEK cells with reduced mtDNA content
Therefore, cells were pre-treated for 4 days with EBr, washed, transfected with pKEX-TFAM and further cultivated in EBr free medium for 5, 15 and 18 h in a first experiment (Figure 6A, lanes 3–5). Untransfected cells were harvested simultaneously at 18 h (Figure 6A, lane 2). The content of TFAM remained almost undetectable in such untransfected cells (2% of control, lane 2 versus 1, see also Figure 5A), while 5 h after transfection, TFAM was clearly increased (Figure 6A, lane 3 versus lane 2). At 15 and 18 h post transfection, levels of mature TFAM had increased ∼2-fold compared with control (Figure 6A, lanes 4 and 5 versus 1) and again the precursor protein was observed. Mitochondrial transcription was almost completely abolished after such long-term treatment with EBr (Figure 5B and Supplementary Figure 5Y), and transcript levels remained low for further 18 h in untransfected cells (Figure 6B, lane 2). However, overexpressed TFAM caused a dramatic elevation of mitochondrial transcripts. The ND1, COI and ATPase 6/8 transcripts and their precursors were increased 5- to 50-fold, respectively, as early as 5 h after transfection; however, again they decreased later when TFAM had reached high levels (Figure 6B, compare lanes 3 with 5).
Quantitative data were obtained from a second, independent experiment (Table 1). In this series, in untransfected cells as well as in cells transfected with empty vector, both the ND1 mRNA and its unprocessed precursor RNA19 had already recovered to a significant extent after 5 h. However, following overexpression of TFAM, transcripts were further increased by 30–40% compared with the empty vector or 45–65% compared with untransfected cells, respectively. Considering again that routinely ∼50% of the cells had been transfected, mitochondrial transcript levels had approximately increased 2-fold in the transfected cells. Surprisingly, also the truncated isoform caused a stimulation of mtDNA transcription under these conditions, while it had failed to do so in untreated cells (compare with Figure 2C).
Table 1. Levels of mitochondrial transcripts 5 h after transient transfection of HEK cells treated for 4 days with EBr or untreated control cells.
| n | RNA19/18S rRNA | ND1 mRNA/18S rRNA | |
|---|---|---|---|
| Untreated control | 5 | 0.73 ± 0.04 | 1.09 ± 0.25 |
| 4 d EBr | 5 | n.d. | n.d. |
| Untransfected | 5 | 0.15 ± 0.05 | 0.30 ± 0.01 |
| pKEX | 10 | 0.16 ± 0.02 | 0.38 ± 0.01 |
| pKEX-TFAM | 10 | 0.22 ± 0.04 | 0.50 ± 0.14 |
| pKEX-ΔC-15-TFAM | 10 | 0.18 ± 0.06 | 0.49 ± 0.18 |
n, number of individual plates; n.d., not detectable; values are mean ± SD.
Cells remained untransfected or were transfected with the indicated plasmids.
In conclusion, also in cells in which mitochondrial transcripts had been depleted, moderate increases of wt-TFAM stimulates transcription, but under these conditions even the ΔC-15-TFAM increases transcript levels, while high concentrations of TFAM again are inhibitory.
In order to cause a more rapid and pronounced increase in TFAM levels, HEK cells with reduced mtDNA content following EBr treatment were transfected in a third experiment by the efficient nucleoporation method, and protein, RNA and DNA were harvested from the same plates after 6, 18 and 30 h. Untransfected cells and cells transfected with the empty vector served as controls. With this procedure, levels of mature TFAM protein were raised to control values (Figure 7A, lane 1) already at 6 h after transfection with pKEX-TFAM (Figure 7A, lane 9) and remained fairly constant up to 30 h. The truncated ΔC-15-TFAM was initially present at about equal levels compared with endogenous wt-TFAM, but was replaced by endogenous wt protein with passing time. Like in the experiment shown in Figure 1C, this demonstrates that also under these conditions, there are a limited number of accessible binding sites freely available to both TFAM proteins. A northern blot of RNA from the same samples demonstrated the spontaneous recovery of mitochondrial transcripts in untransfected or empty vector transfected cells upon EBr removal (Figure 7B). However, at 6 h, the ND1 mRNA level was similar after pKEX-TFAM or pKEX-ΔC-15-TFAM transfection compared with empty vector transfected cells, and ND1 mRNA recovered even slower at 18 and 30 h. Certainly, the lack of a transcription stimulation by TFAM proteins in this experiment was again due to high concentrations of the proteins, or TFAM/mtDNA ratios, at all time points.
A Southern blot of linearized (Figure 7C, upper panel) or undigested, native DNA (Figure 7C, lower panel) showed that depletion of mtDNA by EBr treatment selectively affected the slowly migrating, relaxed circles (see also Supplementary Figure 5X), while the fast migrating, probably supercoiled population was rather unaffected. Upon removal of the drug, mtDNA levels recovered over time, and species with an intermediate mobility appeared; however, neither their levels nor their distribution were affected by the presence of high concentrations of TFAM proteins (compare with Figure 7A).
In conclusion, as shown above, high levels of TFAM, both wt and the truncated form, inhibit mitochondrial transcription. In addition, this experiment shows that high levels of TFAM are not sufficient to accelerate the recovery of mtDNA after EBr treatment or have any effect on mtDNA topology as analysed in these gels. Finally, to exclude that 30 h may not have been sufficient to increase mtDNA copy number by overexpressed TFAM, HEK cells were treated with EBr for 4 days, transfected by the calcium phosphate method and samples were harvested for western and Southern blot analysis at even more extended times. A significant increase of both wt-TFAM and ΔC-15-TFAM was observed at 15 h (Supplementary Figure 8A) and levels remained elevated up to 85 h. However, also after this long period, high TFAM levels were not accompanied by a significant increase in mtDNA copy number (Supplementary Figure 8B).
DISCUSSION
The TFAM gene is a target for important nuclear transcription activators like the nuclear respiratory factors and the cAMP responsive element binding protein, and during stimulation of mitochondrial biogenesis under a variety of physiological and pathological conditions, its enhanced expression is probably coordinated to other nuclear genes encoding mitochondrial proteins by the PPAR γ coactivator 1α (PGC-1α) [recently summarized in (27)]. Thus, upregulation of TFAM seems to be necessary for increasing mitochondrial mass, however since mtDNA copy number remains unchanged in most of these situations (28), TFAM is obviously responsible for elevating mitochondrial transcript levels under such conditions in vivo.
In reconstituted systems, composed of plasmid DNA containing mtDNA promoters and a partially purified mitochondrial polymerase (4,29,30), or recently the pure recombinant proteins (2), the DNA-binding protein TFAM is absolutely required for efficient and correct initiation of transcription. Elevating the concentration of TFAM increases the yield of transcripts, but high concentrations of TFAM have an inhibitory effect on transcription rate. We have previously shown that import of TFAM into rat liver mitochondria in stoichiometric amounts to mtDNA stimulates the rates of synthesis of 7S DNA, which is thought to be transcription primed from the LSP (15) as well as precursor RNAs, mRNAs and rRNAs derived from the HSP (16). Thus, in freshly isolated mitochondria, TFAM seems to be present in submaximal concentrations and elevating its levels is sufficient for increasing transcription rate from both promoters. Therefore, it also meets the criteria of being a bona fide transcription factor in this system.
In order to show whether the changes in intramitochondrial TFAM levels also affect transcription of mtDNA in situ within an intact cell, and whether and how it influences mtDNA replication and copy number on a long term range, in the present study the concentration of this protein was increased in the mitochondrial matrix by transient overexpression in cultured cells. Maximally, a 4-fold increase of TFAM levels could be reached (2-fold increase on western blots in 50% of the cell population) however, only at delayed time points. No change of mtDNA copy number was ever associated with this expansion of the TFAM pool. It is generally accepted that TFAM is stable only when bound to mtDNA (31), so obviously free binding sites are available in HeLa as well as in HEK cells that are accessible to the protein. Binding sites which are only partially occupied by protein have been routinely observed by several groups upstream of HSP and LSP, and also at other sites within the D-loop region, by in organello footprinting in different tissues and cell types (32–34), including HeLa cells (35). Unfortunately, these authors have not analysed extensively sequences outside the non-coding regions, although unprotected mtDNA seems to exist and served as control in the footprinting experiments [P. Fernandez-Silva, personal communication (35)]. Recently, however, Kang and co-workers (14) reported 1700 molecules of TFAM per mtDNA in HeLa cells and postulated that the whole genome is covered with the protein, whose main function therefore would be DNA packaging. This contrasts with the values previously obtained by Clayton's group (36), who measured 15 molecules of TFAM per mitochondrial genome in human KB cells. The former data (14), however, are problematic since the authors measured 1000 molecules of mtDNA per cell, which is ∼10-fold lower than values previously reported for HeLa, 143B osteosarcoma (24) and HEK cells (this study). Thus, we believe that our value of 35 TFAM molecules per mtDNA is much closer to the true value in cultured cells, although the complexity of such measurements precludes that more than a rough estimate can be given. On the other hand, when mitochondria were isolated prior to analysis of the stoichiometry, from human placenta (14) or mouse kidney (12), in both the cases about 1000 TFAM molecules per mtDNA were measured. Whether this striking difference between cultured cells [our data and (36)] and mitochondria isolated from solid tissues is due to methodological problems in one or the other preparation or due to a true and important difference between cultured cell lines and differentiated cells in vivo remains an important task to be solved in the future.
However, full or only partial coverage is rather irrelevant, since additional TFAM molecules derived from transfected plasmids would in any case bind to the unoccupied sites at both promoters (32–35). Even full saturation of these sites, however, would not lead to a measurable expansion of the total TFAM pool initially, irrespective of the total number of TFAM molecules per mtDNA, yet is obviously sufficient to stimulate transcription rate and ultimately transcript levels (Figure 2A and B). At expanded times after transfection, the matrix concentration of the protein was maximally increased 4-fold, a level that inhibits mitochondrial transcription but does not increase mtDNA copy number (Figure 3A and C). Also the truncated versions ΔC-8-TFAM as well as ΔC-15-TFAM were able to stably bind to the same unoccupied sites, although they were shown to have lower DNA affinity compared with wt-TFAM in vitro (4), whereas ΔC-25-TFAM was not stable. In conclusion, within the mitochondrial matrix of cultured cells, the relationship between TFAM concentration and mitochondrial transcript levels is strikingly similar to the in vitro systems discussed above (2,3,37), in which the yield of run-off transcripts from plasmids was analysed. After overexpression of C-terminal truncated versions (−8 and −15 amino acids), the levels of mitochondrial transcripts remained unchanged (Figure 2C), indicating that also within a cell these truncated proteins are only poor activators, since they lack parts of the amino acid sequence essential for transcription stimulation.
Since it was not possible to unequivocally prove that increased transcript levels observed at early time points were causally linked to increased levels of TFAM, in a second approach mtDNA as well as mitochondrial transcripts were reduced to low levels by treatment with EBr prior to transfection. The mechanism of this reduction of mtDNA by EBr is unclear; however, we noted that the fast migrating population of mtDNA, probably supercoiled circles, was almost unaffected, while the slowly migrating molecules, probably covalently closed, relaxed circles, disappeared (Supplementary Figures 5X and 7C). Since the earliest event following EBr addition is depletion of transcripts (Figure 5B and Supplementary Figure 5Y), its mechanism is probably the inhibition of the synthesis of RNA primers, which are necessary for the initiation of replication, irrespective of the mode of this process (7,13). Indeed, EBr has been shown to decrease methylation interference patterns in footprinting experiments in HeLa cell mitochondria as well as in whole cells, however, not directly within the TFAM-binding sites starting around 15 bp upstream, but rather directly at the transcription start sites (35). Our results also indicate that transcription of mtDNA preferentially takes place at the slow migrating, relaxed templates, while the fast migrating supercoiled molecules may be transcriptionally silent, although being replicated in the dividing cells by an obviously different mechanism, and therefore being insensitive to Ebr. These molecules may be more densely packaged by the remaining TFAM protein. After 4 days of EBr treatment, when TFAM, mtDNA and transcripts were almost absent, we were able to increase TFAM levels significantly within 5 h following transfection (Figure 6A), which was accompanied by a dramatic rise of mitochondrial transcripts (Figure 6B and Table 1), while high levels of TFAM again were inhibitory. Also, we can only speculate why under these conditions the ΔC-15-TFAM stimulated transcription (Table 1): either the C-terminally truncated transactivation domain is indeed sufficient to stimulate POLRMT, which can be postulated according to in vitro data (4), or ΔC-15-TFAM prepares the proximal promoter region for interaction with remaining wt-TFAM molecules.
Even at high levels of TFAM, mtDNA copy number remained unchanged, indicating that increasing its matrix concentration alone is not sufficient to elevate mtDNA content, at least in cultured cells. This is in agreement with recent studies showing that probably it is the release of termination of 7S DNA synthesis which controls copy number (25). However, after overexpression of TFAM, discrete nucleic acid species showed up, which were sensitive to RNAse H, indicating that they are rich in RNA–DNA hybrids (Figure 3A and Supplementary Figure 3X). These molecules could result from the escape from termination control of nascent DNA, whose synthesis rate was accelerated by increased TFAM levels, as observed in organello (15), since it is transcription primed at the LSP (38). In contrast, in tissues of transgenic mice overexpressing human TFAM, which is a poor transcriptional activator in a reconstituted system from otherwise mouse components, mtDNA copy number was increased by 50–70% and correlated well with the total TFAM pool (12). The different results may be either due to the completely different time course, up to 4 days in our experiments compared with weeks in the mice, or as discussed above, different coverage of mtDNA in differentiated tissues in vivo versus cultured cells.
In conclusion, we have obtained evidence that transcription of mtDNA is submaximal, in HeLa as well as in HEK cells in culture, and we have shown that increasing intramitochondrial TFAM is sufficient to stimulate mitochondrial transcription and probably initiate the synthesis of replication intermediates, but not sufficient for successful replication of full-length mtDNA copies. Thus, the matrix concentration of TFAM regulates the rate of transcription from mitochondrial promoters, in vitro (1,2), in organello (16) and in situ within cells (this study). Other transcription factors like, for instance, the newly discovered mitochondrial thyroid hormone receptor p43 (39,40) might function as modulators for mitochondrial transcription, e.g. by directing the machinery to different initiation sites (HSP I and II, respectively), modulating the ratio of mRNA to rRNA transcription by thyroid hormone (41). Since transcription of mtDNA is certainly an important step in the biogenesis of the whole organelle and the TFAM gene is a target for main activators of the mitochondrial biogenesis program (27), TFAM indeed seems to be a key player in regulation of the capacity for oxidative phosphorylation of cells, in the steady state as well as during development and adaptation.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
DNA probes for human CO I and 12S rRNA were gifts from Dr E. Sbisa (Bari, Italy). The excellent technical assistance of Bettina Sogl, Maria Bust and Nadine Lottmann, as well as the expert help of Sabrina Eckertz with the figures are gratefully acknowledged. This work was supported by research grants from The Deutsche Forschungsgemeinschaft (DFG Wi 889/3-3 and SFB 589/P18), Landesgraduiertenförderung Baden-Württemberg, Deutscher Akademischer Austauschdienst—Spanish Ministerio de Educacion y Ciencia (DAAD-Acciones Integradas) and the Spanish Ministerio de Ciencia y Tecnologia (BMC 2001-2421).
REFERENCES
- 1.Parisi M.A. and Clayton,D.A. (1991) Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science, 252, 965–969. [DOI] [PubMed] [Google Scholar]
- 2.Falkenberg M., Gaspari,M., Rantanen,A., Trifunovic,A., Larsson,N.G. and Gustafsson,C.M. (2002) Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nature Genet., 31, 289–294. [DOI] [PubMed] [Google Scholar]
- 3.Parisi M.A., Xu,B. and Clayton,D.A. (1993) A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol., 13, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dairaghi D.J., Shadel,G.S. and Clayton,D.A. (1995) Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol., 249, 11–28. [DOI] [PubMed] [Google Scholar]
- 5.Clayton D.A. (1982) Replication of animal mitochondrial DNA. Cell, 28, 693–705. [DOI] [PubMed] [Google Scholar]
- 6.Yang M.Y., Bowmaker,M., Reyes,A., Vergani,L., Angeli,P., Gringeri,E., Jacobs,H.T. and Holt,I.J. (2002) Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell, 111, 495–505. [DOI] [PubMed] [Google Scholar]
- 7.Bowmaker M., Yang,M.Y., Yasukawa,T., Reyes,A., Jacobs,H.T., Huberman,J.A. and Holt,I.J. (2003) Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem., 278, 50961–50969. [DOI] [PubMed] [Google Scholar]
- 8.Larsson N.G., Wang,J., Wilhelmsson,H., Oldfors,A., Rustin,P., Lewandoski,M., Barsh,G.S. and Clayton,D.A. (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genet., 18, 231–236. [DOI] [PubMed] [Google Scholar]
- 9.Wang J.M., Wilhelmsson,H., Graff,C., Li,H., Oldfors,A., Rustin,P., Bruning,J.C., Kahn,C.R., Clayton,D.A., Barsh,G.S., Thoren,P. and Larsson,N.G. (1999) Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nature Genet., 21, 133–137. [DOI] [PubMed] [Google Scholar]
- 10.Li H., Wang,J., Wilhelmsson,H., Hansson,A., Thoren,P., Duffy,J., Rustin,P. and Larsson,N.G. (2000) Genetic modification of survival in tissue-specific knockout mice with mitochondrial cardiomyopathy [In Process Citation]. Proc. Natl Acad. Sci. USA, 97, 3467–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wredenberg A., Wibom,R., Wilhelmsson,H., Graff,C., Wiener,H.H., Burden,S.J., Oldfors,A., Westerblad,H. and Larsson,N.G. (2002) Increased mitochondrial mass in mitochondrial myopathy mice. Proc. Natl Acad. Sci. USA, 99, 15066–15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekstrand M.I., Falkenberg,M., Rantanen,A., Park,C.B., Gaspari,M., Hultenby,K., Rustin,P., Gustafsson,C.M. and Larsson,N.G. (2004) Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet., 13, 935–944. [DOI] [PubMed] [Google Scholar]
- 13.Shadel G.S. and Clayton,D.A. (1997) Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem., 66, 409–435. [DOI] [PubMed] [Google Scholar]
- 14.Takamatsu C., Umeda,S., Ohsato,T., Ohno,T., Abe,Y., Fukuoh,A., Shinagawa,H., Hamasaki,N. and Kang,D. (2002) Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep., 3, 451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gensler S., Weber,K., Schmitt,W.E., Perez-Martos,A., Enriquez,J.A., Montoya,J. and Wiesner,R.J. (2001) Mechanism of mammalian mitochondrial DNA replication: import of mitochondrial transcription factor A into isolated mitochondria stimulates 7S DNA synthesis. Nucleic Acids Res., 29, 3657–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garstka H.L., Schmitt,W.E., Schultz,J., Sogl,B., Silakowski,B., Perez-Martos,A., Montoya,J. and Wiesner,R.J. (2003) Import of mitochondrial transcription factor A (TFAM) into rat liver mitochondria stimulates transcription of mitochondrial DNA. Nucleic Acids Res., 31, 5039–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enriquez J.A., Fernandez,S.P., Perez,M.A., Lopez,P.M. and Montoya,J. (1996) The synthesis of mRNA in isolated mitochondria can be maintained for several hours and is inhibited by high levels of ATP. Eur. J. Biochem., 237, 601–610. [DOI] [PubMed] [Google Scholar]
- 18.Rittner K., Stöppler,H., Pawlita,M. and Sczakiel,G. (1991) Versatile eucaryotic vectors for strong and constitutive transient and stable gene expression. Methods Mol. Cell. Biol., 2, 176–181. [Google Scholar]
- 19.Gossen M. and Bujard,H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber K., Ridderskamp,D., Alfert,M., Hoyer,S. and Wiesner,R.J. (2002) Cultivation in glucose-deprived medium stimulates mitochondrial biogenesis and oxidative metabolism in HepG2 hepatoma cells. Biol. Chem., 383, 283–290. [DOI] [PubMed] [Google Scholar]
- 21.Wiesner R.J., Kurowski,T.T. and Zak,R. (1992) Regulation by thyroid hormone of nuclear and mitochondrial genes encoding subunits of cytochrome-c oxidase in rat liver and skeletal muscle. Mol. Endocrinol., 6, 1458–1467. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual., 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 23.Attardi G. and Montoya,J. (1983) Analysis of human mitochondrial RNA. Meth. Enzymol., 97, 435–469. [DOI] [PubMed] [Google Scholar]
- 24.King M.P. and Attardi,G. (1989) Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science, 246, 500–503. [DOI] [PubMed] [Google Scholar]
- 25.Brown T.A. and Clayton,D.A. (2002) Release of replication termination controls mitochondrial DNA copy number after depletion with 2′,3′-dideoxycytidine. Nucleic Acids Res., 30, 2004–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seidel-Rogol B.L. and Shadel,G.S. (2002) Modulation of mitochondrial transcription in response to mtDNA depletion and repletion in HeLa cells. Nucleic Acids Res., 30, 1929–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly D.P. and Scarpulla,R.C. (2004) Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev., 18, 357–368. [DOI] [PubMed] [Google Scholar]
- 28.Wiesner R.J. (1997) Adaptation of mitochondrial gene expression to changing cellular energy demands. News Physiol. Sci., 12, 178–184. [Google Scholar]
- 29.Fisher R.P., Parisi,M.A. and Clayton,D.A. (1989) Flexible recognition of rapidly evolving promoter sequences by mitochondrial transcription factor 1. Genes Dev., 3, 2202–2217. [DOI] [PubMed] [Google Scholar]
- 30.Dairaghi D.J., Shadel,G.S. and Clayton,D.A. (1995) Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim. Biophys. Acta, 1271, 127–134. [DOI] [PubMed] [Google Scholar]
- 31.Larsson N.G., Oldfors,A., Holme,E. and Clayton,D.A. (1994) Low levels of mitochondrial transcription factor A in mitochondrial DNA depletion. Biochem. Biophys. Res. Commun., 200, 1374–1381. [DOI] [PubMed] [Google Scholar]
- 32.Ghivizzani S.C., Madsen,C.S. and Hauswirth,W.W. (1993) In organello footprinting. Analysis of protein binding at regulatory regions in bovine mitochondrial DNA. J. Biol. Chem., 268, 8675–8682. [PubMed] [Google Scholar]
- 33.Ghivizzani S.C., Madsen,C.S., Nelen,M.R., Ammini,C.V. and Hauswirth,W.W. (1994) In organello footprint analysis of human mitochondrial DNA: human mitochondrial transcription factor A interactions at the origin of replication. Mol. Cell. Biol., 14, 7717–7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cantatore P., Daddabbo,L., Fracasso,F. and Gadaleta,M.N. (1995) Identification by in organello footprinting of protein contact sites and of single-stranded DNA sequences in the regulatory region of rat mitochondrial DNA. Protein binding sites and single-stranded DNA regions in isolated rat liver mitochondria. J. Biol. Chem., 270, 25020–25027. [DOI] [PubMed] [Google Scholar]
- 35.Micol V., Fernandez,S.P. and Attardi,G. (1997) Functional analysis of in vivo and in organello footprinting of HeLa cell mitochondrial DNA in relationship to ATP and ethidium bromide effects on transcription. J. Biol. Chem., 272, 18896–18904. [DOI] [PubMed] [Google Scholar]
- 36.Fisher R.P., Lisowsky,T., Breen,G.A. and Clayton,D.A. (1991) A rapid, efficient method for purifying DNA-binding proteins. Denaturation-renaturation chromatography of human and yeast mitochondrial extracts. J. Biol. Chem., 266, 9153–9160. [PubMed] [Google Scholar]
- 37.Antoshechkin I. and Bogenhagen,D.F. (1995) Distinct roles for two purified factors in transcription of Xenopus mitochondrial DNA. Mol. Cell Biol., 15, 7032–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee D.Y. and Clayton,D.A. (1998) Initiation of mitochondrial DNA replication by transcription and R-loop processing. J. Biol. Chem., 273, 30614–30621. [DOI] [PubMed] [Google Scholar]
- 39.Wrutniak C., Cassar,M.I., Marchal,S., Rascle,A., Heusser,S., Keller,J.M., Flechon,J., Dauca,M., Samarut,J., Ghysdael,J. and Cabello,G. (1995) A 43-kDa protein related to c-Erb A alpha 1 is located in the mitochondrial matrix of rat liver. J. Biol. Chem., 270, 16347–16354. [DOI] [PubMed] [Google Scholar]
- 40.Casas F., Rochard,P., Rodier,A., Cassar-Malek,I., Marchal-Victorion,S., Wiesner,R.J., Cabello,G. and Wrutniak,C. (1999) A variant form of the nuclear triiodothyronine receptor c-ErbAalpha1 plays a direct role in regulation of mitochondrial RNA synthesis. Mol. Cell Biol., 19, 7913–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enriquez J.A., Fernandez,S.P., Garrido,P.N. and Montoya,J. (1999) Direct regulation of mitochondrial RNA synthesis by thyroid hormone. Mol. Cell. Biol., 19, 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.