Abstract
We report here on the identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Immunocytochemical experiments confirm that the enzyme indeed localizes to mitochondrial compartment. Inhibition of expression of the enzyme by RNA interference results in significant shortening of the poly(A) tails of the mitochondrial ND3, COX III and ATP 6/8 transcripts, suggesting that the investigated protein represents a bona fide mitochondrial poly(A) polymerase. This is in agreement with our sequencing data which show that poly(A) tails of several mitochondrial messengers are composed almost exclusively of adenosine residues. Moreover, the data presented here indicate that all analyzed mitochondrial transcripts with profoundly shortened poly(A) tails are relatively stable, which in turn argues against the direct role of long poly(A) extensions in the stabilization of human mitochondrial messengers.
INTRODUCTION
Polyadenylation of messenger RNAs plays important, yet diverse, role in RNA turnover in different biological systems. For nuclear-encoded mRNAs of eukaryotes, the presence of the poly(A) tail stabilizes the transcript, and promotes its nuclear export and translation (1,2). In contrast to this, polyadenylation in bacteria and chloroplasts facilitates the decay of RNA (2–6). Neither occurrence nor the physiological roles of polyadenylation have been conserved between mitochondria of different organisms. In mitochondria of the protist Trypanosoma brucei, most mRNAs contain either short (∼20 nt) or long (120–200 nt) poly(A) tails (7). The function of the long tails is not clear yet, while the presence of the short poly(A) tails seems to promote RNA degradation (8). In plant mitochondria, although polyadenylation is not constitutive with respect to mature messengers, it initiates exonucleolytic degradation of the RNA (9–12). A notable exception in the emerging view of diverse mechanisms of mitochondrial RNA turnover is yeast, where RNA is not polyadenylated at all; instead the DNA-encoded AU-rich dodecamer sequence at the 3′ ends ensures stability and translatability (13).
In human mitochondria, the large transcript encompassing almost the whole mt genome is processed by endonucleolytic cleavage, and subsequently the mRNAs are polyadenylated (14–17). The addition of poly(A) tails to mammalian mitochondrial mRNAs generates proper stop codons, that are absent in some messengers (14,15). However, no detailed studies on the influence of polyadenylation on mitochondrial transcript stability were published to date.
Polyadenylation in different biological systems can be performed by two classes of enzymes. One of them is poly(A) polymerase, which is able to synthesize homopolymeric poly(A) tails, like in the case of eukaryotic nucleus, cytoplasm and in prokaryotes in the exponential phase of growth, as well as in green algae chloroplasts (18–27). Another enzyme possessing polyadenylating activity is the polynucleotide phosphorylase (PNPase) (28,29). Poly(A) tails created by PNPase are heteropolymeric and besides adenosine residues contain a significant fraction of other nucleotides. This is the case of cyanobacteriae, chloroplasts of higher plants and prokaryotes (23,30–32). In plant mitochondria, the polyadenylating enzyme is yet to be identified, but nearly homopolymeric composition of analyzed poly(A) tails indicates that PAP, rather than PNPase, might be engaged in this process (9–11,33,34). On the other hand, it was shown that maize mitochondrial mRNAs frequently terminate with CCA extensions, which could indicate the possible role of terminal nucleotidyltransferase in the posttranscriptional modification of 3′ ends (35).
In eukaryotic cells, several poly(A) polymerases (PAPs) have been described up to date, and it appears that they are the result of gene duplication during evolution (36). In human cells, three genes have been identified whose products display poly(A) polymerase activity (37). Two of them, PAPOLA and PAPOLG, are ubiquitously expressed. Protein product encoded by PAPOLA gene (PAPalpha) is localized only in the nuclear compartment, while PAPgamma, arising from PAPOLG expression displays dual nucleo-cytoplasmic localization (36,38). The third gene, PAPOLB, encodes PAPbeta protein, whose expression is tissue specific, and it functions exclusively in testes (39,40). In addition, the poly(A) polymerase active in the process of tumorigenesis has been recently discovered, displaying 71% homology with respect to PAPalpha (41). Furthermore, human cells contain an enzyme adenylating SRP, U2 snRNA as well as 5S rRNA particles (42). However, no data have hitherto emerged that would indicate a putative protein that could be responsible for the addition of poly(A) tails at the 3′ ends of human mitochondrial messengers. None of the above-mentioned proteins possesses the mitochondrial targeting sequence.
A novel family of cytoplasmic poly(A) polymerases has been recently discovered in Schizosaccharomyces pombe and Caenorhabditis elegans (43,44). Its first identified metazoan member, called GLD-2, is involved in the control of germline development and embryogenesis (45). It has been reported that GLD-2 alone possesses low poly(A) polymerase activity, which is considerably stimulated by another developmental regulator, GLD-3, belonging to the bicaudal-C family of RNA-binding proteins (44,46). While canonical PAPs are monomeric, GLD-2 requires an RNA-binding partner to accomplish its function, and it is probably due to the fact that it does not contain an apparent RNA recognition motif (RRM)-like domain. It has been postulated that GLD-3 may target poly(A) polymerase activity of GLD-2 to specific mRNAs (44). Five human orthologs of the GLD-2 protein named Hs1 to Hs5 were recently described by Kwak et al. (47); only one of them (Hs1) displayed a poly(A) polymerase activity resembling that of the C.elegans enzyme while studied in Xenopus oocyte assay system.
Mammalian mitochondrial poly(A) polymerase was characterized biochemically more than 30 years ago (48–50). Purified enzyme catalyzed the incorporation of ATP into poly(A) and used ATP exclusively as a substrate (51). However, no molecular analysis of the enzyme has been performed. In view of those facts the question arises which gene could code for a human mitochondrial poly(A) polymerase.
We present here the identification of a novel human mitochondrial poly(A) polymerase. Our data indicate that long poly(A) tails are not required for stability of the mitochondrial transcripts.
MATERIALS AND METHODS
Cell cultures
Human HeLa and simian COS-1 cell lines were grown as monolayer at 37°C in a 5% CO2 humidified atmosphere in DMEM (Sigma) supplemented with 10% fetal bovine serum (FCS), 4 mM glutamine with or without antibiotics (Penicillin–Streptomycin; Sigma).
Determination of the sequence of the 3′ ends of mitochondrial RNAs by circularization assay (CR–RT–PCR)
The experimental procedure described by Kuhn and Binder (52) was used with minor modifications. In brief, 10 μg of RNA was circularized with T4 RNA ligase in a total volume of 30 μl, following recommendations of the manufacturer of the enzyme (Fermentas). Resulting circular RNA was submitted to the standard phenol–chloroform extraction, precipitated with ethanol and resuspended in 10 μl of distilled water. Reverse transcription was carried out using half of the self-ligated RNA (usually ∼3 μg), 10 pmol of the appropriate primer (for sequences and positions of the respective primers in human mitochondrial DNA refer to the Supplementary Material 1) and RevertAid™ M-MuLV Reverse Transcriptase (Fermentas). The first-strand cDNA (0.5 μl of the reaction mixture above) was submitted to PCR with external primers (Supplementary Material 1) flanking the 3′→5′ end joining site of the particular RNA. The PCR product was diluted 50-fold and employed in the second round of PCR by using nested primers (Supplementary Material 1). The PCR products were analyzed by 2.5% agarose gel electrophoresis and cloned into pGEM-T®-Easy Vector (Promega). Plasmids with inserts were submitted to sequencing with nested primers (DNA Sequencing Laboratory IBB PAS).
In silico analyses
BLAST searches were performed using protein–protein BLAST available at the NCBI (www.ncbi.nlm.nih.gov/BLAST) and Expasy (http://ca.expasy.org/tools/blast) servers. The presence of mitochondrial targeting sequence was checked with the use of following programs: Mitoprot II 1.0a4 (http://ihg.gsf.de/ihg/mitoprot.html), TargetP V1.0 (www.cbs.dtu.dk/services/TargetP), SignalP v1.1 (www.cbs.dtu.dk/services/SignalP-1.1), PSORT, PSORTII, iPSORT (all at the www.psort.org) and Predotar v1.03 (http://genoplante-info.infobiogen.fr/predotar/predotar.html). Protein analysis was carried out using InterProScan tool at the EBI server (www.ebi.ac.uk/InterProScan). Pairwise BLAST of mtPAPs was carried out with bl2seq tool from the NCBI server (see above). Sequence alignment was carried out using ClustalW program available at the EBI server (www.ebi.ac.uk/clustalw).
Cloning of the full-length hmtPAP open reading frame
The full-length hmtPAP open reading frame (ORF) was cloned by screening the available human cDNA library constructed from HeLa D98-H cells (53). About 75 000 clones were divided into 80 pools by seeding into 96 well plates and tested for the presence of hmtPAP ORF by PCR amplification using DNA isolated from the pools and oligonucleotides PAPL (5′-atatggatccaATGGCGGTTCCCGGCGTGG-3′; BamHI restriction site is bolded and in lowercase; region complementary to the 5′ end of the hmtPAP ORF is in uppercase) and PAPR (5′-tatagtcgacTGTCTGAGTACTAATTGTTCTC-3′; SalI restriction site is bolded and in lowercase; region complementary to the 3′ end of the hmtPAP is in uppercase) as forward and reverse primers, respectively. Product of correct length (1759 bp) was obtained from one pool of plasmids. DNA band was extracted from agarose, cloned into the BamHI and SalI sites of the pBluescript® II KS (+) vector (Stratagene) and sequenced. The differences in the sequence with respect to databases were corrected by site-directed mutagenesis.
Protein localization using immunofluorescence
The plasmid used for subcellular localization of C-terminally c-myc-tagged hmtPAP in COS-1 and HeLa cell lines was constructed as follows: hmtPAP ORF was PCR amplified using forward primer (5′-atagaattcATGGCGGTTCCCGGCGTGGG-3′; region complementary to hmtPAP ORF is in uppercase) introducing an EcoRI site (bolded in lowercase) and the reverse primer (5′-ataggatccCTACAGGTCCTCCTCGGAGATCAGCTTCTGCTGTGTCTGAGTACTAATTGTTC-3′; sequence complementary to hmtPAP ORF is in uppercase), introducing a c-myc (9E10) epitope coding sequence (italic uppercase), artificial termination codon UAG (underlined uppercase) and BamHI site (bolded lowercase). The resulting PCR product was first cloned into pGEM-T®-Easy Vector (Promega), and then recloned into the EcoRI and BamHI sites of the pcDNA3.1(−) vector (Stratagene).
Samples of 2 × 105 cells were seeded per well in 6 well plates (Falcon) onto a coverslip placed at the bottom of the well. The cells were transiently transfected with plasmid DNA using FuGene6 reagent (Roche). Twenty four hours after transfection, Mitotracker CMX-Ros (300 nM) was added to the medium and incubated at 37°C for 45 min. Afterwards, the cells were washed three times with phosphate-buffered saline (PBS) and fixed with 4% ice-cold formaldehyde in PBS for 15 min at room temperature. Then the cells were permeabilized with 1% ice-cold Triton X-100 in PBS or 5 min at room temperature and incubated with 10% FCS in PBS for 1 h. The detection of c-myc-tagged protein was performed using anti-c-myc 9E10 monoclonal antibody (Santa Cruz Biotechnology) (diluted 1:100 in PBS with 10% FCS; 1 h at room temperature), followed by incubation with fluorescein 5-isothiocyanate (FITC)-conjugate goat anti-mouse immunoglobulin G (Molecular Probes) (diluted 1:200 in PBS with 10% FCS; 1 h at room temperature). Fluorescent signals were examined using either fluorescent or confocal microscope.
RNAi experiments
siRNA against hmtPAP were obtained by digestion of long RNA product of in vitro transcription. Templates for RNA synthesis corresponding to the almost full-length hmtPAP ORF (nucleotides 20–1620) or its left (nucleotides 20–947) or right (nucleotides 927–1620) part were obtained by PCR amplifications using plasmid pairs of oligonucleotides PAPT7LE (5′-GAATTAATACGACTCACTATAGGGAGAGGCTCTTGACCCGTTTGAAC-3′) and PAPT7RE (5′-GAATTAATACGACTCACTATAGGGAGAGGACTTTCTGTTTGGAGCAG-3′), PAPT7LE and PAPT7RI (5′-GAATTAATACGACTCACTATAGGGAGAGAGAACCTCACGAGCGGACA-3′) or PAPT7LI (5′-GAATTAATACGACTCACTATAGGGAGAGTGTCCGCTCGTGAGGTTCT-3′) and PAPT7RE, respectively, as primers. Each primer comprised T7 promoter sequence (italics underlined), followed by the fragment complementary to the hmtPAP coding sequence. The PCR products were used for in vitro transcription with T7 RNA polymerase (Fermentas). Double-stranded RNA (dsRNA) was then purified by standard phenol–chloroform extraction and precipitated with isopropanol.
siRNAs were generated by incomplete digestion with Escherichia coli RNase III (New England BioLabs) or by initial cleavage of long dsRNA with RNase III to the fragments of ∼100–400 bp which were subsequently digested with the recombinant Dicer enzyme (Stratagene) into short 21–23 nt siRNAs. siRNAs were obtained by separation of the digestion mixture on 12% native polyacrylamide gel, cutting out the slice of the gel corresponding to appropriate size, elution of RNA from the polyacrylamide by incubation with 1.5 M ammonium acetate overnight at 37°C and recovering by isopropanol precipitation.
siRNAs were transfected into human HeLa cell with the use of Oligofectamine™ Reagent (GibcoBRL), according to the manufacturer's instructions. Seventy two hours after transfection, the cells were collected and RNA was then isolated with TRI Reagent (Sigma), according to the manufacturer's instructions.
Northern blots
For standard northern blot ∼5 μg of total RNA were dissolved in 1× NBC buffer (50 mM boric acid, 1 mM sodium acetate, 5 mM NaOH), containing 5.6% formaldehyde and 50% formamide, heat-denatured for 7 min at 65°C, mixed with the appropriate volume of 10× loading dye (15% Ficoll, 0.25% bromophenol blue, 0.25% xylenecyanol in 0.1 M EDTA, pH = 8.0) and run on 1% denaturing agarose–formaldehyde gel in 1× NBC. High Range RNA Ladder (Fermentas) was used as a marker. Following electrophoresis, RNA was blotted onto Nytran-N membrane (Schleicher & Schuell) by overnight capillary transfer in 20× SSC (3 M sodium chloride, 0.3 M sodium citrate). Filter was then washed with 2× SSC and the RNA was immobilized by ultraviolet-crosslinking. Transfer efficiency was monitored by staining the filter with 0.03% methylene blue in 0.3 M sodium acetate, pH = 5.2. For high-resolution northern blot, 5 μg of total RNA in 50% formamide were run in 4–5% denaturing acrylamide–urea gel in 0.5× TBE. GeneRuler™ 100 bp DNA Ladder Plus (Fermentas) was applied as a marker. Nucleic acids were then blotted onto Nytran-N filter by electrotransfer in 0.5× TBE buffer using Trans-Blot Cell apparatus (Bio-Rad). For expression profile analysis, Human Multiple Tissue Northern Blot (Clontech) was used. All hybridizations were performed in PerfectHyb™ Plus buffer (Sigma). PCR products corresponding to internal regions of appropriate ORFs and GeneRuler™ 100 bp DNA Ladder Plus were labeled with [α-32P]ATP using HexaLabel DNA Labeling Kit (Fermentas) and used as probes. Where necessary, membranes were stripped with the use of boiling 0.1% SDS, and re-hybridized with new probe. Following hybridization, filters were exposed to either Kodak BioMax films using intensifying screens (Sigma), or to the PhosphorImager screens. Results were obtained by developing the films in Curix 60 developer (AGFA) or by scanning the PhosphorImager screens using Storm Scanner (Molecular Dynamics), and analyzed with the use of ImageQuant.
Real-time PCR
Approximately 5 μg of each total RNA sample isolated from human HeLa cells were treated with DNase I (Fermentas), according to the manufacturer's instructions. DNA-free RNA served subsequently as a template for reverse transcription, which was performed with the use of oligo-(dT)18 primer and RevertAid™ M-MuLV Reverse Transcriptase (Fermentas). First-strand cDNA was then used in real-time PCR reaction, which was carried out using LightCycler (Roche) and QuantiTect SYBR Green PCR Kit (Qiagen). Primers for B2M, GAPDH and β-actin, described by Vandesompele et al. (54), were applied for standardization. Primers PAPREALL (5′-CTATTGCTACCATCTGCTCCAA-3′) and PAPREALR (5′-ATTGTTCTCTTCCCACTGGTTT-3′) were used for the determination of hmtPAP mRNA levels. All primers were used at the final concentration of 0.5 μM. All real-time amplifications were performed in the presence of 4 mM Mg2+. The cycling conditions comprised 15 min polymerase activation at 95°C and 45 cycles: 95°C for 15 s, 55°C for 15 s (slope 2°C/s) and 72°C for 15 s. Each assay included three serial dilutions of tested cDNAs and no-template negative control.
RESULTS
Sequence analysis of the 3′ ends of selected human mitochondrial RNAs
Nucleotide composition of poly(A) tails of human mitochondrial transcripts may indicate what kind of enzyme is responsible for their synthesis, as enzymes other than poly(A) polymerases displaying polymerization activity (such as PNPase) frequently incorporate nucleotides other than adenosine (55).
In order to precisely determine nucleotide sequence of the 3′ ends of the selected human mitochondrial RNAs, we employed the technique that was previously successfully applied for the analysis of the editing changes at the extremities of land snail mitochondrial tRNAs, sequences of the 5′ and 3′ termini of pea mitochondrial cox2 mRNA, and most recently to mapping of the 3′ ends of Arabidopsis thaliana atp9 transcripts (33,52,56).
Circularized human mitochondrial RNAs were submitted to reverse transcription followed by two rounds of PCR amplification with the use of specific primers enabling the examination of the joining site of the termini of the selected human mitochondrial RNAs. By that means, we managed to obtain a large number of clones corresponding to ND3 mRNA, CYTB mRNA, ATP6/8 mRNA and COXIII mRNA, encoding subunits of different oxidative chain complexes. In order to document the fidelity of the technique, we analyzed simultaneously cloned cDNAs corresponding to 12S rRNA, which in human mitochondria are known to contain one or two adenosine residues at their 3′ ends (57,58). The prediction of the lengths of the final inserts was carried out with the assumption that poly(A) tail size of mitochondrial mRNAs should be ∼50–60 bp, while 12S rRNA should not contain poly(A) extension [(57,58), unpublished data]. For all mRNAs except CYTB the observed lengths were in agreement with the presence of ∼60 bp long poly(A) tail (Supplementary Material 1; data not shown).
We analyzed in total 51 independent clones corresponding to five different mitochondrial RNAs with respect to the sequence of their 3′ extensions. The results of this analysis are summarized in Table 1. The length of poly(A) tails of all tested mitochondrial mRNAs varied from 10 to 62, with the average of 43 bp (±14) [excluding clones for 12S rRNA with short tails and three clones for CYTB, that lacked poly(A) tails, probably due to mRNA degradation]. As expected, the 3′ terminus of 12S rRNA contained oligo(A) tails comprising only 1–2 adenosine residues, which is in concordance with the data from literature, and confirms the accuracy of the selected method.
Table 1. Results of sequence analysis of the 3′ ends of selected mitochondrial RNAs.
| Mitochondrial RNA | Number of clones analyzed | Analysis of the poly(A) tails | ||
|---|---|---|---|---|
| The sequence of the 3′ end | Average length | Ratio A:G/C | ||
| ND3 | 6 | (A)18; (A)33; (A)45; (A)47; (A)58 | 40 ± 14 | 238:1 |
| (A)14 G(A)23 | ||||
| ATP6/8 | 17 | (A)13; (A)16; (A)29; (A)34; (A)42; (A)44; (A)45; (A)46; (A)48; (A)50; (A)51; (A)62; | 42 ± 13 | 709:10 |
| (A)8 C(A)34 | ||||
| (A)9 C(A)38 | ||||
| (A)13 CC(A)38 | ||||
| (A)39 CC(A)5 | ||||
| (A)25 CCAGG(A)19 | ||||
| COXIII | 17 | (A)10; (A)14; (A)17; (A)49; (A)49; (A)50; (A)51; (A)51; (A)52; (A)53; (A)56; | 47 ± 16 | 801:0 |
| (A)56; (A)58; (A)58; (A)58; (A)58; (A)61 | ||||
| CYTB | 6 | (A)0; (A)0; (A)0; | a | |
| (A)29; (A)36; (A)24 C(A)19 | 36 ± 8 | 108:1 | ||
| 12S rRNA | 5 | (A)1; (A)1; (A)1; (A)2; (A)2 | a | 7:0 |
| Total | 51 | 43 ± 14 | 1863:12 |
(A)n indicates the stretch of n adenines.
(A)0 means no poly(A) tail; the clones probably represent degradation intermediates as they lack mtDNA-encoded nucleotides at their 3′ termini.
aNot taken into account during calculations.
Our results indicate that poly(A) extensions at the 3′ ends of human mitochondrial messengers are essentially homopolymeric. We found only 12 residues other than adenosine (G or C) among 1875 that have been sequenced. Their presence might be explained as the result of amplification error or an activity of an enzyme other than poly(A) polymerase. In plant chloroplasts, where PNPase is the only polyadenylating enzyme, the 3′-extensions contain 30% non-A nucleotides (55). In the case of E.coli, where both PAP and PNPase are active, probably resembling the situation in human mitochondria, the content of nucleotides other than adenosine may vary from 0% (exponential phase of growth) up to 17% (stationary growth) (23,32). In our study, the inclusion of non-A residues was very low (0.6%). This almost 100% homopolymeric composition of the poly(A) tails indicated that neither PNPase nor the CCA adding enzyme, which had been localized in human mitochondria (59,60), was likely to be responsible for the polyA tail synthesis in this organelle. This prompted us to perform an in silico search for the gene encoding a putative human mitochondrial poly(A) polymerase.
In silico identification and analysis of the putative human mitochondrial poly(A) polymerase
None of the three known human poly(A) polymerases, such as PAPalpha, PAPgamma or PAPbeta, possesses a mitochondrial targeting sequence. Therefore, we decided to perform BLAST search of the human genome databases with the first member of the recently discovered family of cytoplasmic poly(A) polymerases, S.pombe Cid13 protein (43).
BLAST search of genome databases using the sequence of S.pombe Cid13 protein as a query resulted in finding of at least five human orthologs, which were independently identified in the recent report concerning the analysis of C.elegans GLD-2 homologs by Kwak et al. (47).
We found that one of these proteins, identical to Hs4 reported previously (47) (accession nos: BAB13981and Q9HA74), was strongly predicted to be imported into the mitochondrial compartment by the following programs: MitoProt II 1.0a4 (score 0.9853), TargetP V1.0 (score 0.914) and PSORT (score 0.73) (61–63). Putative mitochondrial localization of this protein was also indicated with slightly lower probability by iPSORT, PSORTII, Predotar v1.03 and SignalP v1.1 programs (64–66) (http://www.inra.fr/predotar/). Moreover two of the programs, MitoProt II 1.0a4 and SignalP v1.1, detected exactly the same position of the potential cleavage site, located between amino acids 37 and 38. It is worth noting that the results presented above resembled those obtained for the two other proteins investigated in our laboratory, namely hSUV3 and hPNPase, where localization in mitochondria was verified experimentally (60,67). The putative human mitochondrial poly(A) polymerase thus identified will be hereby referred to as hmtPAP.
The gene coding for the putative hmtPAP is localized at chromosome 10 (band 10p12.1). We analyzed the genomic organization of this gene by comparing an available sequence of human chromosome 10 genomic contig (accession no. NT_008705) with the longest cDNA sequence (2571 bp) corresponding to the examined protein (accession no. AK022188). The gene spans over the region of ∼35.5 kb and consists of nine exons, divided by eight introns. All exon/intron junctions fulfill the criteria of GT-AG rule (Supplementary Material 2).
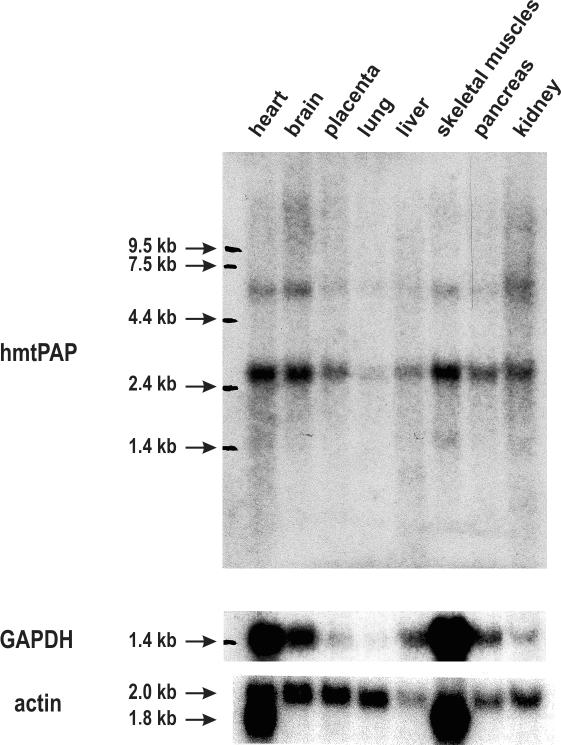
The gene coding for the putative hmtPAP is expressed in humans, as confirmed by the availability of numerous ESTs in databases (data not shown). Furthermore, we were able to detect an mRNA of length ∼2.6 kb by performing hybridization to Human Multiple Tissue Northern Blot, containing poly(A)+ mRNA samples isolated from eight different human tissues, using PCR product corresponding to the ORF of hmtPAP as a probe and β-actin as a standard to normalize RNA input (Figure 1). The value of 2.6 kb is very close to that of the above-mentioned cDNA sequence. As can be seen in the Figure 1, the hmtPAP mRNA is expressed in all analyzed tissues, nonetheless the levels of its expression vary considerably between tissues when compared with actin mRNA expression. The highest level of expression is observed in the tissues displaying high energy requirements: heart, brain as well as skeletal muscles. The pattern of hmtPAP expression in various tissues resembles that which is characteristic for another mitochondrial protein under examination in our laboratory, namely hSUV3 (68), as well as for the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (Figure 1). Interestingly, a fainter hybridization signal was observed, representing a larger RNA species of length ∼6.0 kb. It could rather be excluded that this band resulted from unspecific hybridization, as the pattern of its expression displays significant similarity to that of the main hmtPAP mRNA species recognized by the probe corresponding to the full-length ORF. The nature and identity of the larger RNA remain to be investigated. It is likely that it could contain a longer 3′-untranslated region, as in the case of human PNPase mRNA (69,70).
Figure 1.
Analysis of hmtPAP expression in eight normal human tissues. Human Multiple Tissue Northern Blot (Clontech) was employed to analyze the pattern of hmtPAP gene expression in various human tissues. Sources of the samples are indicated above all lanes. Upper panel shows that the hmtPAP probe (DNA corresponding to the full-length ORF) recognizes two transcripts; the 2.6 kb RNA species is much more abundant than the second ∼6.0 kb transcript, being in agreement with the size of the longest cDNA sequence available in databases (see text for details). The filter was stripped and reprobed with GAPDH cDNA as a probe. Results of this rehybridization are shown in the middle panel. Bottom panel presents the results of hybridization of the same filter with β-actin probe. Positions of the RNA molecular weight marker are indicated at the left side of each panel.
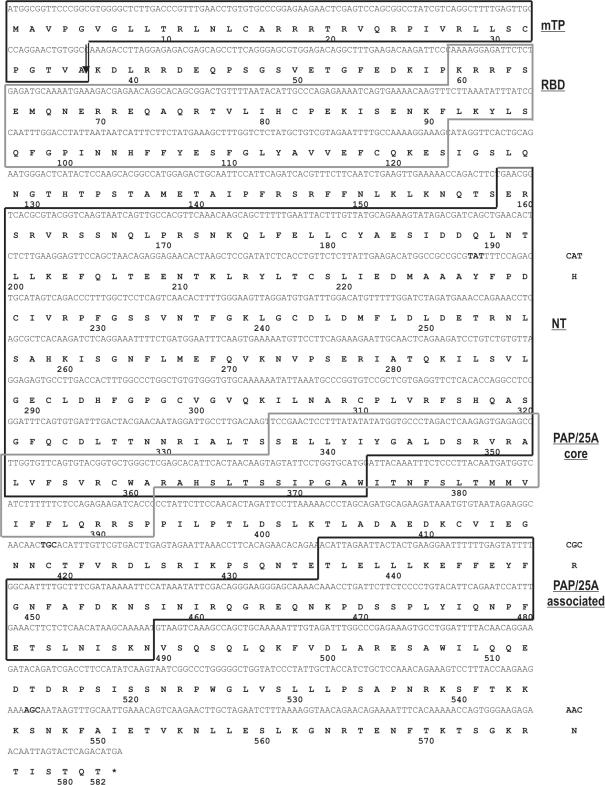
Members of the novel family of cytoplasmic poly(A) polymerases display relatively low level of homology with canonical PAPs and this homology is confined to the predicted catalytic domains (43). Kwak and coworkers (47) showed that GLD-2 and its homologs possess central and catalytic domains, resembling the classical PAPs; however, in contrast to them they lack C-terminal RRM. We performed our own search for the predictable conserved domains within the putative hmtPAP using InterProScan tool available at the EBI Server. Results of this analysis revealed that the protein encompasses several conserved domains, among which the most significant with respect to the its putative function were PAP/25A core domain (Interpro accession no. IPR001201), PAP/25A-associated domain (Interpro accession no. IPR002058), as well as two IPR non-integrated domains, SSF54928 RNA-binding domain (RBD) and SSF81301 nucleotidyltransferase domain, which partially overlaps with PAP/25A core domain. The complete coding sequence and the derived polypeptide sequence of the putative hmtPAP, together with its modular structure, are shown in Figure 2.
Figure 2.
Sequence of the putative hmtPAP and its domain organization. Presented are cDNA and amino acid sequences available in the human genomic databases under accession nos: AK022188 and Q9HA74, respectively. Protein sequence is shown below corresponding DNA coding sequence using one-letter code. Asterisk indicates termination codon. Numbering refers to the amino acid sequence. The coding region is 1746 nt long and corresponds to 582 amino acid residues. The positions of protein domains detected by using bioinformatics tools (see text for details) are indicated on the right as follows: mTP, mitochondrial targeting peptide; RBD, SSF54928 RNA binding domain; NT, SSF81301 nucleotidyltransferase domain; PAP/25A core, PAP/25A core domain; PAP/25 associated, PAP/25A-associated domain. Note that NT and PAP/25A core domains partially overlap. The most probable cleavage site of mTP is marked with an arrow. Three discrepancies between genomic and cDNA sequence are specified on the right.
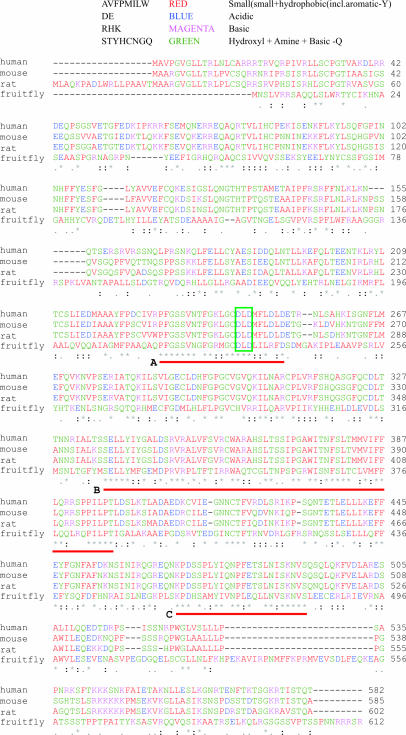
To verify whether our protein of interest might represent the first member of mitochondrially targeted subgroup of the novel family of poly(A) polymerases, we carried out another BLAST search using hmtPAP amino acid sequence as query. Our aim is to find its homologs among other model eukaryotic organisms that would possess in silico predictable mitochondrial transition signal at the N-terminus and comparable domain organization. We managed to detect proteins that fulfilled those two criteria in Mus musculus (mmtPAP), Rattus norvegicus (rmtPAP) and Drosophila melanogaster (fmtPAP) [accession nos: Q9D0D3 (BAB27689), XP_225468 and O46102 (NP_569904), respectively], but we were unable to confirm the presence of such orthologs in C.elegans or A.thaliana. The results of pairwise BLAST between hmtPAP and its mouse, rat and fruitfly counterparts are presented in Table 2, and their aligned sequences are shown in Figure 3. The only significant difference of mmtPAP in comparison with the human protein is the presence of three additional amino acid residues (PSS) inserted between N and Q in positions 155/156 in hmtPAP. rmtPAP is almost identical to the mouse protein, but it possesses slightly extended N-terminus (18 amino acids longer), which constitutes mitochondrial targeting sequence. Fruitfly ortholog displays the highest level of divergence, especially in terms of the composition of both N- and C- terminal parts. Alignment of all four sequences allows to distinguish three regions of highest homology marked A, B and C in Figure 3. Region A is located in the middle of putative nucleotidyltransferase domain and encloses conserved DXD motif, characteristic for members of the class II nucleotidyltransferase family, encompassing CCA-adding enzymes and eubacterial poly(A) polymerases (59). It is also worth noting that this motif together with one of glycine residues located upstream and other aspartates positioned downstream may consitute G-D-D-D signature sequence, characteristic of the nucleotidyltransferase superfamily (71,72). Region B corresponds almost perfectly to the putative PAP/25A core domain, while region C is localized within the putative PAP/25A-associated domain. The putative RBD, detected with the lowest probability by InterProScan tool, is conserved only between human, mouse and rat homologs, therefore its existence appears to be the most questionable. Nevertheless, general structural similarity of all mtPAPs is relatively high.
Table 2. Results of pairwise BLAST comparison between hmtPAP and its mouse, rat and fruitfly homologs.
| Identity | Homology | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Human | Mouse (%) | Rat (%) | Fruitfly (%) | Human | Mouse (%) | Rat (%) | Fruitfly (%) | ||
| Human | — | 72 | 72 | 31 | Human | — | 82 | 80 | 49 |
| Mouse | — | — | 87 | 31 | Mouse | — | — | 91 | 51 |
| Rat | — | — | — | 30 | Rat | — | — | — | 49 |
| Fruitfly | — | — | — | — | Fruitfly | — | — | — | — |
The left part shows the level of identity between analyzed proteins, while the right part presents the level of their mutual homology. Amino acid sequences of human, mouse, rat and fruitfly mtPAPs available under accession nos: Q9HA74, Q9D0D3, XP_225468 and NP_569904, respectively, were applied to the analysis.
Figure 3.
Alignment of amino acid sequences of hmtPAP and its closest orthologs. Protein sequences of human, mouse, rat and fruitfly mtPAPs were aligned using ClustalW program. Small and hydrophobic residues (AVFPMILW) are colored red, acidic residues (DE), blue; basic residues (RHK), magenta; and other residues (STYHCNGQ), green. Residues identical or conserved in all aligned sequences are marked with an asterisk; conserved substitutions, with a colon; and semi-conserved substitutions, with a dot. Three regions of highest homology are marked with red lines and designated with letters A, B and C. Conserved DXD motif is indicated with the green rectangle. For further details refer to text.
All proteins described above have very similar physico-chemical parameters, such as length (582–612 amino acids), molecular mass (65–68 kDa), isoelectric point (8,64–9,24), total number on negatively (55–61 amino acids) as well as positively (67–77 amino acids) charged residues, instability index, aliphatic index, etc. It is worth mentioning that the estimated molecular mass of the rmtPAP is in agreement with the biochemical data obtained in early 70's (48–50).
Cloning of the ORF coding for the putative hmtPAP
In order to clone the hmtPAP ORF, we took the advantage of possessing human cDNA library, which was previously successfully used in our laboratory for the isolation of full-length cDNA clone encoding human SUV3 protein (68). The library was divided into pools, which were screened for the presence of full-length hmtPAP ORF by PCR amplification, by using appropriate primers. We were able to limit the presence of the adequate clone to the single well (∼2000 clones) out of 384 subjected to the analysis. The hmtPAP ORF has been subsequently cloned into pBluescript® II KS (+) vector, and two out of the seven positive clones have been submitted to sequencing. One of the examined clones, named KS5, harbored two point mutations in comparison with the cDNA sequence present in databases (accession nos: FLJ12126 and AK022188), which were not the result of known discrepancies between genomic and mRNA sequence (Figure 2); transversion T1201G resulted in the change of serine in position 401 into alanine, and transversion A1513C caused replacing of serine in position 505 into arginine. Owing to this fact two rounds of site-directed mutagenesis were performed.
Experimental verification of mitochondrial localization of hmtPAP
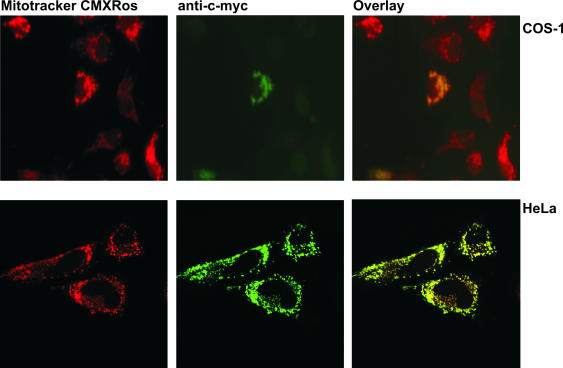
In order to determine the subcellular localization of hmtPAP within mammalian cells, we transiently transfected both human HeLa and simian COS-1 cells with the construct encompassing the full-length hmtPAP tagged on the C-terminus with the c-myc epitope 9E10. The subcellular distribution of such overexpressed fusion protein was then assessed by comparing the position of green fluorescent signal arising from FITC-conjugated secondary antibodies used in the last step of the detection of c-myc epitope, with the red fluorescence of the mitochondrial marker MitoTracker CMX-Ros. As can be seen in the Figure 4, both kinds of fluorescence showed almost perfect co-localization. It can therefore be concluded that the hmtPAP is indeed localized in mitochondrial compartment. Further studies are required to determine whether this localization is dependent upon the presence of the N-terminal, in silico predictable mitochondrial targeting sequence.
Figure 4.
Mitochondrial localization of hmtPAP in mammalian cells. COS-1 and HeLa cells were grown on coverslips and transiently transfected with pcDNA + hmtPAP plasmid, encoding c-myc-tagged hmtPAP. After incubation with the mitochondria-specific dye MitoTracker CMXRos (300 nM), fixation with 4% formaldehyde and permeabilization with 1% Triton X-100, the cells were immunostained with anti-c-myc monoclonal antibody, which was then visualized with FITC-conjugated antibody. Fluorescent images of MitoTracker (red) and c-myc-tagged hmtPAP (green) were taken by either a fluorescent (COS-1 cells) or a confocal (HeLa cells) microscope. Co-localization of hmtPAP and mitochondria appears yellow in digitally overlaid images.
RNA interference study of the hmtPAP function
In order to confirm that the in vivo role of hmtPAP is indeed the synthesis of poly(A) extensions at the 3′ ends of mitochondrial mRNAs, we decided to attempt an RNA interference experiment and see whether the ablation of hmtPAP expression would result in shortening of the poly(A) tail of the selected mitochondrial mRNAs.
It is often necessary to try several different siRNAs to exert the desired effect (73,74). Therefore, we decided to employ a modified approach described by Yang and co-workers (75–77), which relies on enzymatic generation of siRNAs from long dsRNAs corresponding to the mRNA target. By this means, we intended to produce a heterogeneous siRNA pool that could potentially target numerous sites per mRNA molecule, which should at least in theory increase the probability of successful inhibition of hmtPAP activity.
We generated three PCR products corresponding to the almost full-length hmtPAP ORF (PCR F), as well as its left (PCR L) and right (PCR R) part. They served as templates for the generation of relatively long double-stranded RNA (dsRNA F, dsRNA L and dsRNA R, respectively) by an RNA in vitro transcription. Those dsRNA molecules were initially cleaved with RNAse III and the resulting 100–300 nt long dsRNA species were further processed by recombinant DICER into 21–23 bp fragments and purified. Anti-hmtPAP siRNAs were then pooled and introduced into human HeLa cells by liposome-mediated transfection and after collection of the control (C) as well as siRNA-treated (RNAi) cells, we isolated total RNA from both samples.
In order to check whether our siRNAs exert the expected effect on hmtPAP expression, the level of siRNA-mediated repression was assayed by real-time PCR. The amount of hmtPAP mRNA remaining in siRNA-inhibited cells was 11–26% of the level observed in mock-treated cells, depending on which of the three reference genes (B2M, GAPDH or β-actin) was applied for standardization (data not shown).
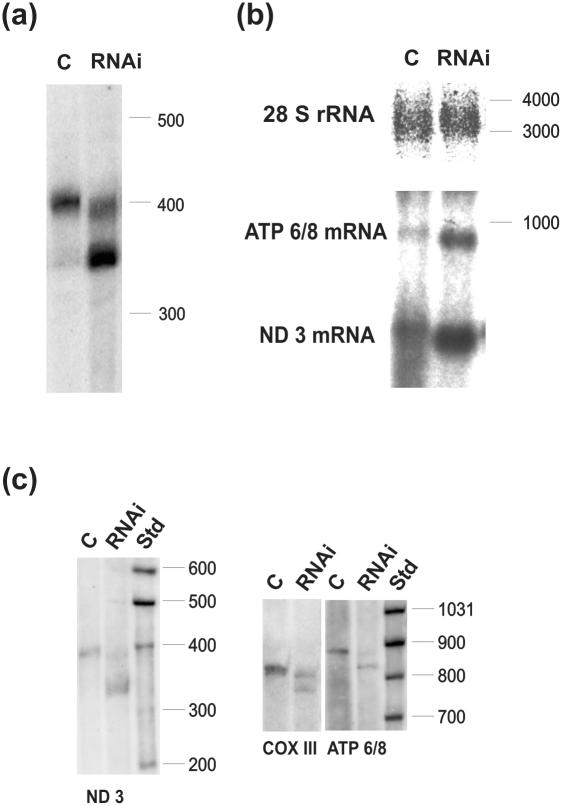
We started the analysis of the effects of hmtPAP silencing from the shortest mitochondrial mRNA species, ND3, using high-resolution northern blot. As can be seen in Figure 5a, the majority of the mature ND3 mRNA migrates much faster in the lane corresponding to the siRNA-treated cells (lane RNAi) when compared with control cells (lane C). The difference in size corresponds to ∼50 nt, which is in agreement with the lack of poly(A) tails in the lower fraction of ND3 mRNA. This result appears to confirm the hypothesis that hmtPAP participates in mitochondrial polyadenylation and that the ablation of its expression leads to the profound shortening of poly(A) extensions at the 3′ ends of mitochondrial messengers.
Figure 5.
Effects of RNA interference against hmtPAP on selected mitochondrial transcripts. (a) High-resolution northern blot for ND3 mRNA on samples isolated from control (lane C) and siRNA-treated (lane RNAi) human HeLa cells; (b) standard northern blot with the same amounts of RNA as in (a) probed with DNA fragments corresponding to ND3 and ATP 6/8 transcripts; 28S rRNA-specific probe was used as a loading control; (c) high-resolution northern blots of RNA samples derived from an independent RNAi experiment, probed with DNA fragments corresponding to for ND3, ATP6/8 and COX III. Molecular weight markers (Std) in nucleotides are indicated on the right. A total of 4 and 5% PAGE was used for high-resolution northern analysis of ND3 and other transcripts, respectively.
Quite unexpectedly, we found that the steady-state level of ND3 mRNA lacking the long poly(A) extension was relatively high. High-resolution northern blot was applied to visualize qualitative changes of transcript length, but it turned out not to be particularly quantitative in our hands. In order to demonstrate the quantitative change in the steady-state level of ND3 mRNA more accurately, exactly the same amounts of both analyzed samples were subjected to conventional northern blot analysis using large ribosomal cytoplasmic RNA as the loading control. By this approach, we were able to observe a significant increase in the level of ND3 following RNA interference against hmtPAP comparing to mock-treated cells. Similar result was also obtained for another mitochondrial mRNA, ATP6/8 (Figure 5b). Combining data gained from high-resolution and standard northern blots, it appears that the long poly(A) tails do not act directly as the determinants of mitochondrial mRNA stability.
We extended our analysis of the effects of siRNA-mediated suppression of hmtPAP expression using high-resolution northern blots for two other mitochondrial transcripts, COXIII and ATP 6/8. As this set of hybridizations was performed using RNA samples obtained from the experiment independent from the previous one, we repeated the analysis for ND3 mRNA as the positive control. The degree of silencing was similar in both experiments, as judged by real-time PCR results (data not shown). The respective northern blots are presented in Figure 5c. Results for COXIII and ATP 6/8 were comparable to those observed in the case of ND3 in terms of the appearance of a substantial fraction of shortened mRNA species after siRNA treatment. In all the cases, the observed difference in length between poly(A) and lower fractions was ∼50 nt, which is in agreement with the reported sizes of poly(A) tails of human mitochondrial mRNAs. The effect of hmtPAP silencing on mitochondrial mRNAs analyzed was slightly divergent when we looked at the ratios of longer:shorter fraction using ImageQuant. They were 25%:75%, 41%:59% and 56%:44% for ND3, ATP 6/8 and COXIII, respectively, and 90%:10% in the control cells. Nevertheless, the general trend of quantitative changes between two fractions was similar for all three messengers. It may be also noticed that in the siRNA-treated cells, the polyadenylated fraction migrates slightly faster than in the control cells.
In order to prove unequivocally that the lower, abundant mRNA fraction after inhibition of hmtPAP indeed results from shortening of the poly(A) tails of mitochondrial mRNAs and to study the shortened tail structure, we determined the sequence of the 3′-5′ joining sites for ND3 and ATP 6/8 mRNAs, employing the CR–RT–PCR procedure as described in Materials and Methods. The results are summarized in Table 3. We found that all sequences corresponded to mRNA species containing oligo(A) tails ranging from 1 to 17 residues in length with the average of 6 nt (±3). Moreover, sequencing data allowed us to indisputably exclude the possibility that shortening of ND3 and ATP6/8 transcripts observed in high-resolution northern blots is due to the truncations affecting coding sequence, as both ends of transcripts were intact in all cases. In conclusion, inhibition of hmtPAP activity in HeLa cells resulted in shortening of the poly(A) tails of the mitochondrial ND3 and ATP 6/8 mRNAs from about 43 residues on average to ∼6 nt. It is worth mentioning that in normal HeLa cells the fraction of RNA lacking the long poly(A) tails is also oligoadenylated (J. Piechota, personal communication).
Table 3. Results of sequence analysis of the 3′-extensions of ND3 and ATP 6/8 transcripts after RNA interference against hmtPAP.
| Mitochondrial mRNA | Total number of clones analyzed | Sequence of 3′-extension | Number of clones | Average length |
|---|---|---|---|---|
| ND3 | 33 | (A)2 | 4 | 6 ± 4 |
| (A)3 | 3 | |||
| (A)4 | 4 | |||
| (A)5 | 8 | |||
| (A)6 | 3 | |||
| (A)7 | 1 | |||
| (A)8 | 3 | |||
| (A)10 | 2 | |||
| (A)12 | 1 | |||
| (A)13 | 1 | |||
| (A)17 | 2 | |||
| (A)1C(A)2 | 1 | |||
| ATP 6/8 | 12 | (A)1 | 1 | 4 ± 2 |
| (A)3 | 2 | |||
| (A)4 | 5 | |||
| (A)5 | 2 | |||
| (A)6 | 1 | |||
| (A)7 | 1 | |||
| Total | 45 | — | 6 ± 3 |
The results presented above indicate that the long poly(A) tails are not indispensable for transcript stability in human mitochondria, but the oligo(A) tails may be sufficient for transcript stabilization.
DISCUSSION
In this paper, we demonstrate the identification of a novel human nuclear-encoded poly(A) polymerase, and we provide the experimental data indicating that this enzyme indeed localizes in mitochondria.
In order to study the physiological function of the hmtPAP, we decided to look at the effects of its silencing in human HeLa cells. To this end, we applied RNA interference technology. The decrease of the hmtPAP mRNA level to ∼11–26% of that in the non-treated cells resulted in a dramatic change in proportion of the steady-state levels of polyadenylated mitochondrial mRNAs to the oligoadenylated ones. Unpublished data obtained in our laboratory indicate that under physiological conditions, 90% of a given mitochondrial mRNA contain poly(A) tails of ∼50 bp, while only about 10% contain short oligo(A) tails varying in length between 2 and 13 nt, and both fractions are stable (J. Piechota, personal communication). This is in concordance with the situation observed in our mock-transfected cells. In contrast, inhibition of hmtPAP leads to a decrease in the level of polyadenylated fraction to 25–56% depending on mRNA, while the oligoadenylated fraction is increased accordingly to 44–75%. We interpret this result as the proof that hmtPAP is indeed responsible for the synthesis of long poly(A) tails in human mitochondria.
The role of polyadenylation in the mRNA decay pathway in human mitochondria may differ from the other known systems. In E.coli polyadenylation triggers degradation of RNA, and the same is true for plant chloroplasts and mitochondria (2–6,9–13,78). In all these systems, only a small fraction of mRNA is polyadenylated, in bacteria at any given time <2% of total RNA bear poly(A) tails (2,18,22,79), while non-polyadenylated molecules are more stable and translatable. In contrast to this, in eukaryotic cells almost all translationally active mRNA molecules are polyadenylated, while deadenylation stimulates degradation [reviewed in (80–81)]. As in human mitochondria, the majority of transcripts is polyadenylated, and hmtPAP suppression resulted in strong accumulation of the oligo(A)-tailed fraction, it would be interesting to determine whether both mRNA fractions are equally good substrates for translation.
The physiological role of polyadenylation in human mitochondria has been studied recently by Temperley et al. (82), who used a cell line from a patient with a micro-deletion in the ATP6/8 mitochondrial gene. The deletion resulted in the loss of the terminal 3′ uridine, from which polyadenylation would generate a proper UAA stop codon. In the patient cell line processing and polyadenylation still occurred, but the resulting mRNA lacked the stop codon. Translation was unaffected, but the mRNA was rapidly degraded in a deadenylation-dependent manner (83). The data were interpreted as consistent with the role of poly(A) tails in promoting mRNA stability, similar to the eukaryotic cytosol. However, it should be noted that the results of Temperley et al. were obtained in a system with a potentially pathogenic mitochondrial mutation, where polyadenylation was not affected. Therefore, the question that what would be the effect of disturbance in the synthesis of poly(A) tails on mitochondrial translation is still unanswered.
The abundance of the oligoadenylated mRNA species in the cells treated with siRNAs against hmtPAP is striking. Moreover, the total level of transcripts [poly(A) plus oligo(A)] is increased in samples obtained after RNA interference. This effect is more clearly visible on standard northern blots, which in our hands appear to be much more quantitative that the high-resolution northern blots, that we applied for qualitative analyses. The accumulation of the oligo(A) fraction could indicate either that short oligo(A)-tailed mRNAs are stable [in other words long poly(A) tails are not the prerequisite for the stability of mitochondrial mRNAs] or that the transcription rate is much higher in the siRNA-inhibited cells, so that it compensates for the degradation of oligoadenylated mRNAs. Although it seems unlikely to us that the observed increased steady-state level of mitochondrial mRNAs could be achieved if the oligoadenylated species were rapidly degraded, experiments are under way to address this issue and to discriminate which of the two scenarios envisaged above is true.
In principle, the oligo(A) tails may be either an intermediate in the synthesis of long poly(A) tails, or an intermediate of the deadenylation process, leading to RNA degradation. Recent unpublished results from our laboratory, obtained from experiments with inhibitors of polyadenylation, transcription and translation, indicate that human mitochondrial oligo(A) tails are intermediates in the polyadenylation pathway (J. Piechota, personal communication). This possible two-step mechanism of human mitochondrial polyadenylation appears to be similar to the respective processes in both E.coli and eukaryotic nucleus (84–86).
At present, it is unclear which enzymatic activity is responsible for the synthesis of the short oligo(A) extensions in HeLa cells treated with siRNAs against hmtPAP. It seems possible that the residual activity of hmtPAP is sufficient for the synthesis of short oligo(A) extensions in siRNA-treated cells. Under these conditions, the poly(A)-containing fraction is slightly shorter (∼10 nt) than in the control; this might also be explained as the result of the low concentration of the poly(A) polymerase, which abandons not fully extended tails. On the other hand, we cannot exclude that when hmtPAP level is low, both oligo(A) and poly(A) tails are synthesized by another, yet undiscovered, enzyme. In the E.coli strain devoid of PAP activity, the percentage of heteropolymeric tails increased up to 83%, due to the activity of PNPase (28). We did not observe such phenomenon in the case of human mitochondrial system following ablation of hmtPAP expression, as only one clone for ND3 represented heteropolymeric tail. Research is in progress to obtain statistically significant number of sequences of poly(A) tails after hmtPAP inhibition; our preliminary data indicate that these tails are homopolymeric. The above facts argue against the hypothesis that hPNPase is responsible for oligo(A) or poly(A) synthesis in conditions when hmtPAP activity is lowered.
It is important to note that no mRNA species completely devoid of post-transcriptionally added adenosine residues could be observed either in siRNA-inhibited cells or in the control. This could have two explanations: (i) oligoadenylation is a very rapid reaction coupled to the RNA processing and thus no such intermediates could be detected; and (ii) deadenylated mRNAs (in contrast to oligoadenylated ones) are very rapidly degraded.
Identification of hmtPAP should facilitate the development of the model of human mtRNA decay. It seems plausible that the poly(A) tails and oligo(A) tails play double roles in both stabilizing and destabilizing mRNAs, depending on which proteins are bound to mRNA and whether the transcripts are engaged in translation or not. So far we have shown the presence of three important enzymes in human mitochondria, which are known to participate in RNA turnover in other systems: RNA helicase hSUV3, which is also one of the components of yeast mitochondrial degradosome (87), PNPase (60) and poly(A) polymerase hmtPAP (this study). It seems that the interplay of these enzymes plus the involvement of yet unidentified putative exoribonuclease(s), endoribonuclease(s), poly(A) binding proteins and finally translational machinery [as suggested by Temperley et al. (82)] regulate in concert mitochondrial RNA decay. Research is in progress to identify the missing gene products and to develop a model for the regulation of mitochondrial RNA stability.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Z. M. A. Chrzanowska-Lightowlers and R. N. Lightowlers for encouragement, discussions and sharing their results prior to publication. We thank J. Piwowarski for his help in designing the siRNA experiments. M. Minczuk is acknowledged for valuable suggestions concerning immunofluorescence studies of hmtPAP localization. We are indebted to Prof. E. Bartnik for critical reading of the manuscript and to J. Piechota for helpful discussions. This work was supported by the Polish Ministry of Scientific Research and Information Technology, Grant no PBZ-KBN-091/P05/2003/40, the Centre of Excellence for Multi-Scale Biomolecular Modelling, and through the intramural grants BW-1601/20 and BW-1636/54 in years 2003 and 2004 from the Faculty of Biology, Warsaw University to R.T. The authors are members of an EU-funded MitEuro Network. R.T. is the recipient of a scholarship from the Postgraduate School of Molecular Medicine affiliated with the Medical University of Warsaw.
REFERENCES
- 1.Manley J.L. and Proudfoot,N.J. (1994) RNA 3′ ends: formation and function–meeting review. Genes Dev., 8, 259–264. [DOI] [PubMed] [Google Scholar]
- 2.Dreyfus M. and Regnier,P. (2002) The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell, 111, 611–613. [DOI] [PubMed] [Google Scholar]
- 3.Regnier P. and Arraiano,C.M. (2000) Degradation of mRNA in bacteria: emergence of ubiquitous features. Bioessays, 22, 235–244. [DOI] [PubMed] [Google Scholar]
- 4.Schuster G., Lisitsky,I. and Klaff,P. (1999) Polyadenylation and degradation of mRNA in the chloroplast. Plant Physiol., 120, 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes R., Kudla,J. and Gruissem,W. (1999) Degrading chloroplast mRNA: the role of polyadenylation. Trends Biochem. Sci., 24, 199–202. [DOI] [PubMed] [Google Scholar]
- 6.Monde R.A., Schuster,G. and Stern,D.B. (2000) Processing and degradation of chloroplast mRNA. Biochimie, 82, 573–582. [DOI] [PubMed] [Google Scholar]
- 7.Militello K.T. and Read,L.K. (1999) Coordination of kRNA editing and polyadenylation in Trypanosoma brucei mitochondria: complete editing is not required for long poly(A) tract addition. Nucleic Acids Res., 27, 1377–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan C.M., Militello,K.T. and Read,L.K. (2003) Polyadenylation regulates the stability of Trypanosoma brucei mitochondrial RNAs. J. Biol. Chem., 278, 32753–32762. [DOI] [PubMed] [Google Scholar]
- 9.Gagliardi D. and Leaver,C.J. (1999) Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J., 18, 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupold D.S., Caoile,A.G. and Stern,D.B. (1999) Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell, 11, 1565–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagliardi D., Perrin,R., Marechal-Drouard,L., Grienenberger,J.M. and Leaver,C.J. (2001) Plant mitochondrial polyadenylated mRNAs are degraded by a 3′- to 5′-exoribonuclease activity, which proceeds unimpeded by stable secondary structures. J. Biol. Chem., 276, 43541–43547. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn J., Tengler,U. and Binder,S. (2001) Transcript lifetime is balanced between stabilizing stem-loop structures and degradation-promoting polyadenylation in plant mitochondria. Mol. Cell. Biol., 21, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butow R.A., Zhu,H., Perlman,P. and Conrad-Webb,H. (1989) The role of a conserved dodecamer sequence in yeast mitochondrial gene expression. Genome, 31, 757–760. [DOI] [PubMed] [Google Scholar]
- 14.Montoya J., Ojala,D. and Attardi,G. (1981) Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature, 290, 465–470. [DOI] [PubMed] [Google Scholar]
- 15.Ojala D., Montoya,J. and Attardi,G. (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature, 290, 470–474. [DOI] [PubMed] [Google Scholar]
- 16.Clayton D.A. (1984) Transcription of the mammalian mitochondrial genome Annu. Rev. Biochem., 53, 573–594. [DOI] [PubMed] [Google Scholar]
- 17.Shadel G.S. and Clayton,D.A. (1997) Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem., 66, 409–435. [DOI] [PubMed] [Google Scholar]
- 18.Cao G.J. and Sarkar,N. (1992) Poly(A) RNA in Escherichia coli: nucleotide sequence at the junction of the lpp transcript and the polyadenylate moiety. Proc. Natl Acad. Sci. USA, 89, 7546–7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao G.J. and Sarkar,N. (1992) Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl Acad. Sci. USA, 89, 10380–10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wahle E. and Keller,W. (1992) The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Annu. Rev. Biochem., 61, 419–440. [DOI] [PubMed] [Google Scholar]
- 21.Sachs A. and Wahle,E. (1993) Poly(A) tail metabolism and function in eucaryotes. J. Biol. Chem., 268, 22955–22958. [PubMed] [Google Scholar]
- 22.Hajnsdorf E., Braun,F., Haugel-Nielsen,J. and Regnier,P. (1995) Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc. Natl Acad. Sci. USA, 92, 3973–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarkar N. (1997) Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem., 66, 173–197. [DOI] [PubMed] [Google Scholar]
- 24.Colgan D.F. and Manley,J.L. (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev., 11, 2755–2766. [DOI] [PubMed] [Google Scholar]
- 25.Komine Y., Kwong,L., Anguera,M.C., Schuster,G. and Stern,D.B. (2000) Polyadenylation of three classes of chloroplast RNA in Chlamydomonas reinhardtii. RNA, 6, 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickson K.S., Thompson,S.R., Gray,N.K. and Wickens,M. (2001) Poly(A) polymerase and the regulation of cytoplasmic polyadenylation. J. Biol. Chem., 276, 41810–41816. [DOI] [PubMed] [Google Scholar]
- 27.Komine Y., Kikis,E., Schuster,G. and Stern,D. (2002) Evidence for in vivo modulation of chloroplast RNA stability by 3′-UTR homopolymeric tails in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA, 99, 4085–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohanty B.K. and Kushner,S.R. (2000) Polynucleotide phosphorylase functions both as a 3′→5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 11966–11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yehudai-Resheff S., Hirsh,M. and Schuster,G. (2001) Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol., 21, 5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rott R., Zipor,G., Portnoy,V., Liveanu,V. and Schuster,G. (2003) RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli. J. Biol. Chem., 278, 15771–15777. [DOI] [PubMed] [Google Scholar]
- 31.Lisitsky I., Klaff,P. and Schuster,G. (1996) Addition of destabilizing poly (A)-rich sequences to endonuclease cleavage sites during the degradation of chloroplast mRNA. Proc. Natl Acad. Sci. USA, 93, 13398–13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao G.J. and Sarkar,N. (1997) Stationary phase-specific mRNAs in Escherichia coli are polyadenylated. Biochem. Biophys. Res. Commun., 239, 46–50. [DOI] [PubMed] [Google Scholar]
- 33.Perrin R., Meyer,E.H., Zaepfel,M., Kim,Y.J., Mache,R., Grienenberger,J.M., Gualberto,J.M. and Gagliardi,D. (2004) Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J. Biol. Chem., 279, 25440–25446. [DOI] [PubMed] [Google Scholar]
- 34.Perrin R., Lange,H., Grienenberger,J.M. and Gagliardi,D. (2004) AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res., 32, 5174–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams M.A., Johzuka,Y. and Mulligan,R.M. (2000) Addition of non-genomically encoded nucleotides to the 3′-terminus of maize mitochondrial mRNAs: truncated rps12 mRNAs frequently terminate with CCA. Nucleic Acids Res., 28, 4444–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nordvarg H. (2002) Functional significance of multiple poly(A) polymerases (PAPs). Acta Univ. Ups., 1140, 1–51. [Google Scholar]
- 37.Thuresson A.C., Astrom,J., Astrom,A., Gronvik,K.O. and Virtanen,A. (1994) Multiple forms of poly(A) polymerases in human cells. Proc. Natl Acad. Sci. USA, 91, 979–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kyriakopoulou C.B., Nordvarg,H. and Virtanen,A. (2001) A novel nuclear human poly(A) polymerase (PAP), PAP gamma. J. Biol. Chem., 276, 33504–33511. [DOI] [PubMed] [Google Scholar]
- 39.Kashiwabara S., Zhuang,T., Yamagata,K., Noguchi,J., Fukamizu,A. and Baba,T. (2000) Identification of a novel isoform of poly(A) polymerase, TPAP, specifically present in the cytoplasm of spermatogenic cells. Dev. Biol., 228, 106–115. [DOI] [PubMed] [Google Scholar]
- 40.Kashiwabara S., Noguchi,J., Zhuang,T., Ohmura,K., Honda,A., Sugiura,S., Miyamoto,K., Takahashi,S., Inoue,K., Ogura,A. and Baba,T. (2002) Regulation of spermatogenesis by testis-specific, cytoplasmic poly(A) polymerase TPAP. Science, 298, 1999–2002. [DOI] [PubMed] [Google Scholar]
- 41.Topalian S.L., Kaneko,S., Gonzales,M.I., Bond,G.L., Ward,Y. and Manley,J.L. (2001) Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol. Cell. Biol., 21, 5614–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinha K., Perumal,K., Chen,Y. and Reddy,R. (1999) Post-transcriptional adenylation of signal recognition particle RNA is carried out by an enzyme different from mRNA Poly(A) polymerase. J. Biol. Chem., 274, 30826–30831. [DOI] [PubMed] [Google Scholar]
- 43.Saitoh S., Chabes,A., McDonald,W.H., Thelander,L., Yates,J.R. and Russell,P. (2002) Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell, 109, 563–573. [DOI] [PubMed] [Google Scholar]
- 44.Wang L., Eckmann,C.R., Kadyk,L.C., Wickens,M. and Kimble,J. (2002) A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature, 419, 312–316. [DOI] [PubMed] [Google Scholar]
- 45.Kadyk L.C. and Kimble,J. (1998) Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development, 125, 1803–1813. [DOI] [PubMed] [Google Scholar]
- 46.Eckmann C.R., Kraemer,B., Wickens,M. and Kimble,J. (2002) GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C.elegans. Dev. Cell, 3, 697–710. [DOI] [PubMed] [Google Scholar]
- 47.Kwak J.E., Wang,L., Ballantyne,S., Kimble,J. and Wickens,M. (2004) Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl Acad. Sci. USA, 101, 4407–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacob S.T. and Schindler,D.G. (1972) Polyriboadenylate polymerase solubilized from rat liver mitochondria. Biochem. Biophys. Res. Commun., 48, 126–134. [DOI] [PubMed] [Google Scholar]
- 49.Jacob S.T., Rose,K.M. and Morris,H.P. (1974) Expression of purified mitochondrial poly(A) polymerase of hepatomas by an endogenous primer from liver. Biochim. Biophys. Acta, 361, 312–320. [DOI] [PubMed] [Google Scholar]
- 50.Gallerani R., De Giorgi,C., De Benedetto,C. and Saccone,C. (1976) Contemporaneous isolation of deoxyribonucleic acid-dependent ribonucleic acid polymerase and poly(A) polymerase from rat liver mitochondria. Biochem. J., 157, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rose K.M., Morris,H.P. and Jacob,S.T. (1975) Mitochondrial poly(A) polymerase from a poorly differentiated hepatoma: purification and characteristics. Biochemistry, 14, 1025–1032. [DOI] [PubMed] [Google Scholar]
- 52.Kuhn J. and Binder,S. (2002) RT–PCR analysis of 5′ to 3′-end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria. Nucleic Acids Res., 30, 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minet M. and Lacroute,F. (1990) Cloning and sequencing of a human cDNA coding for a multifunctional polypeptide of the purine pathway by complementation of the ade2-101 mutant in Saccharomyces cerevisiae. Curr. Genet., 18, 287–291. [DOI] [PubMed] [Google Scholar]
- 54.Vandesompele J., De Preter,K., Pattyn,F., Poppe,B., Van Roy,N., De Paepe,A. and Speleman,F. (2002) Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol., 3, research 0034.1–0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bollenbach T.J., Schuster,G. and Stern,D.B. (2004) Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol., 78, 305–337. [DOI] [PubMed] [Google Scholar]
- 56.Yokobori S. and Paabo,S. (1995) Transfer RNA editing in land snail mitochondria. Proc. Natl Acad. Sci. USA, 92, 10432–10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ojala D. and Attardi,G. (1974) Expression of the mitochondrial genome in HeLa cells. XIX. Occurrence in mitochondria of polyadenylic acid sequences, ‘free’ and covalently linked to mitochondrial DNA-coded RNA. J. Mol. Biol., 82, 151–174. [DOI] [PubMed] [Google Scholar]
- 58.Dubin D.T., Montoya,J., Timko,K.D. and Attardi,G. (1982) Sequence analysis and precise mapping of the 3′ ends of HeLa cell mitochondrial ribosomal RNAs. J. Mol. Biol., 157, 1–19. [DOI] [PubMed] [Google Scholar]
- 59.Nagaike T., Suzuki,T., Tomari,Y., Takemoto-Hori,C., Negayama,F., Watanabe,K. and Ueda,T. (2001) Identification and characterization of mammalian mitochondrial tRNA nucleotidyltransferases. J. Biol. Chem., 276, 40041–40049. [DOI] [PubMed] [Google Scholar]
- 60.Piwowarski J., Grzechnik,P., Dziembowski,A., Dmochowska,A., Minczuk,M. and Stepien P.P. (2003) Human polynucleotide phosphorylase, hPNPase, is localized in mitochondria. J. Mol. Biol., 329, 853–857. [DOI] [PubMed] [Google Scholar]
- 61.Claros M.G. and Vincens,P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem., 241, 779–786. [DOI] [PubMed] [Google Scholar]
- 62.Emanuelsson O., Nielsen,H., Brunak,S. and von Heijne,G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol., 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- 63.Nakai K. and Kanehisa,M. (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics, 14, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bannai H., Tamada,Y., Maruyama,O., Nakai,K. and Miyano,S. (2002) Extensive feature detection of N-terminal protein sorting signals. Bioinformatics, 18, 298–305. [DOI] [PubMed] [Google Scholar]
- 65.Nakai K. and Horton,P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–36. [DOI] [PubMed] [Google Scholar]
- 66.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural. Syst., 8, 581–599. [DOI] [PubMed] [Google Scholar]
- 67.Minczuk M., Piwowarski,J., Papworth,M.A., Awiszus,K., Schalinski,S., Dziembowski,A., Dmochowska,A., Bartnik,E., Tokatlidis,K., Stepien,P.P. and Borowski,P. (2002) Localisation of the human hSuv3p helicase in the mitochondrial matrix and its preferential unwinding of dsDNA. Nucleic Acids Res., 30, 5074–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dmochowska A., Kalita,K., Krawczyk,M., Golik,P., Mroczek,K., Lazowska,J., Stepien,P.P. and Bartnik,E. (1999) A human putative Suv3-like RNA helicase is conserved between Rhodobacter and all eukaryotes. Acta Biochim. Pol., 46, 155–162. [PubMed] [Google Scholar]
- 69.Sarkar D., Leszczyniecka,M., Kang,D.C., Lebedeva,I.V., Valerie,K., Dhar,S., Pandita,T.K. and Fisher,P.B. (2003) Down-regulation of Myc as a potential target for growth arrest induced by human polynucleotide phosphorylase (hPNPase-old35) in human melanoma cells. J. Biol. Chem., 278, 24542–24551. [DOI] [PubMed] [Google Scholar]
- 70.Leszczyniecka M., Kang,D.C., Sarkar,D., Su,Z.Z., Holmes,M., Valerie,K. and Fisher,P.B. (2002) Identification and cloning of human polynucleotide phosphorylase, hPNPase old-35, in the context of terminal differentiation and cellular senescence. Proc. Natl Acad. Sci. USA, 99, 16636–16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin G. and Keller,W. (1996) Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J., 15, 2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 72.Raynal L.C. and Carpousis,A.J. (1999) Poly(A) polymerase I of Escherichia coli: characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol. Microbiol., 32, 765–775. [DOI] [PubMed] [Google Scholar]
- 73.Harborth J., Elbashir,S.M., Bechert,K., Tuschl,T. and Weber,K. (2001) Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci., 114, 4557–4565. [DOI] [PubMed] [Google Scholar]
- 74.Holen T., Amarzguioui,M., Wiiger,M.T., Babaie,E. and Prydz,H. (2002) Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res., 30, 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang D., Buchholz,F., Huang,Z., Goga,A., Chen,C.Y., Brodsky,F.M. and Bishop,J.M. (2002) Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 9942–9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Calegari F., Haubensak,W., Yang,D., Huttner,W.B. and Buchholz,F. (2002) Tissue-specific RNA interference in postimplantation mouse embryos with endoribonuclease-prepared short interfering RNA. Proc. Natl Acad. Sci. USA, 99, 14236–14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang D., Goga,A. and Bishop,J.M. (2004) RNA interference (RNAi) with RNase III-prepared siRNAs. Methods Mol. Biol., 252, 471–482. [DOI] [PubMed] [Google Scholar]
- 78.Gagliardi D., Stepien,P.P., Temperley,R., Lightowlers,R.N. and Chrzanowska-Lightowlers,Z.M.A. (2004) Messenger RNA stability in mitochondria: different means to an end. Trends Genet., 20, 260–267. [DOI] [PubMed] [Google Scholar]
- 79.O'Hara E.B., Chekanova,J.A., Ingle,C.A., Kushner,Z.R., Peters,E. and Kushner,S.R. (1995) Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc. Natl Acad. Sci. USA, 92, 1807–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Munroe D. and Jacobson,A. (1990) Tales of poly(A): a review. Gene, 91, 151–158. [DOI] [PubMed] [Google Scholar]
- 81.Beelman C.A. and Parker,R. (1995) Degradation of mRNA in eukaryotes. Cell, 81, 179–183. [DOI] [PubMed] [Google Scholar]
- 82.Temperley R.J., Seneca,S.H., Tonska,K., Bartnik,E., Bindoff,L.A., Lightowlers,R.N. and Chrzanowska-Lightowlers,Z.M. (2003) Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Hum. Mol. Genet., 12, 2341–2348. [DOI] [PubMed] [Google Scholar]
- 83.Chrzanowska-Lightowlers Z.M., Temperley,R.J., Smith,P.M., Seneca,S.H. and Lightowlers,R.N. (2004) Functional polypeptides can be synthesized from human mitochondrial transcripts lacking termination codons. Biochem. J., 377, 725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hajnsdorf E. and Regnier,P. (2000) Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl Acad. Sci. USA, 97, 1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Le Derout J., Folichon,M., Briani,F., Deho,G., Regnier,P. and Hajnsdorf,E. (2003) Hfq affects the length and the frequency of short oligo(A) tails at the 3′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res., 31, 4017–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bienroth S., Keller,W. and Wahle,E. (1993) Assembly of a processive messenger RNA polyadenylation complex. EMBO J., 12, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dziembowski A., Piwowarski,J., Hoser,R., Minczuk,M., Dmochowska,A., Siep,M., van der Spek,H., Grivell,L. and Stepien,P.P. (2003) The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem., 278, 1603–1611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.