Summary
In the ciliated protozoan Paramecium tetraurelia, Piwi-associated small RNAs are generated upon the elimination of tens of thousands of short transposon-derived DNA segments as part of development. These RNAs then target complementary DNA for elimination in a positive feedback process, contributing to germline defense and genome stability. In this work, we investigate the formation of these RNAs, which we show to be transcribed directly from the short (length mode 27 bp) excised DNA segments. Our data support a mechanism whereby the concatenation and circularization of excised DNA segments provides a template for RNA production. This process allows the generation of a double-stranded RNA for Dicer-like protein cleavage to give rise to a population of small regulatory RNAs that precisely match the excised DNA sequences.
Video Abstract
Keywords: small RNA, transcription, DNA concatemers, circular DNA, Ligase IV, Dicer, DNA elimination, DNA repair, transposable elements, Piwi-interacting RNA, ciliates, Paramecium
Graphical Abstract
Highlights
-
•
In Paramecium, pieces of deleted DNA are transcribed to form regulatory RNAs
-
•
Ultra-short DNA segments are concatenated and circularized, allowing transcription
-
•
This concatenation is carried out by Ligase IV, which also repairs DNA ends
-
•
Concatenation is random, which leads to diversity in the resulting sRNA population
“Junk” DNA can be ligated into circles and transcribed to generate regulatory RNAs.
Introduction
The functions of small RNAs in the eukaryotic cell are many and varied and their study continues to yield new insights into biological processes. The most well-characterized small RNA pathways are involved in silencing, either on the DNA level where transposons are silenced by Piwi-interacting RNAs (piRNAs) during germline development, or post-transcriptionally where small interfering RNAs (siRNAs) and microRNAs (miRNAs) control gene expression through mRNA degradation and translational inhibition (Kawaji and Hayashizaki, 2008). In the nucleus, small nuclear RNAs (snRNAs) are involved in post-transcriptional RNA processing including splicing and RNA editing (Matera et al., 2007). More recently, additional small RNA pathways have been discovered with roles in processes such as DNA repair (Wei et al., 2012) and transcriptional enhancement (Kim et al., 2010). It seems likely that additional classes of small RNAs await discovery as sequencing technologies continue to improve.
The mechanisms by which small RNAs are produced are as diverse as the functions of the RNAs themselves. In many cases, the RNAs are produced from specific genes that have evolved separately from the RNA targets—this is the case for most miRNAs and for the snRNAs (Ha and Kim, 2014). In other cases, the small RNAs are generated directly from the sequence that they target, such as when siRNAs are generated from invading double-stranded RNA (dsRNA). piRNAs are generated from large piRNA transcription units, similar to miRNAs, but these clusters are mostly generated from ancient transposons, which are thus in some senses promoting their own silencing (Czech and Hannon, 2016). An important phenomenon found in more than one small RNA pathway is that of signal amplification by feedforward processes, whereby a secondary wave of RNAs is generated as a consequence of the action of the initial subset. An example of this is the production of secondary siRNAs from primary siRNAs by RNA-dependent RNA polymerases (RDRPs) in C. elegans and plants (Pak and Fire, 2007, Sijen et al., 2007). Another such process is the so-called “ping-pong” mechanism for the generation of secondary piRNAs (Brennecke et al., 2007, Gunawardane et al., 2007, Aravin et al., 2008). Here, a secondary wave of transposon-targeting RNAs is generated as a result of piRNA cleavage of transcribed transposons, ensuring that a large pool of piRNAs is generated which precisely match the target sequence.
DNA deletion is a widespread phenomenon in most organisms. On the genome-wide level, it occurs in a precisely programmed, controlled way in a wide variety of organisms. Examples include the removal during development of germline-specific DNA in ciliates and certain parasitic nematodes and the removal of an entire chromosome in a wide variety of organisms including certain insects, birds, and mammals (Wang and Davis, 2014). DNA deletion also occurs in a programmed, but less precisely determined way in the immune system of jawed vertebrates, where V(D)J recombination and class switch recombination involve the rearrangement of the immunoglobulin heavy chain gene (Dudley et al., 2005). Deletion of genomic regions is of course also associated with various genetic diseases as well as cancer, where deletions and rearrangements are commonplace and in many cases are believed to drive oncogenesis. An extreme example is that of chromothripsis, whereby an entire chromosome or region of a chromosome is shattered in a single catastrophic event and then re-ligated in an apparently random order, often with the loss of chromosomal regions (Stephens et al., 2011). It should be noted, however, that genomic deletions in a cell are generally only identified when associated with disease. It is thus possible that DNA deletion in healthy cells occurs at a higher frequency than thought.
All of these examples involve the initiation of double-stranded DNA breaks and the ligation of the resulting ends via one of a number of DNA repair pathways. In the case of programmed DNA deletion, the Ligase IV-dependent non-homologous end joining (NHEJ) pathway is frequently used. While much work has been done to determine the breakpoint initiation processes and the mechanisms for dsDNA breakpoint repair, not much is known about what happens to the pieces of excised DNA after they are removed from the genome. It is assumed in most cases that they are degraded and lost to the cell, but this is not known with certainty. In some cases, however, it is clear that deleted DNA is maintained apart from the genome and transcribed. An example is the so-called “double-minute chromosomes” that have been identified in some cancers that have undergone chromothripsis (Stephens et al., 2011, Rausch et al., 2012). Here, fragments of broken chromosomes are ligated and circularized and can be maintained as oncogene-producing units.
Paramecium tetraurelia is a species of ciliate: single-celled organisms that separate their germline from their soma by way of nuclear dimorphism. Each individual contains both a germline nucleus, kept transcriptionally silent, and a somatic nucleus that is transcriptionally active and highly polyploid. The germline nucleus contains transposons, repeats and other parasitic DNA elements, while in the somatic nucleus all such unwanted elements are removed (Jahn and Klobutcher, 2002). During the sexual reproductive phase of the ciliate life cycle, a new somatic nucleus is generated from a meiotic product of the germline nucleus. In Paramecium, development of a new somatic nucleus involves the highly precise deletion of tens of thousands of parasitic DNA elements, known as internal eliminated sequences (IESs). These sequences, derived from ancient transposons, are excised by a domesticated piggyBac-like transposase called PiggyMac (Baudry et al., 2009). Because the excisase is expressed by the host, IESs have been free to evolve to very short lengths, with the shortest being only 26 bp in length.
The removal of IESs depends on two classes of Piwi-interacting small RNAs, which target their complementary sequences for deletion. The first class of Piwi-interacting RNAs are known as scan RNAs (scnRNAs). scnRNAs are generated via a germline genome-wide transcription event in early development and are then sorted in the maternal somatic nucleus so that genome-matching RNAs are degraded and only IES-matching RNAs remain. These IES-matching scnRNAs then target their complementary sequences for excision in the newly developing somatic nucleus. Once excision has begun, the second class of Piwi-interacting RNAs is generated. These are termed iesRNAs, as they map exclusively to IESs, and they are between 26 and 31 nt in length (Sandoval et al., 2014). Due to the IES mapping, and to the fact that inhibition of IES excision leads to the loss of iesRNAs, it is believed that they are transcribed from IESs after excision. This would provide a means for amplifying the signal targeting IESs for excision, which might be necessary given that the DNA in the developing somatic nucleus is at the same time being amplified to high levels of polyploidy. The dual small RNA pathways in Paramecium development are thus reminiscent of the primary and secondary piRNA pathways in animals, whereby a first wave of general transposon-directed piRNAs initiates silencing, then leads to a second wave of piRNAs generated from the precise sequences that are being silenced. This both amplifies and specifies the silencing signal.
The generation of iesRNAs has been a mystery due to the very short length of the supposed template. Here, we demonstrate that Paramecium has evolved a previously undescribed mechanism for transcribing short DNA pieces—their concatenation and circularization.
Results
Excised IESs Enter the iesRNA Pathway and Lead to Excision in the New Macronucleus
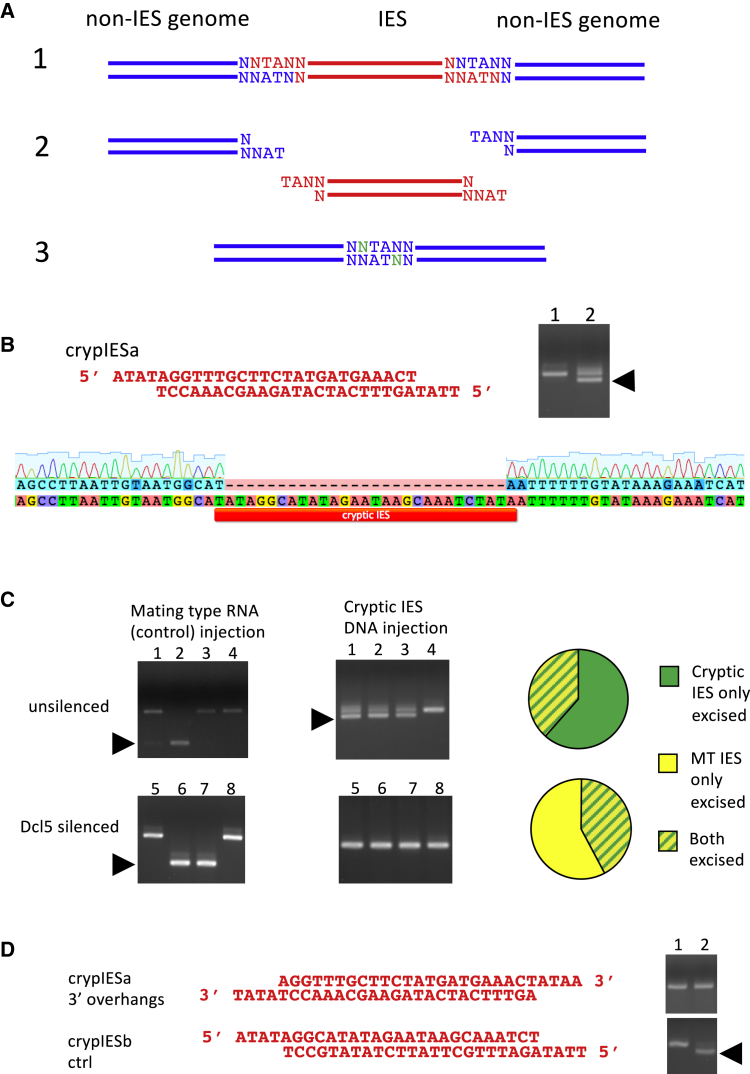
The short transposon-derived IESs that are eliminated in the somatic nucleus have certain hallmarks that are related to their excision. They all contain a TA dinucleotide within an inverted repeat at either end. These TAs, one copy of which is excised and one retained in the genomic sequence, are necessary for the IESs’ recognition by the excision machinery. IESs also have end consensus sequences, thought to aid in their recognition, and exhibit an interesting length distribution (Arnaiz et al., 2012, Swart et al., 2014). The shortest IESs, of 26–28 bp, form the largest group and then the distribution peaks every 10 bp with the exception of 36–38 bp. The current model for the excision of IESs is depicted in Figure 1A. The excisase PiggyMac cuts DNA with 4 nt 5′ overhangs centered on the TA repeat. The 5′-most nucleotide of the overhang is resected, and the ends are joined together at the TAs (Gratias and Bétermier, 2003). The missing nucleotide is then filled in and the ends are ligated with Ligase IV and Xrcc4 (Kapusta et al., 2011).
Figure 1.
Injection of DNA Resembling an Excised IES Is Sufficient to Trigger the iesRNA Pathway and Lead to Positive Feedback-Mediated DNA Excision
(A) The current model for IES excision. An IES (red) with the TA repeats that mark its boundaries is represented in panel 1. The excisase PiggyMac cleaves at the ends leaving 4 nt 5′ overhangs centered on the TA (panel 2). The 5′ most nucleotide is resected (panel 2) and the broken DNA ends are annealed at the TA, then ligated. The missing nucleotide is filled in during the annealing-ligation process (green nucleotides) and a single TA dinucleotide is left in the genome (blue, panel 3).
(B) A cryptic IES oligonucleotide corresponding to crypIESa with its 4 nt 5′ overhangs and 5′ phosphate. The gel represents the product of PCR performed with primers flanking the targeted region. PCR was performed after injected cells had been left to expand vegetatively for four divisions, and the PCR reactions were set up with ten cells each. Lane 1 is uninjected, and lane 2 is successfully injected as is seen from the presence of a lower band representing amplicon with a 27 bp deletion (arrowhead). Sequencing of the band demonstrates the precise excision of the crypIESa, with one TA removed and one retained.
(C) Left: representative gels demonstrating excision of mating type IES (injection control) and crypIESb following co-injection of mating type IES-matching small RNAs (do not require Dcl5) and crypIESb DNA in control (lanes 1–4) versus Dcl5-silenced (lanes 5–8) cells. Arrowheads indicate bands corresponding to excision products. Right: co-injection of control RNA (mating type) and cryptic IES DNA was repeated to quantify the relative excision efficiencies in unsilenced (n = 13) and Dcl5-silenced (n = 19) cells. Charts are shown quantifying the relative efficiencies of excision.
(D) Cryptic IES DNA with 3′ overhangs does not induce excision. DNA oligonucleotides for crypIESa (3′ overhangs) and crypIESb (control, 5′ overhangs) are shown. PCR products corresponding to excised cryptic IES are only found for crypIESb after coinjection of both oligonucleotides. This experiment was repeated with successful co-injection of crypIESa (3′ overhangs) and crypIESb (control, 5′ overhangs) in 12 cells, none of which exhibited excision of crypIESa.
See also Figure S1.
The production of iesRNAs is dependent on the excision of IESs, as is evident from the fact that silencing of proteins involved in IES excision prevents the production of iesRNAs (Sandoval et al., 2014, Maliszewska-Olejniczak et al., 2015). One explanation for this dependence is that the excised IESs act as a template for iesRNA transcription. Alternately, the transcription could occur pre-excision but be dependent on the assembly of the excisase complex on the DNA strands. We wished to determine whether excised IES DNA was sufficient in and of itself to initiate iesRNA production. To do this, regions of genomic DNA were identified whereby the IES end consensus inverted repeat of TATAG is separated by 27 nt, meaning that the locus has the length and sequence hallmarks of an IES, without being an IES. These loci were termed “cryptic IESs.” They are parts of genes or regulatory sequences and in normal cells are not excised during development. DNA oligonucleotides with 4 nt 5′ overhangs were designed and annealed to generate the theoretical excision product of these cryptic IESs. These oligonucleotides were injected into cells during the late stages of autogamy, at the point when iesRNAs are produced in the developing somatic nucleus. Cells were left to complete autogamy and go through four divisions before they were checked for excision by PCR using primers flanking the cryptic IES. PCR products were seen that correspond to amplified region following excision of a 27-bp segment (Figure 1B). These products were sent for sequencing that confirmed that the cryptic IES had been excised precisely, with one TA repeat left in the genome and one removed (Figure 1B). Three separate cryptic IESs of 27 nt were successfully excised following injection of DNA corresponding to their excision products. This supports the idea of a positive feedback loop, whereby excised transposon-like sequences trigger further excision of their own identical sequences. Co-injection of three cryptic IES oligonucleotides led to excision of all three in the same injected cells (Figure S1B). We named these cryptic IESs crypIESa, crypIESb, and crypIESc (see the Supplemental Information for sequences and primers).
Figure S1.
Related to Figure 1
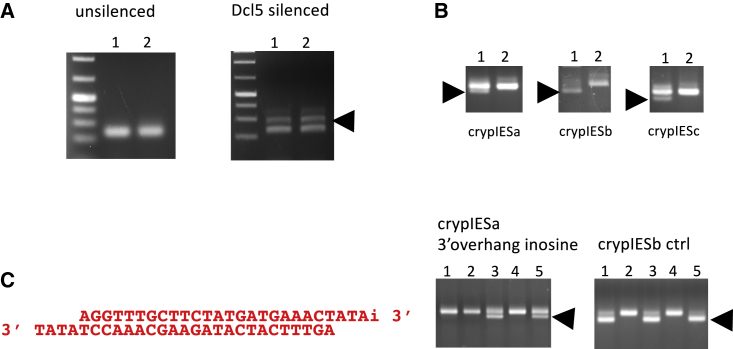
(A) silencing of Dcl5 was confirmed by PCR across an IES that is dependent on iesRNAs, and thus Dcl5, for complete excision. The upper band (arrowhead) corresponds to the PCR product when the IES is retained. Retention is seen in the Dcl5 silenced cells, demonstrating that Dcl5 silencing was successful. (B) co-injection of three cryptic IESs leads to excision of all three in the same cells. Lanes 1 correspond to successfully injected cells and lanes 2 correspond to uninjected cells. (C) Left: crypIESa modified with 3 overhangs and inosine replacing the 3′ most A on the upper strand. Right: gel from PCR to check excision. In two out of three injected cells, inosine rescues the excision.
We wished to ascertain that the excision of cryptic IESs following oligonucleotide injection is due to the effect of iesRNAs. To do this, Dcl5 was silenced through RNAi-mediated gene knockdown by feeding (Beisson et al., 2010). Dcl5 is the Dicer-like protein responsible for iesRNA production, so if iesRNAs are required for DNA-induced cryptic IES excision, then Dcl5 silencing should abrogate this effect. Following Dcl5 silencing, cells were starved to initiate autogamy and injected with dsDNA corresponding to the excision product of crypIESb. As an injection control, 27 nt RNAs corresponding to the ends of the mating type IES were co-injected with the cryptic IES oligos. The mating type IES is only excised in mating type O cells and is retained in mating type E, with the excision or retention being epigenetically inherited (Singh et al., 2014). Injections were carried out in mating type E cells so that excision of the mating type IES was indicative of a successful injection. Dcl5 silencing will not affect excision following such an RNA injection, as the small RNAs are already present and thus do not need to be produced by Dcl5. Successful silencing of Dcl5 was confirmed by PCR to establish retention of IESs that require iesRNAs for their excision (Figure S1A). In control (unsilenced) cells, cryptic IES DNA injection was significantly more efficient at initiating excision than mating type (MT) RNA injection, with 13 successfully injected cells exhibiting crypIESb excision and only 5 cells exhibiting MT IES excision (Figure 1C). All cells that exhibited MT IES excision also exhibited crypIESb excision. In contrast, on Dcl5 silencing, the MT RNA injection was much more efficient at initiating excision than the cryptic IES DNA injection (Figure 1C). Very few injected cells excised the cryptic IES, and for those where excision occurred, the efficiency was low (Figure 1C). The silencing of Dcl5 thus has a strong effect on DNA-mediated cryptic IES excision, confirming that the excision occurs via the iesRNA pathway.
iesRNA Production from Excised IESs Is Dependent on Their Ability to Ligate Together
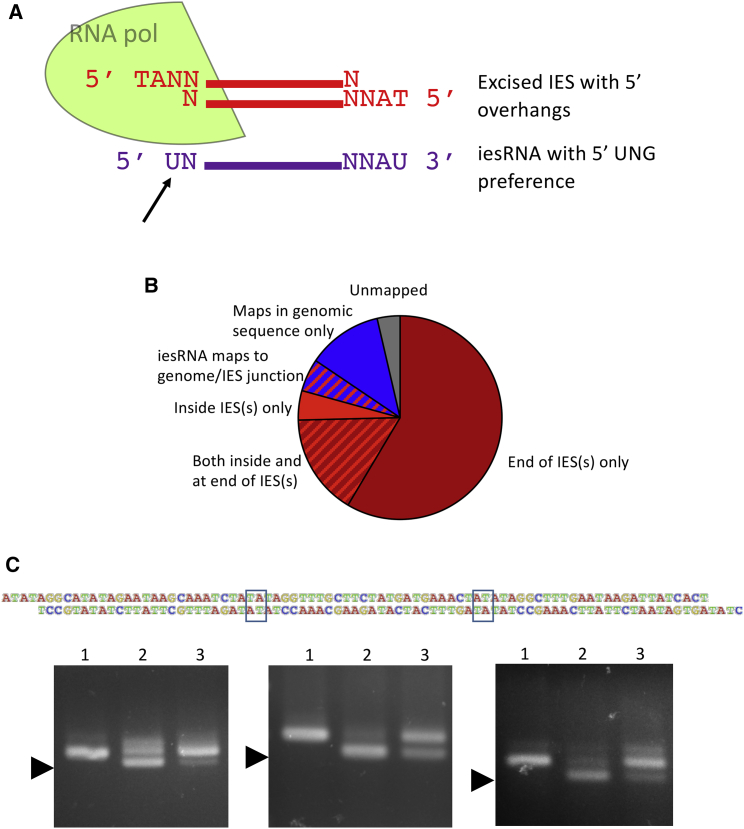
The transcription of iesRNAs from excised IESs explains several observations, including the fact that iesRNAs map uniquely to IESs, are dependent on IES excision, and are only found in the developing somatic nucleus (Sandoval et al., 2014). However, it poses serious problems for the transcription of the iesRNA template. As mentioned, the majority of IESs are <100 bp in length, and the length mode is 27 bp. If the iesRNA template is generated from these excised IESs, an RNA polymerase complex would have to assemble on these tiny fragments of DNA. RNA polymerase II has a footprint in the order of 30 nt (Brabant and Acheson, 1995) that makes this impossible. Even if the polymerase responsible for iesRNA template transcription were capable of transcribing from such short molecules, there is a logistical problem with transcribing a complete iesRNA from an excised IES (see Figure 2A). iesRNAs have a 5′ UNG signature, with the 5′ U mapping to the nucleotide directly downstream of the TA repeat at the 5′ end of the IES (a T by consensus) (see Sandoval et al., 2014). If the excised IES acts as a template for iesRNA transcription, then the polymerase will transcribe in a 5′-3′ direction from the lower strand. Due to the 4 nt 5′ overhangs, this strand does not contain the nucleotide that corresponds to the 5′ U in the iesRNA. Thus, there is a missing nucleotide in the hypothetical template. Mapping of iesRNAs demonstrates that this nucleotide is present in iesRNAs corresponding to the ends of IESs, which raises the question of how it is transcribed (Sandoval et al., 2014). It is known that long IESs (over 200 nt) circularize following excision, with the 5′ most nucleotide being resected and then filled in upon ligation in a manner identical to that of the genomic DNA ends (Gratias et al., 2008). This circularization would solve the problem of the missing templated nucleotide, as the circle junction would provide a complete template (see Figure 2A). Instead of having single-stranded 5′ overhangs whereby the antisense strand—from which transcription occurs—is missing, such circles would have complete double-stranded junctions with transcription possible from both strands. However, the shortest IESs are unable to form circles due to limits on the bending of the DNA double helix (Gratias and Bétermier, 2003).
Figure 2.
Can iesRNAs Be Generated from Ligated Excised IESs?
(A) Representation of the logistical problem of iesRNA transcription from excised IESs. A schematic excised dsDNA IES is shown in red, with its 4 nt 5′ overhangs. The corresponding iesRNA as deduced from small RNA mapping (Sandoval et al., 2014) is shown below in purple. The assembly of an RNA polymerase complex on a short (27 bp) DNA molecule is probably not possible. Even if it were, the RNA (purple) that corresponds to the 5′ end of the excised IES lacks a template for its 5′ nucleotide (indicated with a black arrow).
(B) Two hundred iesRNAs that map only partially to IES ends, with overhangs of at least 12 bp, were identified, and the overhangs were mapped. These iesRNAs were chosen randomly by the software Geneious 8.1.8. Overhangs were sorted into groups according to where they map. The majority map at the ends of other IESs, which is a huge overrepresentation considering that the number of possible mapping sites for a random 12 nt sequence within an IES is much greater than at its ends.
(C) The oligonucleotide corresponding to three ligated cryptic IESs is depicted, with the TAs that form the theoretical ligation points outlined. Gels of PCR products for each individual 27 bp cryptic IES (all on different chromosomes) are shown. Arrowheads indicate bands corresponding to excision products. Successful injection of the double-stranded oligonucleotide led to excision of all three cryptic IESs from the genome. This was successfully repeated in eight independent injected cells. All cells that exhibited excision of one cryptic IES following concatemer injection also exhibited excision of the other two.
To investigate the dependence of the cryptic IES excision on the ends of the injected DNA, modified oligonucleotides with 4 nt 3′ overhangs were designed, annealed, and injected into autogamous cells. The cryptic IES in question had previously been successfully excised using dsDNA oligonucleotides with normal (4 nt 5′) overhangs. As an injection control, a second cryptic IES dsDNA oligonucleotide with 4 nt 5′ overhangs was co-injected with the modified oligonucleotide. Excision was seen for the control cryptic IES, but not for the 3′ overhang cryptic IES (Figure 1D). We hypothesized that the compatibility of the ends of the injected DNA was important for the formation of iesRNAs. The overhangs of the injected cryptic IES DNA are not complementary to each other (see Figures 1D and S1C), except at one end where a dimer could potentially form. Thus, they cannot ligate without prior end processing. In the normal situation, the 5′ most nucleotide from the 5′ overhang is removed, leaving a complementary TA on each overhang. The exonuclease responsible for this nucleotide removal can presumably only work in a 5′-3′ direction, meaning that the 3′ most nucleotide of the 3′ overhangs on the injected DNA will not be removed. To test whether complementary ends are necessary for iesRNA formation from injected DNA, modified dsDNA oligonucleotides with 3′ overhangs were injected, where the 3′ most nucleotide was replaced with an inosine nucleotide that can base pair with any other nucleotide. In two out of three injected cells, this injection led to efficient excision of the cryptic IES (Figure S1C). We conclude that complementarity of excised IES ends is an important factor in the formation of iesRNAs.
iesRNAs Are Found that Map across IES-IES Junctions
If excised IESs are annealed via their 5′ overhangs in the same manner as the genomic DNA ends are ligated together, this provides a possible solution to the problem of the templating of the iesRNA template. Long dsDNA molecules could be formed from the annealing of multiple excised IESs and could thus provide a suitable template for an RNA polymerase to transcribe. The junctions, if annealed in the same way as the genomic DNA ends, could be read through by the polymerase and would solve the problem of the missing 5′ ribonucleotide template. One prediction from this model is that the Dcl5-mediated cleavage of the dsRNA template would be expected to occasionally produce iesRNAs that map across IES-IES junctions. To search for such RNAs, reads from a small RNA sequencing experiment were analyzed. The sequencing was carried out on Paramecium cells in the late stages of sexual development when iesRNAs are abundant (Sandoval et al., 2014). RNAs were identified that map partially to the ends of IESs with overhangs of at least 12 nt. The overhangs were then mapped to the Paramecium genome and IESs. The majority of such overhangs were found to map to the ends of other IESs (Figure 2B), which is what would be expected if IES ends are ligated together to form the template for iesRNA production. Many overhangs mapped inside IESs, but the majority of these were also found to map to other IES ends. It is likely that the highly repetitive nature of IES sequences means that there is a fairly high chance that a 12 nt sequence will map in multiple locations within the ∼45,000 IESs in the genome. However, if the overhangs mapped randomly, then it would be expected that the number that mapped inside IESs would be much greater than the number that mapped at ends, as there are far more possibilities for the mapping of a random 12 nt sequence within a ≥27 nt sequence than at the ends. We thus conclude that iesRNAs mapping to IES-IES junctions are present at significant levels and reflect the presence of such junctions within the iesRNA template.
iesRNAs Can Be Formed from Concatenated IESs
We hypothesized that concatenated IESs form a template for iesRNA generation and wished to investigate the ability of such a concatemer to produce iesRNAs. To do this, a dsDNA oligonucleotide was designed to correspond to the theoretical ligation product of three excised 27 bp cryptic IESs. These three sequences are in different genomic loci, on separate scaffolds and had all been previously demonstrated to be excised upon individual dsDNA oligonucleotide injection. The oligonucleotide was 77 bp of dsDNA with 4 nt 5′ overhangs similar to the individual oligonucleotides. On injection of the oligonucleotide, all three cryptic IESs were excised (Figure 2C), demonstrating that iesRNAs can be formed from concatenated IESs.
Long RNA Molecules Transcribed from IES Concatemers Are Present In Vivo
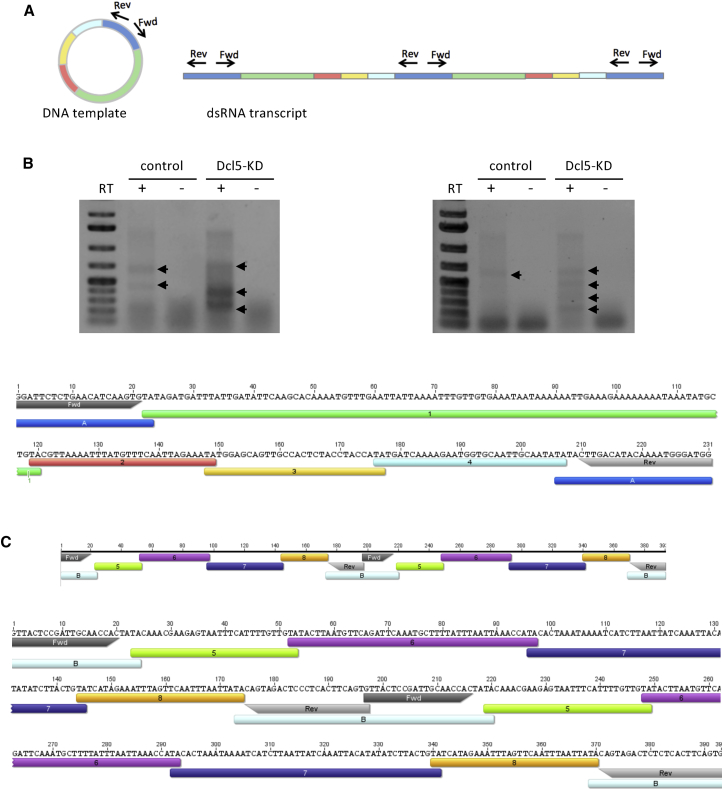
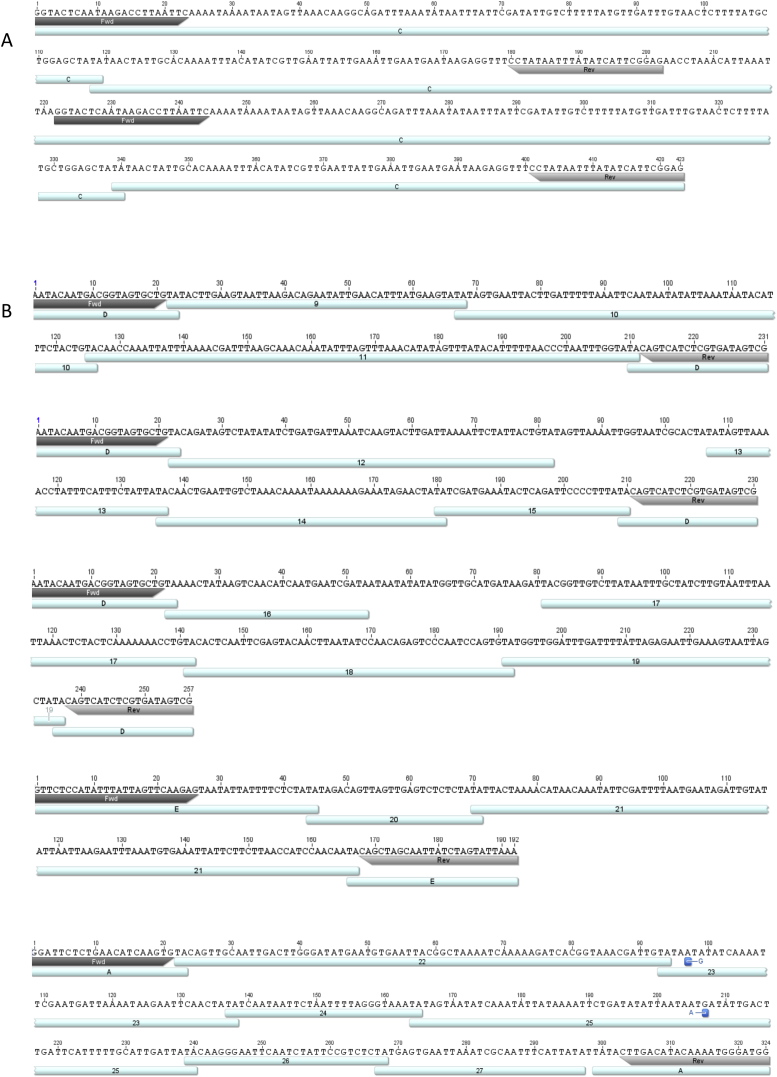
To establish whether or not iesRNA precursors are formed from IES concatemers and/or circles in vivo, total RNA was extracted from cells undergoing autogamy. Both Dcl5-silenced cells and control (empty vector silencing) cells were used. Following DNase treatment, RNA was reverse transcribed and PCR-amplified using primer pairs situated inside IESs but directed outward, toward the ends. We hypothesized that if IESs concatenate and circularize, such primer pairs will amplify the segment of the circle not included in the chosen IES (Figure 3A). A selection of IESs was chosen for primer design, both longer ones (>200 nt) and shorter ones (<50 nt). Longer IESs are expected to form circles by themselves but shorter IESs cannot do so alone due to bending constraints on the DNA. However, if several short IESs are ligated together, they could reach the required length for circle formation, thought to be ∼200 bp (Gratias and Bétermier, 2003). Following PCR, bands were excised, cloned into vectors, and sequenced. Stronger bands were detected in the Dcl5-silenced cells, which was expected as lack of Dcl5 cleavage should mean that iesRNA precursors are more abundant in the cells. Representative sequencing results are shown in Figures 3B, 3C and S2. For long IESs, primer pairs amplified circle junctions as expected (Figure S2A). For shorter IESs, amplicons were detected that contained up to six additional IESs apart from the primer binding site containing IES (Figures 3B, 3C, and S2B). The IESs were joined with overlapping TAs, as would be expected from a ligation product formed in the same manner as the ligated genomic DNA ends (Figure 1A). In one case, an amplicon was detected consisting of two identical products joined at the primer-containing IES. This represents the expected RNA transcript that would result if the IES concatemer were transcribed continuously in a circular manner.
Figure 3.
Detection of RNA Corresponding to Concatenated IESs
(A) Schematic representation of concatenated IESs forming a DNA circle. The corresponding RNA transcript is a repetitive sequence of the concatenated IESs. Outward-directed primers Fwd and Rev on IES A amplify one repeat of the RNA transcript. Individual IESs are shown in different colors.
(B) Following reverse transcription (RT) of total RNA during late development, cDNA was amplified using primer pairs as depicted in (A). Upper: representative gels showing bands (arrows) amplified. Lower: representative result of sequencing. The amplicon contains four different concatenated IESs (1–4) in addition to the primer-containing IES, resulting from transcribed circular DNA. Annotated IESs overlap with their terminal TA dinucleotide, resulting from the concatenation event.
(C) Sequence of amplicon from Fwd and Rev primers as in (B), including one additional circle repeat. Primer binding site- containing IESs are annotated in letters A and B, and internal IES are numbered (1–8). Additional concatenated IES transcripts and annotation of IESs are shown in Figure S2 and Table S2.
Figure S2.
RNA Transcripts of Concatenated IESs, Related to Figure 3
(A) RNA transcript of a repetitive IES sequence, presumably deriving from a circularised IES (> 200nt in length). Primer sites on the 221bp long IES are annotated, which amplify each IES end followed by the sequence of the opposite IES end. (B) Sequencing results of transcripts from concatenated IES. Primer binding site containing IESs are annotated in letters (A-E) and internal IESs in numbers (1-27). Gaps in sequence annotation result from lack of corresponding sequences in the database. Sequencing the Paramecium MIC genome is expected to reveal additional IESs that are not documented yet, but are present in these concatemers. iesRNAs exclusively map IESs and also map the gap sequences, suggesting these are non-annotated IESs. Point substitutions in the IES sequence are marked in blue and likely derive from sequence amplification with GoTaq® polymerase that lacks proofreading activity. In several cases, only partial IES sequences are detected, suggesting additional cleavage of these IESs occurs either while excision from the genome or after concatenation and circle formation. One example is IES C in A, where nucleotides 5-227 of the total 287bp long IES are forming the repeat.
IES Concatenation Is Dependent on Ligase IV and Is Necessary for iesRNA Production
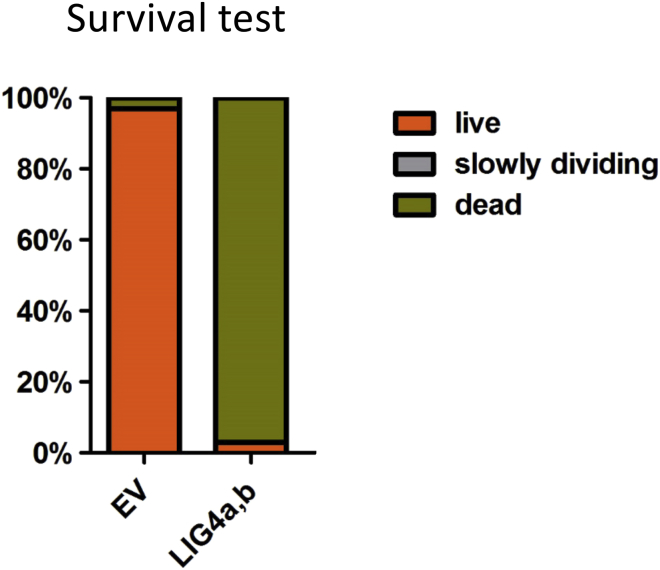
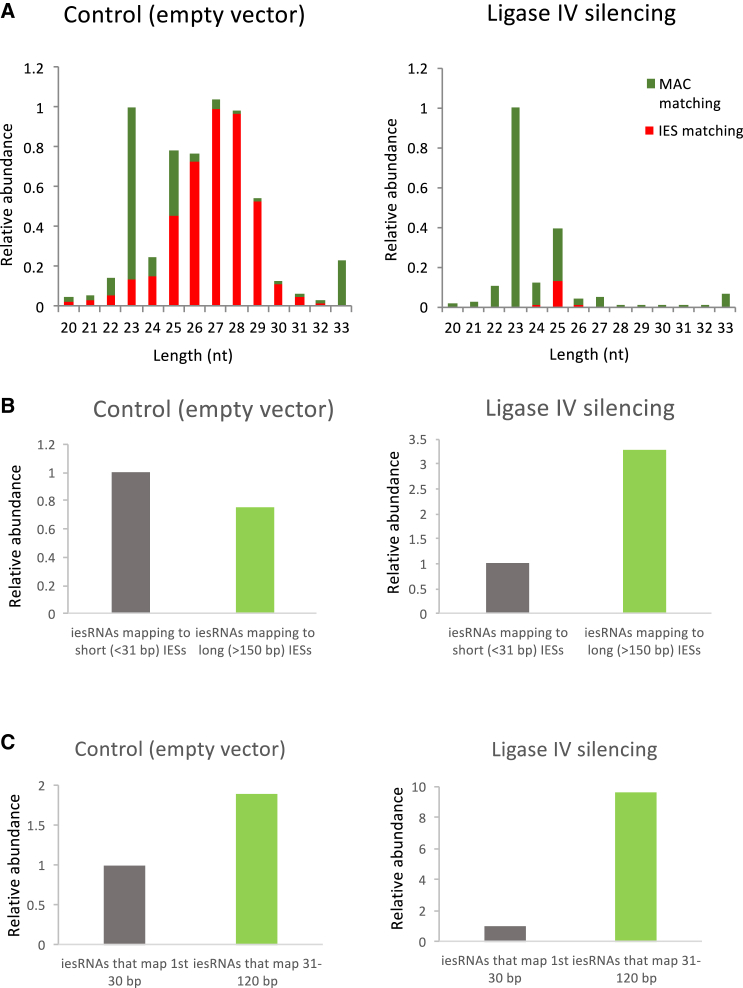
Thus far, the accumulated evidence points toward the formation of IES concatemers for transcription of iesRNA template. It is known that the enzyme responsible for the annealing of broken DNA ends upon IES excision is Ligase IV, which works with its partner Xrcc4 to join ends via their 5′ overhangs (Kapusta et al., 2011). To test whether the same enzyme is required to ligate excised IESs, Ligase IVa and IVb were co-silenced via RNAi by feeding. The silencing of Ligase IV has been shown to prevent the repair of genomic DNA following IES excision during Paramecium development, leading to a massively fragmented genome and cell death (Kapusta et al., 2011). Silencing was confirmed with a survival test following autogamy, where almost all cells died following refeeding (Figure S3). RNA was extracted during the late stages of autogamy, when cells were still alive and were undergoing genome rearrangement. Small RNA sequencing was then carried out to establish whether the silencing had an effect on iesRNA production. Ligase IV silencing almost completely abrogated iesRNA production (Figure 4A). This is consistent with a model whereby ligation of excised IESs is required for the formation of iesRNAs. As discussed, the problem of iesRNA precursor formation from unaltered excised IESs is 2-fold. First, for short IESs, the assembly of an RNA polymerase complex on the DNA molecule is incompatible with the size of RNA polymerase. Second, even if a polymerase can assemble, the first nucleotide downstream of the TA will be untemplated, and thus the formation of iesRNAs that map precisely to the ends of IESs, as is observed in normal cells, should be impossible. We wished to determine whether there was a difference in the mapping of iesRNAs between control and Ligase IV-silenced cells. In the Ligase IV-silenced cells, very few iesRNAs were detected compared to control (Figure 4A, ∼15,000 reads versus ∼2,000,000, a more than 100-fold reduction). The iesRNAs that could be detected were mapped to two groups of IESs, long (>150 nt) and short (<30 nt). In the Ligase IV-silenced cells, the number of iesRNAs that mapped to long IESs was proportionally much higher than in the control cells (Figure 4B), which is consistent with the notion that short IESs are more severely limited in terms of their ability to act as templates for iesRNA production than longer IESs. A second mapping was then carried out to establish whether there was a reduction in the number of iesRNAs that map to the ends of IESs in the Ligase IV-silenced cells versus in the control. iesRNAs were mapped to the first 30 nt of IESs over 150 nt in length and to the 31st–120th nt. The numbers that mapped in each group were compared (Figure 4C). In control cells, the number of iesRNAs that mapped to the middle portions (30th–120th nt) of the IESs was 1.9 times greater than the number that mapped to the end, but in the Ligase IV-silenced cells this ratio increased more than 5-fold to 9.6. This supports the idea that Ligase IV is necessary in particular for the formation of iesRNAs that match to the ends of IESs. Considering that iesRNAs are produced from the cleavage by Dcl5 of a dsRNA template, this makes sense. In order to produce a dsRNA template from an excised IES, an RNA polymerase must transcribe in both directions along the DNA molecule. Given the restricted length of a linear excised IES, the likelihood of generating dsRNA from the middle section is much greater than from the ends. While RNA polymerase may transcribe all the way to the end of a linear molecule, this will only produce a single-stranded RNA unless the opposite strand is primed at the very end, due to the directionality of transcription. As discussed, the assembly of an RNA polymerase complex to begin transcribing from the very end of a broken DNA molecule is unlikely. The Ligase IV silencing RNA sequencing data strongly supports a model whereby ligation of IESs allows transcription to occur through IES ends and short IESs, giving rise to iesRNA precursors.
Figure S3.
Related to Figure 4
Survival test following Ligase IVa and IVb silencing. Cells (n = 30) were scored for growth and survival two days after re-feeding following autogamy. In the Ligase IV silenced cells, almost all cells died, confirming that silencing had taken place.
Figure 4.
Sequencing of Small RNAs in Control versus Ligase IV-Silenced Cells
(A) RNA reads by length, normalized to 23 nt reads for each sample. Green bars are RNAs that map to macronuclear sequences not including IESs (mature MAC sequence). Red bars are RNAs that map to IES sequences. iesRNAs are IES matching RNAs (red columns) of around 26–31 nt, evident in the control but not in the Ligase IV silencing experiment. Ligase IV silencing leads to an ∼100-fold reduction in iesRNAs (see text).
(B) Mapping of 27–31 nt RNAs (iesRNAs) to long (>150 bp) versus short (<31 bp) IESs in control and Ligase IV silencing. Reads are normalized to the number that map to short IESs, for the purposes of clarity as the number of iesRNAs in the Ligase IV silencing is very small. Proportionally more iesRNAs map to long IESs in the Ligase IV-silenced cells than in the control, which is consistent with a model whereby RNA polymerase assembly is a prerequisite of iesRNA production and unligated <31 bp IESs are too short to allow for polymerase assembly.
(C) The ratio of iesRNAs that map to middle portions (31st–150th bp) versus ends (1st–30th bp) of IESs increases drastically in the Ligase IV-silenced cells. This is consistent with a model whereby IES ligation allows transcription through IEs ends.
See also Figure S3.
Discussion
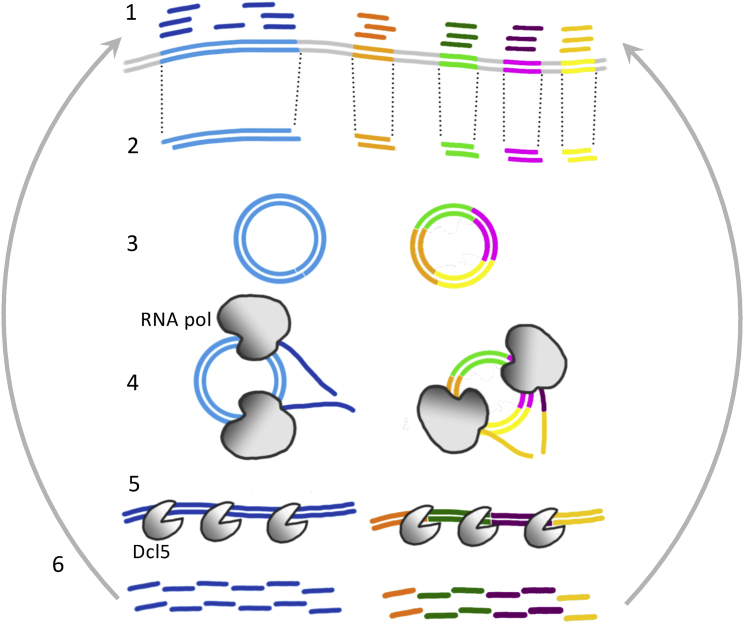
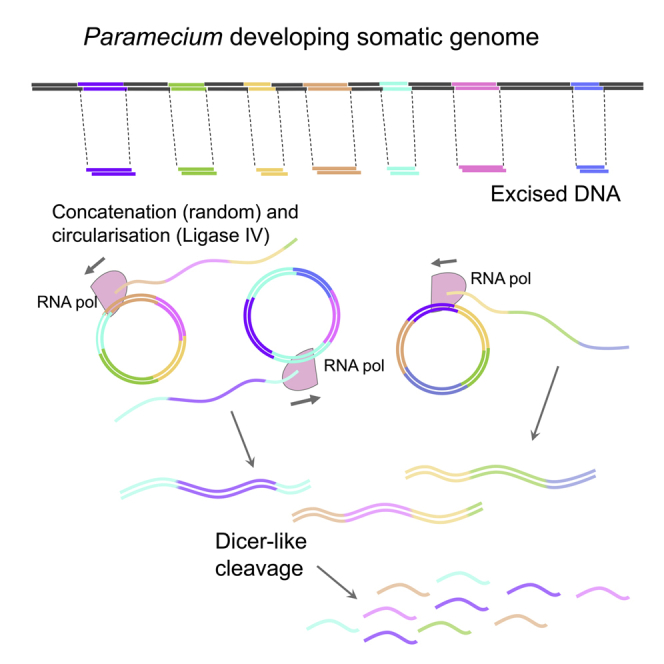
We demonstrate that Paramecium has evolved an innovative way to produce small RNAs from excised pieces of DNA, the concatenation and circularization of the short DNA pieces. This means that DNA thought to be non-coding “junk,” degraded after being removed from the genome, is actually a functional template for a biologically important class of small RNAs. The proposed model for iesRNA generation is shown in Figure 5. IESs are excised from the genome with 4 nt 5′ overhangs. The 5′ most nucleotide is resected and the IESs are ligated together with Ligase IV to form concatemers. This provides a DNA template from which RNA polymerase may seamlessly transcribe complete IESs. Concatenation and circularization thus allow transcription of IES ends and of short IESs that would otherwise not be transcribable.
Figure 5.
A Model for Small RNA Production from Short DNA Fragments in Paramecium
(1) Developing macronuclear genomic DNA is targeted for excision by short RNAs. (2) Excised DNA fragments of different lengths. Blue: >200 bp, other colors: <200 bp. (3) Circularization of longer excised DNA fragments (blue), concatenation and circularization of short fragments (multicolored) by Ligase IV. (4) Bidirectional transcription to form dsRNA. (5) Cleavage of long dsRNA by Dcl5 to produce iesRNAs. (6) iesRNAs target their complementary sequences for excision in a positive feedback loop.
As described, the Dcl5 protein cleaves dsRNA, which according to our model, is transcribed from circularized concatenated excised IESs. The IES joining process is predicted to be random, which means that each IES circle will most likely be present in only a single copy. This raises a question about the bidirectional transcription of such circles: in order to obtain RNA from both strands of a single-copy circle, transcription must occur in both directions from the same molecule. We do not know whether simultaneous transcription from both DNA strands is possible or whether the polymerases complexes would collide and/or interfere with one another. Theoretically, it should be possible for such transcription to occur because each RNA polymerase complex will be using a separate strand of the DNA double helix, and the DNA is melted at the point of polymerase binding. However, other possibilities for the generation of dsRNA from unique circles also exist. For example, one strand could be transcribed, then an RNA-directed RNA polymerase could form the second RNA strand from this template. Alternately, the two strands could be transcribed sequentially from the DNA, with the polymerase dropping off the template after a certain length of transcription, and a second polymerase then joining and initiating transcription from the opposite strand.
A second question relating to the transcription of the IES circles is how transcription is initiated on randomized short pieces of DNA without conventional promoters. In ciliates, such non-specific transcription initiation is known to occur at defined points in development: during meiosis in the germline when the scnRNAs are produced (Mochizuki and Gorovsky, 2004) and in the parental macronucleus during the “scanning” process whereby scnRNAs are compared to the old somatic genome (Aronica et al., 2008). Such generalized transcription could be initiated with the help of a specialized transcription factor, which allows polymerase initiation on any DNA sequence. A candidate for such a transcription factor in Paramecium is the recently-identified TFIIS4 (Maliszewska-Olejniczak et al., 2015), which localizes to both parental and developing macronuclei and that is necessary both for iesRNA generation and for scnRNA sorting.
While the experiments in this study strongly support the model shown in Figure 5, it should be pointed out that there are limitations to the data acquired. For example, it has not yet proven possible to isolate and sequence IES concatemers on the DNA level. The reason for this is, we believe, the low quantities at which each individual circle is present. Our model predicts that the IESs are ligated essentially at random, meaning that each circular concatemer may only be present in a single copy. A further limitation is the fact that small RNAs from the injected cells were never sequenced, and hence it could not be directly shown that iesRNAs are generated from the injected concatemers (Figure 2C). This is due to the fact that that injection is performed on single cells, and we currently do not have the ability to sequence small RNAs from single Paramecium cells. We anticipate that as sequencing technology improves, the DNA circles in normal developing cells will be detected.
We believe that the data in this study supports our theory; however, it is important to consider other possible models. While it has been shown that long IESs do circularize (Gratias and Bétermier, 2003), our data do not unequivocally show that all IES concatemers are circularized. It is possible that long linear concatemers exist, although we predict that their transcription would be less efficient. Additionally, while we show that Ligase IV is necessary for the production of iesRNAs, it should be pointed out that it is not proven beyond doubt that this is a direct effect. The possibility remains that another protein involved in iesRNA biogenesis is compromised in the Ligase IV-silenced cells. For example, any gene expressed from the developing MAC will be severely affected in the Ligase IV knockdown. Nonetheless, we believe that the relative abundance of iesRNAs mapping to different portions of IESs in the Ligase IV-silenced cells (Figures 4B and 4C), along with the rest of the data presented in this study, strongly supports the model that we propose.
As described, there are many mechanisms by which DNA is eliminated in the genomes of eukaryotic cells. Usually it is assumed that eliminated DNA is lost, except in rare cases such as the double-minute chromosomes associated with chromothripsis in cancer. Here, we provide an example of a case in which eliminated DNA is used to generate long templates for RNA transcription. It is an example of a positive feedback loop in small RNA generation, previously described examples of which include piRNA generation in animals and siRNA generation in plants and animals. In these examples, however, the feedback operates at the RNA level rather than involving an entirely new DNA template.
Questions arising from this research include the regulation of the IESs joining: is this a random process or is it controlled by the cell? We do not detect any kind of pattern in the IES concatemers we have identified (Figure 3), the IESs that are joined together are from different chromosomes and do not have any obvious similarities, but further work will need to be done to establish if this is correct. If IESs are joined randomly, it raises questions as to how this process is controlled and more generally about what happens to excised DNA in a Ligase IV-rich environment. A second question relates to the longevity of the concatemers and circularized IESs: how long do they survive in the cell and are they actively degraded? Any such active degradation will require a means for the cell to recognize concatenated IESs and for determining when they are no longer needed, neither of which are currently known.
A third and interesting point relates to the Dicer-like enzyme Dcl5, which is responsible for producing iesRNAs from the dsRNA template transcribed from the concatenated IESs. While iesRNAs that map to IES-IES junctions can be found (Figure 2), they are far less abundant than iesRNAs that map wholly within IESs, even short IESs of only 27 nt (Sandoval et al., 2014). This suggests that there is a mechanism by which Dcl5 can recognize IES-IES junctions in the transcribed template and reliably cut RNAs that map completely to IESs. Whether Dcl5 has a sequence preference or whether other proteins are involved remains to be determined.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Endura competent E. Coli cells | Lucigen | 60242-0 |
| Feeding bacteria HT115 E. Coli cells | Gift from Eric Meyer (ENS, Paris) | HT115 |
| Klebsiella pneumoniae non-virulent strain, food source for Paramecium | Gift from Eric Meyer (ENS, Paris) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Wheat Grass Powder | Pines International, Lawrence, KS | N/A |
| β-sitosterol | Calbiochem, Millipore | CAS 83-46-5 |
| 567152 | ||
| T4 DNA Ligase | New England Biolabs | M0202 |
| Critical Commercial Assays | ||
| TRI Reagent | Sigma-Aldrich | T3809 |
| GoScript Reverse Transcription kit | Promega | A5000 |
| GoTaq G2 DNA polymerase | Promega | M8296 |
| Wizard® SV Gel and PCR Clean-Up System | Promega | A9281 |
| pGEM®-T Easy Vector System | Promega | A3600 |
| Illumina TruSeq small RNA kit | Illumina | RS-200-0012 |
| Deposited Data | ||
| Ligase IV silencing and empty vector control silencing small RNA data | This work | GenBank, accession number ENA: SUB2107094 |
| Experimental Models: Organisms/Strains | ||
| Paramecium tetraurelia strain 51 | Gift from Eric Meyer (ENS, Paris) | N/A |
| Oligonucleotides | ||
| See Table S1 for complete list of oligonucleotides | This paper | N/A |
| Recombinant DNA | ||
| Plasmid L440 for feeding bacteria | Gift from Eric Meyer (ENS, Paris) | N/A |
| Dcl5 silencing construct | Sandoval et al., 2014 | N/A |
| Ligase IV silencing constructs | This work, see Table S1 | N/A |
| Software and Algorithms | ||
| Geneious 8.1.8 | http://www.geneious.com/ | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mariusz Nowacki (mariusz.nowacki@izb.unibe.ch).
Experimental Model and Subject Details
Paramecium tetraurelia strain 51, mating type 7, was used in all experiments. Cultivation and autogamy were carried out at 27°C as previously described (Beisson et al., 2010). Cells were grown in a wheat grass powder (WGP; Pines International, Lawrence, KS) infusion medium bacterized with Klebsiella pneumonia, supplemented with 0.8mg/l of β-sitosterol (Calbiochem, Milipore). Cells were diluted daily into fresh media during vegetative growth to ensure a constant food supply. After 25 vegetative cell divisions, autogamy was induced by starvation whereby cells were left to deplete the media of bacteria.
Method Details
Injection of dsDNA Corresponding to Cryptic IESs
Cells were starved to induce autogamy and stained with DAPI to check for developmental progression. When 100% of cells showed fragmented parental macronuclei, cells were microinjected with 100uM dsDNA oligonucleotides as described (Beisson et al., 2010). Single cells were immobilised in droplets of mineral water (Volvic) with 0.5% BSA under mineral oil (Sigma) using a micropipette. Microinjections were performed using a Femtojet microinjector and an InjectMan NI 2 micromanipulator along with a CellTram oil aspirator (Eppendorf). Since no macronuclei are visible at the selected developmental stage, DNA was injected into the cytoplasm. Cells were then refed in WGP and grown for eight divisions before being harvested for PCR.
Silencing Specific Genes by dsRNA Feeding
The dsRNA feeding method was used for specific gene silencing as previously described (Beisson et al., 2010). E.coli feeding bacteria containing the silencing constructs for Dcl5, Ligase 4a and 4b or empty vector EV, were inoculated overnight with shaking at 37°C in LB media supplemented with 0.0125mg/ml tetracycline and 0.1mg/ml ampicillin. The preculture was diluted 1:100 in WGP Paramecium medium containing 0.1mg/ml ampicillin and let grow overnight with shaking at 37°C. The next day, the culture was diluted 1:5 into the same medium and incubated 45 min in 37°C shaker. dsRNA synthesis was induced by the addition of IPTG at a final concentration of 0.4mM and the culture was incubated overnight shaking at 37°C. The feeding medium was then cooled to 27°C and supplemented with 0.8mg/ml of β-sitosterol. Paramecium cells (minimum 25 vegetative divisions) are added to the feeding medium at a concentration of 200 cells/ml. The culture is incubated at 27°C until Paramecium cells starve and autogamy is induced.
Efficient silencing of Dcl5 was confirmed by PCR on retained IESs, which specifically depend on Dcl5 for excision. RNA transcripts of concatenated IESs derive from late developmental stage (100% of cells with fragmented MACs plus the presence of a new MAC).
RNA Extraction, DNase I Treatment, and Reverse Transcription
Total RNA was extracted from 400ml Paramecium culture pellets frozen in liquid nitrogen, according to the TRI Reagent® BD protocol (catalog # T3809, Sigma-Aldrich). Cells were washed by centrifuging at 279 g for two minutes in 100 mL pear-shaped tubes and resuspending in 10 mM Tris-HCl pH 7.4 twice. They were then harvested by a further centrifugation step at 279 g for 2 min and the pellet was snap-frozen in liquid nitrogen. The frozen cell pellets were then resuspended in 5 mL TRI Reagent and the manufacturer’s protocol for RNA extraction was followed using chloroform and isopropanol, and RNA was resuspended in nuclease-free water (Ambion). Potential DNA contamination was prevented by DNase I (NEB) treatment of the RNA sample. RNA (4 μg) was reverse transcribed using GoScript™ Reverse Transcription System (Promega) with random primers. A negative control without reverse transcriptase was processed simultaneously to detect DNA contamination. Concatenated IES were amplified with GoTaq® G2 DNA Polymerase (Promega). Reaction mix consisted of 2 μL cDNA, GoTaq® G2 DNA Polymerase (0.625 units), Green GoTaq® Reaction Buffer, PCR nucleotide mix (0.2mM each dNTP) and IES-specific primers (0.5 μM) in a final reaction volume of 25 μl.
Cloning of PCR Products for Sequencing
Amplified DNA from PCR was run on agarose gel (0.8% in 0.5x TAE) and extracted, using Wizard® SV Gel and PCR Clean-Up System (Promega). Alternatively for small DNA amounts, gel extraction was performed with low melting point agarose (Invitrogen, 0.8% in 0.5x TAE), according to protocol. DNA was ligated into pGEM-T vector using the pGEM®-T Easy Vector System (Promega) and T4 DNA Ligase (400 units, NEB). Plasmids were transformed into electro-competent E.coli cells ‘Endura’, and positive clones were selected through ampicillin resistance and colony PCR for right sized inserts. Plasmids from positive bacterial clones were extracted, using Wizard® Plus SV Minipreps DNA Purification System (Promega). Sequencing of the cloned PCR product was performed by Microsynth.
Sequencing of Small RNA
Library preparation was performed according to Illumina’s protocol (TruSeq small RNA kit).
3 μg of Paramecium total RNA was used to ligate Illumina adapters (ATP, HML, RA3, RA5, RNase Inhibitor, STP, T4 RNA Ligase, Ultrapure Water). RNA was then reverse transcribed and PCR amplified (dNTP Mix, PML, RP1, RNA Primer Index, RTP, RNase Inhibitor, Ultrapure Water). cDNA was purified using CRL and HRL reagents. The library was checked using Agilent DNA 1000 kit, normalized with Tris-HC1 reagent, and sequenced on an Illumina HiSeq 2500 machine. Sequencing was performed by Fasteris, Geneva.
Data and Software Availability
The accession number for the RNA sequencing data from the Ligase IV silencing experiment reported in this paper is ENA: SUB2107094.
Quantification and Statistical Analysis
Experimental Replicates
For the injection experiments, replicates are described in the figure legends and n corresponds to the number of individually injected cells. Each individually injected cell was then allowed to progress through the end of development and commence vegetative growth, where all offspring are clonal.
Mapping of iesRNA Ends
iesRNAs (small RNAs of length 27-31 nt) from the control (unsilenced) subset of RNAs generated in the Ligase IV silencing experiment were mapped to the ends of IESs using Geneious 8.1.8 software. Reads were mapped to the ends of IESs with a minimum of 12 nt identity and zero mismatches. The reads with overhangs of 12 or more nt were then put in a random order and the 200 first reads chosen for mapping of the unmapped end. These were manually mapped to separate databases of IESs, Paramecium MAC including IESs, and Paramecium MAC with IESs removed, from the ParameciumDB (Arnaiz and Sperling, 2011).
Small RNA Quantification and Mapping
RNA reads from the sequencing experiment were binned by length and mapped to the databases for Paramecium MAC and IESs. Unmapped reads were removed from the analysis. Reads were normalized to 23 nt reads. Charts were made in Microsoft Excel.
Author Contributions
S.E.A. and M.N. designed the experiments. S.E.A performed the majority of the experiments. I.H. performed the experiments shown in Figures 3 and S2 (RT-PCR identifying RNA-level concatemers). S.P. and I. R. performed silencing of Ligase IV. C.H. helped I.H. and contributed to the design of the RT-PCR experiment. S.E.A. wrote the manuscript with input from M.N. M.N. supervised the project.
Acknowledgments
We would like to thank Eric Meyer for inspiring discussions regarding the experiments and Nasikhat Stahlberger for technical support. This research was supported by grants from the European Research Council (ERC) (260358 “EPIGENOME” and 681178 “G-EDIT”), the Swiss National Science Foundation (31003A_146257 and 31003A_166407), and the National Center of Competence in Research (NCCR) RNA and Disease.
Published: March 9, 2017
Footnotes
Supplemental Information includes three figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2017.02.020.
A video abstract is available at http://dx.doi.org/10.1016/j.cell.2017.02.020#mmc2.
Supplemental Information
References
- Aravin A.A., Sachidanandam R., Bourc’his D., Schaefer C., Pezic D., Toth K.F., Bestor T., Hannon G.J. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O., Sperling L. ParameciumDB in 2011: new tools and new data for functional and comparative genomics of the model ciliate Paramecium tetraurelia. Nucleic Acids Res. 2011;39(Database issue):D632–D636. doi: 10.1093/nar/gkq918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O., Mathy N., Baudry C., Malinsky S., Aury J.-M., Denby Wilkes C., Garnier O., Labadie K., Lauderdale B.E., Le Mouël A. The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genet. 2012;8:e1002984. doi: 10.1371/journal.pgen.1002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronica L., Bednenko J., Noto T., DeSouza L.V., Siu K.W.M., Loidl J., Pearlman R.E., Gorovsky M.A., Mochizuki K. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008;22:2228–2241. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry C., Malinsky S., Restituito M., Kapusta A., Rosa S., Meyer E., Bétermier M. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 2009;23:2478–2483. doi: 10.1101/gad.547309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson J., Bétermier M., Bré M.-H., Cohen J., Duharcourt S., Duret L., Kung C., Malinsky S., Meyer E., Preer J.R. Silencing specific Paramecium tetraurelia genes by feeding double-stranded RNA. Cold Spring Harb. Protoc. 2010;2010 doi: 10.1101/pdb.prot5363. pdb.prot5363. [DOI] [PubMed] [Google Scholar]
- Brabant F., Acheson N.H. RNA footprint mapping of RNA polymerase II molecules stalled in the intergenic region of polyomavirus DNA. J. Virol. 1995;69:4423–4430. doi: 10.1128/jvi.69.7.4423-4430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Czech B., Hannon G.J. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem. Sci. 2016;41:324–337. doi: 10.1016/j.tibs.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley D.D., Chaudhuri J., Bassing C.H., Alt F.W. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. In: Alt F.W., editor. Advances in Immunology. Academic Press; 2005. pp. 43–112. [DOI] [PubMed] [Google Scholar]
- Gratias A., Bétermier M. Processing of double-strand breaks is involved in the precise excision of paramecium internal eliminated sequences. Mol. Cell. Biol. 2003;23:7152–7162. doi: 10.1128/MCB.23.20.7152-7162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratias A., Lepère G., Garnier O., Rosa S., Duharcourt S., Malinsky S., Meyer E., Bétermier M. Developmentally programmed DNA splicing in Paramecium reveals short-distance crosstalk between DNA cleavage sites. Nucleic Acids Res. 2008;36:3244–3251. doi: 10.1093/nar/gkn154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Jahn C.L., Klobutcher L.A. Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 2002;56:489–520. doi: 10.1146/annurev.micro.56.012302.160916. [DOI] [PubMed] [Google Scholar]
- Kapusta A., Matsuda A., Marmignon A., Ku M., Silve A., Meyer E., Forney J.D., Malinsky S., Bétermier M. Highly precise and developmentally programmed genome assembly in Paramecium requires ligase IV-dependent end joining. PLoS Genet. 2011;7:e1002049. doi: 10.1371/journal.pgen.1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H., Hayashizaki Y. Exploration of small RNAs. PLoS Genet. 2008;4:e22. doi: 10.1371/journal.pgen.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliszewska-Olejniczak K., Gruchota J., Gromadka R., Denby Wilkes C., Arnaiz O., Mathy N., Duharcourt S., Bétermier M., Nowak J.K. TFIIS-dependent non-coding transcription regulates developmental genome rearrangements. PLoS Genet. 2015;11:e1005383. doi: 10.1371/journal.pgen.1005383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., Terns R.M., Terns M.P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Gorovsky M.A. RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryot. Cell. 2004;3:1233–1240. doi: 10.1128/EC.3.5.1233-1240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J., Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Rausch T., Jones D.T.W., Zapatka M., Stütz A.M., Zichner T., Weischenfeldt J., Jäger N., Remke M., Shih D., Northcott P.A. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval P.Y., Swart E.C., Arambasic M., Nowacki M. Functional diversification of Dicer-like proteins and small RNAs required for genome sculpting. Dev. Cell. 2014;28:174–188. doi: 10.1016/j.devcel.2013.12.010. [DOI] [PubMed] [Google Scholar]
- Sijen T., Steiner F.A., Thijssen K.L., Plasterk R.H.A. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Singh D.P., Saudemont B., Guglielmi G., Arnaiz O., Goût J.-F., Prajer M., Potekhin A., Przybòs E., Aubusson-Fleury A., Bhullar S. Genome-defence small RNAs exapted for epigenetic mating-type inheritance. Nature. 2014;509:447–452. doi: 10.1038/nature13318. [DOI] [PubMed] [Google Scholar]
- Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart E.C., Wilkes C.D., Sandoval P.Y., Arambasic M., Sperling L., Nowacki M. Genome-wide analysis of genetic and epigenetic control of programmed DNA deletion. Nucleic Acids Res. 2014;42:8970–8983. doi: 10.1093/nar/gku619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Davis R.E. Programmed DNA elimination in multicellular organisms. Curr. Opin. Genet. Dev. 2014;27:26–34. doi: 10.1016/j.gde.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S., White C.I., Rendtlew Danielsen J.M., Yang Y.-G., Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.