Myeloid STAT3 promotes type I interferon response, and DC and NK cell activation, to protect mice against HSV-1 infection.
Keywords: innate immunity, viral infection, type I interferons, dendritic cells, natural killer cells
Abstract
Signal transducer and activator of transcription 3 (STAT3) mediates cellular responses to multiple cytokines, governs gene expression, and regulates the development and activation of immune cells. STAT3 also modulates reactivation of latent herpes simplex virus-1 (HSV-1) in ganglia. However, it is unclear how STAT3 regulates the innate immune response during the early phase of HSV-1 lytic infection. Many cell types critical for the innate immunity are derived from the myeloid lineage. Therefore, in this study, we used myeloid-specific Stat3 knockout mice to investigate the role of STAT3 in the innate immune response against HSV-1. Our results demonstrate that Stat3 knockout bone marrow-derived macrophages (BMMs) expressed decreased levels of interferon-α (IFN-α) and interferon-stimulated genes (ISGs) upon HSV-1 infection. In vivo, knockout mice were more susceptible to HSV-1, as marked by higher viral loads and more significant weight loss. Splenic expression of IFN-α and ISGs was reduced in the absence of STAT3, indicating that STAT3 is required for optimal type I interferon response to HSV-1. Expression of TNF-α and IL-12, cytokines that have been shown to limit HSV-1 replication and pathogenesis, was also significantly lower in knockout mice. Interestingly, Stat3 knockout mice failed to expand the CD8+ conventional DC (cDC) population upon HSV-1 infection, and this was accompanied by impaired NK and CD8 T cell activation. Collectively, our data demonstrate that myeloid-specific Stat3 deletion causes defects in multiple aspects of the immune system and that STAT3 has a protective role at the early stage of systemic HSV-1 infection.
Introduction
HSV-1 is estimated to infect more than two thirds of the population worldwide. Common symptoms of HSV-1 infection, including local skin or mucosa membrane lesions, usually resolve within a few days to several weeks [1]. Localized HSV-1 infections are also often accompanied by viremia [2, 3]. Following lytic infection, the virus retreats to neurons where it establishes latency. Latency is lifelong and may be asymptomatic with occasional relapses [1]. HSV-1 infection seldom causes severe disease in healthy individuals. However, newborns, the elderly, and immunocompromised individuals are prone to HSV-1–induced diseases, such as herpes simplex keratitis and life-threatening herpes simplex encephalitis [1]. These susceptible populations also are more likely to exhibit HSV-1 viremia in the blood [2], and it is now becoming more apparent that primary HSV-1 infection and reactivation can occur beyond the mucocutaneous areas in both immunocompetent and immunocompromised individuals [2]. It is, therefore, important to understand the mechanism by which the immune system controls HSV-1 infection.
Both innate and adaptive immunity are critical in controlling HSV-1 infection. Several major cellular mediators of the innate immunity implicated in limiting HSV-1 pathogenesis are of the myeloid lineage, including macrophages, DCs, and neutrophils. Macrophages are intrinsically resistant to productive HSV-1 infection [4, 5]. Depletion of macrophages in mice leads to higher viral replication and exaggerated pathology [6, 7], whereas adoptive transfer of adult macrophages protects susceptible neonates from lethal HSV-1 infection [8]. Macrophages also limit the replication and dissemination of HSV-1 in the peripheral nervous system [9]. The protective effects of macrophages are partially mediated through secreted factors, including IL-12, TNF-α, and nitric oxide. These factors inhibit viral replication, recruit leukocytes to the site of infection, promote the cellular immune response, and further activate macrophages to protect mice from HSV-1 and other herpesvirus infections [9–13]. Specialized macrophages, such as MZMΦs and MMMΦs, produce substantial amounts of type I IFNs to elicit antiviral response during HSV-1 viremia [14]. In addition to macrophages, DCs have a pivotal role in immunity against herpesvirus infections. Mice depleted of DCs have shortened survival and higher viral loads in their nervous systems during HSV-1 infection, and the susceptibility is attributed to impaired NK and T cell activation in the absence of DCs [15]. Major subsets of DCs include CD11b+ cDCs, CD8+ cDCs, and pDCs. Both CD8+ cDCs and pDCs are involved in promoting NK cell activation and anti-HSV CTL immunity [16–18], but only CD8+ cDC is able to cross-present and directly activate CD8 T cells [19, 20]. Importantly, pDCs are capable of producing large quantities of type I IFNs during HSV-1 infection [17]. Finally, neutrophils have a dual role in HSV-1 infection. It has been suggested that neutrophils inhibit HSV-1 replication [21], but further examination revealed that neutrophils mediate HSV-1–induced tissue injury and immunopathology [22, 23].
Signal transducer and activator of transcription 3 (STAT3) is a transcription factor that mediates the cellular responses to multiple cytokines by promoting the expression of genes implicated in survival, inflammation, proliferation, and migration [24]. STAT3 modulates HSV-1 reactivation in the ganglia [25] and governs various aspects of myeloid cell proliferation, differentiation, and responses to infections [26–29]. For example, STAT3-null macrophages display heightened immune responses in response to LPS and bacterial infections, as marked by the overproduction of proinflammatory cytokines [26, 30]. Additionally, STAT3-null neutrophils are defective in their bactericidal capacity [30]. Nonetheless, the role of STAT3 in the myeloid lineage during viral infections has not been fully investigated.
Given the critical role of macrophages and DCs in limiting HSV-1 pathogenesis at the early phase of infection, we investigated whether STAT3 in the myeloid lineage modulates the innate immune response to HSV-1. Here, we show that the type I IFN response is partially defective in STAT3-null BMMs during HSV-1 infection. In vivo, myeloid Stat3 KO mice exhibited greater viral loads and more significant weight loss during HSV-1 infection compared with WT mice. Importantly, KO mice have decreased splenic expression of IFN-α and ISGs, as well as decreased expression of IL-12 and TNF-α, cytokines known to be protective against HSV-1 infection. Finally, Stat3 KO mice failed to expand CD8+ cDCs during HSV-1 infection, and activation of NK and CD8 T cells was also impaired. In summary, our results indicate that STAT3 in the myeloid lineage contributes to the resistance to HSV-1 infection at the early stage by modulating multiple aspects of innate immunity.
MATERIALS AND METHODS
Animals
This study was performed under protocols approved by Institutional Animal Care and Use Committee guidelines of the UNC (Chapel Hill, NC, USA). Mice were kept and bred in sterile facilities under conditions in accordance with the U.S. National Institutes of Health (Bethesda, MD, USA) Guide for the Care and Use of Laboratory Animals and UNC Institutional Animal Care and Use Committee guidelines. STAT3fl/fl and LysM-Cre C57BL/6 mice were obtained from the Jackson laboratory (Bar Harbor, ME, USA). Mice were bred to each other to generate myeloid-specific STAT3 KO mice. Eight to 12-wk-old animals were used for HSV-1 infection studies. For infections, mice were i.v. injected via the tail vein with HSV-1 (KOS strain) and monitored 1 to 3 times daily after injection. To collect tissues for molecular studies, mice were euthanized by CO2 followed by cervical dislocation. For survival studies, mice that lost more than 20% of their original weight were immediately euthanized, whereas all other mice were euthanized at the end of the study.
Virus, cells, and plaque assay
Vero cells (ATCC, Manassas, VA, USA) were propagated in DMEM with 10% FBS. HSV-1 (KOS strain) was amplified on Vero cells. At 3 dpi, supernatants were harvested and centrifuged at 2500 rpm to remove cell debris. The supernatants were then ultracentrifuged at 25,000 rpm to concentrate the virus. Virus pellets were resuspended in plain DMEM.
To isolate BMMs, bone marrow was collected from the femur and tibia of age- and sex-matched 8–10-wk-old mice. The cells were cultured in RPMI 1640 with 10% FBS and 10% L929-conditioned media. After 5–6 d of culture, the adherent cells contained about 95% F4/80+ macrophages as determined by flow cytometry. BMMs were infected with HSV-1 at an MOI of 3, incubated at 37°C for 30 min, washed with DMEM twice, and cultured in DMEM with 2% FBS.
Infectious viral titer was determined by plaque assay using Vero cells. Supernatants from infected macrophages were serial-diluted in DMEM 1640 before infecting confluent Vero cells for 1 h at 37°C; after which, the supernatants were replaced with DMEM with 1% methylcellulose and 2% serum. Three technical replicates were prepared for each dilution. Infected Vero cells were kept at 37°C with 5% CO2 for 72 h before staining with 0.5% crystal violet in 50% ethanol to reveal the plaques. Titers were determined as PFUs per milliliter.
Quantitative real-time PCR
Total RNA from the cultured macrophages or mouse tissues were purified with Qiagen (Germantown, MD, USA) RNeasy Plus kit, according to manufacturer’s protocol. RNA (1–2 μg) was used to synthesize cDNA using M-MLV reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA). For gene expression assay, standard TaqMan probes, primers, and reagents were used (Thermo Fisher Scientific). To determine the relative genomic copy number of HSV-1 in mouse tissues, real-time PCR was performed using Maxima SYBR green master mix (Thermo Fisher Scientific) with the following primers: Actb forward: 5′-GGCTGTATTCCCCTCCATCG-3′; Actb reverse: 5′-CCAGTTGGTAACAATGCCATGT-3′; HSV-1 pol forward: 5′-CATCACCGACCCGGAGAGGGAC-3′; HSV-1 pol reverse: 5′-GGGCCAGGCGCTTGTTGGTGTA-3′.
Immunoblotting
Cells or tissues were lysed in radioimmunoprecipitation assay buffer (10 mM Tris, pH 8.0; 1 mM EDTA; 1 mM EGTA; 1% Triton X-100; 0.1% sodium deoxycholate; 0.1% SDS; 140 mM NaCl) with a protease inhibitor cocktail (Promega, Madison, WI, USA) and a phosphatase inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA). Lysates were centrifuged to remove cell debris, and the supernatant were mixed with lithium dodecyl sulfate loading buffer (Thermo Fisher Scientific) containing 100 mM DTT and then, heated before running SDS-PAGE with bis-Tris NuPAGE gel and MOPS buffer (Thermo Fisher Scientific). Proteins were transferred onto 0.45-μm polyvinylidene difluoride membrane, blocked with 5% milk in TBST, and incubated in primary Abs at a dilution of 1:500–1:2000 in 5% milk in TBST overnight. Blots were then washed and incubated in the appropriate HRP-conjugated secondary Abs (Promega). Chemiluminescence substrate (Thermo Fisher Scientific) was used for film exposure. STAT1 (9172), p-STAT1 (9167), STAT3 (9139), p-STAT3 (9145), ISG15 (2758), p-p65 (3033), p65 (3034), p-IκBα (9246), and IκBα (4812) Abs were from Cell Signaling Technology (Danvers, MA, USA).
Flow cytometry
Mice were euthanized to collect the spleens and bone marrow. Spleens were mashed using syringe plungers and 100-μm cell strainers. The cells were resuspended in FACS buffer (3% FBS, 1 mM EDTA in PBS) and passed through 40-μm cell strainer, and red blood cells were lysed with ACK buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA). The white blood cell suspensions were blocked with an anti-mouse CD16/CD32 Ab (14-0161-86; Thermo Fisher Scientific) at 1:100 dilution for 30 min before being stained with the appropriate primary and secondary Abs for 1 h each at 1:100 dilution in FACS buffer. CD8-FITC (11-0081-81), CD4-PE (12-0042-81), CD19-PE-Cy7 (250193-81), CD3-APC (17-0032-82), F4/80-Alexa488 (53-4801-82), CD11b-PE-Cy7 (250112-82), CD11c-APC (17-0114-82), CD209-PE (12-2091-82), and streptavidin-eF450 (48-4317-82) Abs were from Thermo Fisher Scientific. Ly6G-PE (108407), Ly6C-APC (128015), B220-biotin (103204), CD169-PE-Cy7 (142412), CD69-APC (104513), and NK1.1-PE-Cy7 (108713) Abs were from BioLegend (San Diego, CA, USA). Flow cytometry was performed on a CyAn machine and analyzed with FlowJo software (Tree Star, Ashland, OR, USA).
Immunohistochemistry
Mouse tissues were fixed in 10% buffered formalin for 24 h, washed with PBS 3 times, and incubated in 30%, 50%, and 70% ethanol for 30 min each. Fixed tissues were embedded in paraffin and sectioned into slices. Immunohistochemistry was performed with a Bond Immunostainer (Leica Biosystems, Wetzlar, Germany). Slides were dewaxed in Bond dewax solution (AR9222) and hydrated in Bond wash solution (AR9590). Ag retrieval was performed for 30 min at 100°C with Bond-Epitope Retrieval solution1 pH-6.0 (AR9961), followed by a 10-min protein block (X0909; Dako, Carpinteria, CA, USA). Rodent block M reagent (RB961G; Biocare Medical, Concord, CA, USA) was applied for 20 min only to STAT3 slides. After pretreatment, slides were incubated for 30 min with CD45 and Ly-6G (25386, 25337; Abcam, Cambridge, MA, USA) Abs at 1:100 and 1:200 dilution, respectively, for 1 h with the STAT3 (9139; Cell Signaling) Ab at 1:750 dilution. Detection of all Abs was performed with Bond Polymer Refine Detection (DS9900; Leica Biosystems), and for CD45 and Ly-6G, the secondary Ab from the kit was replaced with ImmPRESS HRP anti-rat Ig (MP-7444-15, Vector Laboratories, Burlingame, CA, USA). Stained slides were dehydrated and coverslipped. Positive and negative controls (no primary Ab) were also used with each Ab to ensure the specificity of staining.
Statistical analyses
Three to 5 technical replicates were used for each data point. Data points are shown as means ± sd, and the 2-tailed Student’s t test was used to calculate P values and significance, unless otherwise noted.
RESULTS
Characterization of Stat3fl/fl LysM-Cre+/+ mice
To investigate whether STAT3 in the myeloid lineage affects the innate immune response to HSV-1, we crossed Stat3fl/fl mice to LysM-Cre mice to generate myeloid-specific Stat3 KO mice. Cre recombinase driven by the lysozyme (LysM) promoter is specifically expressed in the myeloid lineage, leading to conditional deletion of loxP-flanked target genes in granulocytes, monocytes, and macrophages [31]. In agreement with the literature, BMMs from Stat3fl/fl LysM-Cre+/+ mice (hereafter, referred to as KO mice) showed >90% reduction of STAT3 protein compared with BMMs from Stat3wt/wt LysM-Cre+/+ mice (hereafter, referred to as WT mice). No noticeable reduction of STAT3 protein levels were observed in the brain, liver, or muscle as determined by immunoblotting (Supplemental Fig. 1).
The Stat3fl/fl mice used in this study (see Materials and Methods) differed from the strain used in the previous LysM-Cre/Stat3 mouse study [26]. Thus, to determine the effect of myeloid Stat3 deletion with regard to this specific strain, we examined the composition of lymphocytes and myeloid cells in the KO mice. Lack of Stat3 in the myeloid lineage did not affect the composition of major cell populations in the spleen. Specifically, the percentages of B cells, T cells, neutrophils, monocytes, red pulp macrophages, MMMΦs, and MZMΦs in the spleen were similar between WT and KO mice (Table 1). In the bone marrow, Stat3 KO mice had more SSChi granulocytes and CD11b+ myeloid cells because of a significant expansion of CD11b+ Ly6G+ neutrophils within the granulocyte compartment (Table 2). An increase of the CD11b+ Ly6Chi monocytes was also observed (Table 2). The expansion of neutrophils was accompanied by a mild decrease in B cell and T cell populations in the bone marrow. Neutrophilia has been reported by other groups using hematopoietic-specific or inducible Stat3 KO mice [28, 29]. Our results suggest that myeloid-specific deletion of Stat3 was sufficient to cause the neutrophil expansion phenotype. The number of CD11blo F4/80+ macrophages in the bone marrow did not differ between WT and Stat3 KO mice (Table 2). Similar to the previous study [26], we did not observe a differentiation block of STAT3-null BMMs cultured in vitro (data not shown), suggesting that STAT3 is not crucial in normal macrophage differentiation. Taken together, our data agreed with previous studies and independently verified that myeloid Stat3 deletion leads to neutrophil expansion in the bone marrow but has minimal effects on splenic cells or BMM differentiation by using a different Stat3fl/fl strain.
TABLE 1.
Cellular composition of the spleena
| Cells | WT (n = 3) | KO (n = 3) |
|---|---|---|
| Myeloid cellsb | 12.3 ± 4.7 | 11.8 ± 2.6 |
| Neutrophilsc (% myeloid) | 16.9 ± 5.9 | 19.6 ± 7.4 |
| Ly6Chi monocytesd (% myeloid) | 28.7 ± 2.0 | 32.6 ± 10.5 |
| Ly6Clo monocytese (% myeloid) | 51.9 ± 7.6 | 45.3 ± 17.9 |
| Red pulp macrophagesf | 2.42 ± 0.40 | 2.77 ± 0.12 |
| MZMΦsg | 0.24 ± 0.03 | 0.28 ± 0.08 |
| MMMΦsh | 0.40 ± 0.11 | 0.51 ± 0.18 |
| B cellsi | 56.7 ± 4.2 | 51.6 ± 3.2 |
| T cellsj | 26.4 ± 1.0 | 27.4 ± 5.7 |
| CD4+ T cellsk (% T cells) | 50.6 ± 3.5 | 52.7 ± 3.4 |
| CD8+ T cellsl (% T cells) | 41.3 ± 1.7 | 39.0 ± 1.7 |
All numbers are expressed as the percentages of the total cells, except where noted.
CD11b+
CD11b+Ly6G+Ly6C+
CD11b+Ly6G−Ly6C+
CD11b+Ly6G−Ly6Clo
CD11bloF4/80+
CD11c−CD209+
CD11c−CD169+
CD19+
CD3+
CD3+CD4+
CD3+CD8+
TABLE 2.
Cellular composition of the bone marrowa
| Cells | WT (n = 3) | KO (n = 2) |
|---|---|---|
| Granulocytesb | 54.8 ± 1.2 | 68.2 ± 4.5* |
| Myeloid cellsc | 45.7 ± 0.8 | 58.9 ± 6.2* |
| Neutrophilsd | 27.9 ± 0.3 | 40.4 ± 5.0* |
| Neutrophils (% myeloid cells) | 61.3 ± 1.2 | 68.6 ± 1.3** |
| Macrophagese | 1.3 ± 0.2 | 1.1 ± 0.5 |
| Ly6Chi monocytesf | 5.3 ± 0.4 | 6.4 ± 0.3* |
| Ly6Clo monocytesg | 10.2 ± 0.8 | 10.2 ± 0.9 |
All numbers are expressed as the percentages of the total cells, except where noted.
SSChi
CD11b+
CD11b+Ly6G+Ly6C+
CD11bloF4/80+
CD11b+Ly6G−Ly6Chi
CD11b+Ly6G−Ly6Clo
P < 0.05; **P < 0.01
Stat3 KO BMMs show attenuated type I IFN response to HSV-1 infection
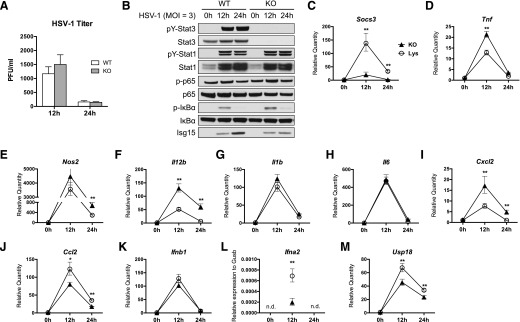
HSV-1 is able to enter and infect BMMs and peritoneal macrophages, but few infectious viral particles are produced after virus entry, indicating that macrophages are nonpermissive to productive HSV-1 infection [4, 5]. To determine whether STAT3 has a role in the intrinsic resistance of macrophages to HSV-1, BMMs from WT or KO mice were infected with HSV-1 at an MOI of 3, and infectious viral particles in the supernatant were measured at 12 and 24 h after infection by plaque assay. Similar to WT BMMs, KO BMMs produced limited amounts of infectious viral particles, and the viral titer was reduced over time (Fig. 1A), indicating that KO BMMs were capable of restricting HSV-1 replication. No statistical difference was observed between WT and KO macrophages in their susceptibility to HSV-1, as measured by infectious viral particles produced (Fig. 1A). Thus, Stat3 deletion in the macrophages does not impair their intrinsic resistance to HSV-1.
Figure 1. STAT3 KO BMMs show attenuated type I IFN response to HSV-1 infection.
WT or Stat3 KO BMMs were infected with HSV-1 at an MOI of 3. (A) Viral titer in the supernatants at 12 and 24 h after infection was determined by plaque assay on Vero cells with 3 technical replicates. (B) Western blotting of lysates from HSV-1–infected BMMs at the indicated times. (C–M) Gene expression of HSV-1–infected BMMs was determined by quantitative real-time PCR with 3 technical replicates for each sample. All values were normalized to Gusb and compared with the expression of WT BMMs at 0 h, except for IFN-α, which is shown as its relative expression to Gusb. All values are means with 95% confidence intervals. Statistical significance was determined by Student’s t test. *P < 0.05, **P < 0.01, n.d., not determined (below detection threshold).
To further investigate the response of BMMs to HSV-1, we examined the signaling events and host gene expression after HSV-1 infection. HSV-1 induced robust tyrosine phosphorylation and activation of STAT1 and STAT3 in WT BMMs, and STAT1 activation appeared to be normal in the absence of STAT3 (Fig. 1B). p-IκBα was induced, and p-p65 was induced to a lesser extent, indicating activation of the NF-κB pathway by HSV-1 infection (Fig. 1B). Importantly, p-IκBα levels were higher in KO BMMs, indicating enhanced NF-κB activity, whereas the induction of ISG15, an IFN-stimulated gene, was reduced in KO BMMs (Fig. 1B). HSV-1 also induced the expression of several genes involved in signaling and immune regulation (Fig. 1C–M). Of note, induction of SOCS3, a STAT3 inhibitor that is highly up-regulated by active STAT3 [32], was blunted in KO macrophages (Fig. 1C), confirming the lack of STAT3 functionality in KO BMMs.
It has been reported that STAT3-null macrophages express higher levels of proinflammatory genes in response to LPS and bacterial infection [26, 30]. Similarly, we found that, during HSV-1 infection, KO BMMs expressed more TNF-α, iNOS, and IL-12b than did WT BMMs (Fig. 1D–F) but not other inflammatory cytokines, such as IL-1β and IL-6 (Fig. 1G and H). KO BMMs also expressed more CXCL2 (Fig. 1I), a neutrophil chemoattractant responsible for neutrophil mobilization to the site of HSV-1 infection [33]. In contrast, induction of CCL2 a monocyte chemoattractant and a STAT3 target gene, was decreased in KO BMMs (Fig. 1J). These data show that Stat3 KO BMMs induced greater expression of a subset of proinflammatory cytokines upon viral infection. We also examined the expression of type I IFNs and ISGs. Although expression of IFN-β was unaffected (Fig. 1K), HSV-1–induced IFN-α expression was severely diminished in the absence of STAT3 (Fig. 1L). Moreover, expression of 2 ISGs, ISG15 and USP18, was decreased in the KO BMMs (Fig. 1B and M), indicating that, although STAT3 is dispensable for IFN-β expression, it is required for IFN-α expression and for optimal expression of ISGs in response to HSV-1 infection.
Taken together, our data show that in vitro HSV-1 infection activates STAT1 and STAT3 in BMMs. Loss of STAT3 did not impair STAT1 activation or the intrinsic ability of BMMs to limit HSV-1 replication. However, Stat3 KO BMMs had significantly lower expression of IFN-α and ISGs, suggesting that effective induction of IFN-α and ISGs in BMMs during HSV-1 infection is dependent on STAT3.
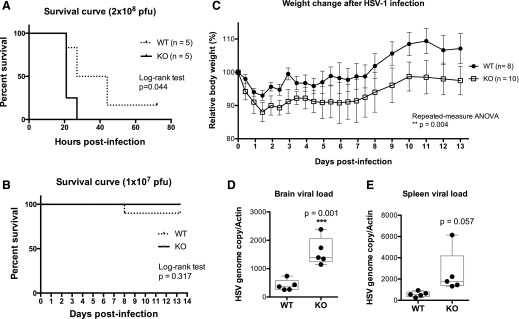
Myeloid Stat3 KO mice are more susceptible to HSV-1 infection
Type I IFNs, including IFN-α and IFN-β, are critical in mounting an antiviral defense at the early stage of viral infection by inducing expression of ISGs [34]. The attenuated type I IFN response in Stat3 KO macrophages may have profound effects on the outcome of systemic HSV-1 infection. Thus, we investigated whether Stat3 KO mice were more susceptible to systemic HSV-1 infection. WT or KO mice were infected with 2 × 108 PFU of HSV-1 via tail-vein injection. Most mice succumbed to death within 2 d, but the WT mice survived longer (Fig. 2A). Viral loads, as determined by HSV-1 genome copy number relative to mouse β-actin copy number, showed a trend of increase in the brain, liver, and spleen of KO mice, but the difference was not statistically significant (Supplemental Fig. 2). These results suggest that STAT3 in myeloid cells has a protective role during lethal, systemic HSV-1 infections and prompted further investigation.
Figure 2. Myeloid Stat3 KO mice are more susceptible to HSV-1 infection.
(A) Sex- and age-matched WT or KO mice (n = 5 each) were i.v. injected with 2 × 108 PFU of HSV-1. Survival of infected mice was monitored 3 times daily. Statistical analysis was performed by the log-rank test. (B) Sex- and age-matched WT or KO mice (n = 10 each) were i.v. injected with 1 × 107 PFU of HSV-1. Survival of infected mice was monitored and analyzed as described in panel A. (C) Sex- and age-matched WT or KO mice were i.v. injected with 1 × 107 PFU of HSV-1. Each mouse was weighted immediately before infection and twice daily after infection. All measurements were normalized to the starting weight of each mouse and are expressed as percentages of the starting weight. Statistical analysis was performed by repeated-measure ANOVA using PRISM (GraphPad Software, La Jolla, CA, USA). (D–E) Mice of each genotype (n = 5 each) were infected as described in panel C and were sacrificed 2 dpi. HSV-1 viral loads in the brain or the spleen were determined by quantitative real-time PCR with 3 technical replicates for each mouse. Data are shown as HSV-1 copy number relative to mouse Actb expression. Statistical significance was determined by Student’s t test. **P < 0.01.
Next, we challenged the mice with a lower dose of HSV-1. Mice were infected with 1 × 107 PFU of HSV-1 by tail-vein injection. Both groups of mice displayed symptoms, including hunched posture, signs of hind limb weakness, and significant weight loss starting at 2 dpi. No statistical difference in survival was observed (Fig. 2B). However, STAT3 KO mice experienced significantly worse weight loss during the course of infection (Fig. 2C). To determine the effect of myeloid-specific STAT3 deletion on the innate immunity against HSV-1 at the early stage of infection, we examined the viral loads in the brain and spleen at 2 dpi. STAT3 KO mice had significantly higher viral loads in the brain at 2 dpi (Fig. 2D). Viral loads were also marginally higher in the spleens of KO mice at 2 dpi (Fig. 2E). These data show that Stat3 KO mice were more susceptible to HSV-1 infection, and the differences are discernible at earlier stages of infection.
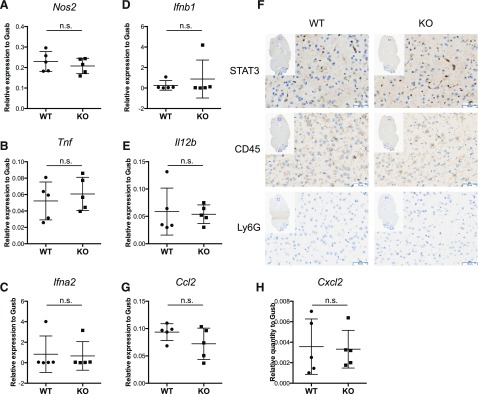
Susceptibility of Stat3 KO mice is not caused by immunopathology in the brain
To investigate the mechanisms contributing to the higher viral loads in the brains of KO mice, we examined the expression of immunoregulatory genes in the brain. Factors including IFN-α, IFN-β, IFN-γ, IL-12, TNF-α, and nitric oxide are known to be critical in inhibiting HSV-1 replication in various settings in vitro and in vivo [10–13, 35, 36]. Although IFN-γ expression in the brain was below the detection threshold (data not shown), iNOS, TNF-α, IFN-α, IFN-β, and IL-12 were not differentially expressed in the brains of WT and KO mice (Fig. 3A–E) and, thus, cannot account for the higher viral loads in the brains of Stat3 KO mice. Microglia, macrophage-like cell types in the brain, are intrinsically resistant to HSV-1 infection [37] and elicit significant immune responses in the brain during HSV-1 infection [23, 38, 39]. Given that LysM-Cre has been reported to result in partial deletion of the loxP-flanked gene in microglia [40], it is possible that LysM-Cre causes Stat3 deletion in microglia, thereby altering their responses to HSV-1 infection and undermining their ability to restrict HSV-1 replication in the brain. However, we found that WT microglia expressed low levels of STAT3 and the expression was not noticeably reduced in the microglia of Stat3 KO animals (Fig. 3F). Previous studies also suggested that immune cell infiltrates were the major cause of herpes simplex encephalitis immunopathology [23, 41]. In this setting, CCL2 and CXCL2 produced by microglia are the primary macrophage and neutrophil chemoattractants [38, 42]. To probe the possibility of immunopathology in Stat3 KO mice, we further examined the expression of CCL2 and CXCL2. We found that CCL2 and CXCL2 were not differentially expressed between WT and KO mice (Fig. 3G and H). Moreover, there was no evidence of CD45+, hematopoietic-origin infiltrating cells or Ly6G+ neutrophils in the brains of either WT or KO mice (Fig. 3F). Taken together, our data show that the higher viral loads in the brain and the exaggerated symptoms in Stat3 KO mice are not due to immune cell infiltration or microglia defects in the brain. Rather, they are more likely caused by the failure of the immune system in Stat3 KO mice to restrain the replication and spread of HSV-1 to the central nervous system.
Figure 3. Susceptibility of Stat3 KO mice is not caused by immunopathology in the brain.
WT or KO mice were i.v. injected with 1 × 107 PFU of HSV-1 as described in Fig. 3. Mice of each genotype (n = 5 each) were sacrificed 2 dpi to collect brain tissue. (A–E) Relative expression of iNOS, TNF-α, IFN-α, IFN-β, and IL-12 in the brain was determined by quantitative real-time PCR with 3 technical replicates for each mouse. All values were normalized to Gusb expression and are shown as means with 95% confidence intervals. Statistical analysis was performed with the Student’s t test. n.s., not significant. (F) Brain hemispheres from WT or KO mice sacrificed at 2 dpi were fixed and stained with STAT3, CD45, or Ly6G Abs, as described in Materials and Methods. Microglia showed low levels of CD45 staining. Representative figures from each genotype are shown. (G and H) Relative expression of CCL2 and CXCL2 in the brain was determined by quantitative real-time PCR, as mentioned above.
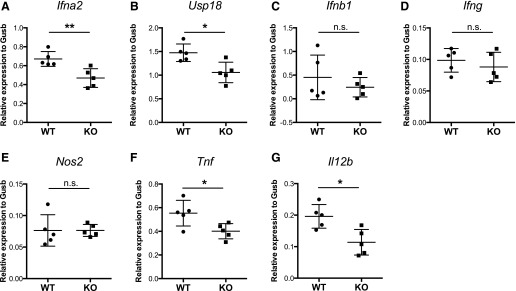
Stat3 KO mice express reduced levels of antiviral cytokines in the spleen
Next, we examined the immune responses in the spleens of infected mice. Infection (i.v.) of HSV-1 in mice is known to elicit robust immune responses in the spleen, and splenic MMMΦs and MZMΦs are the major producer of type I IFNs in that setting [14]. Myeloid Stat3 deletion did not affect the percentage of MMMΦs and MZMΦs in the spleen (Table 1). However, similar to the findings with Stat3 KO BMMs (Fig. 1), splenic expression of IFN-α and USP18 at 2 dpi was significantly lower in the KO mice, whereas expression of IFN-β and IFN-γ was unaffected (Fig. 4A–D), indicating a partially defective type I IFN response. In contrast, splenic expression of iNOS was not affected by myeloid Stat3 deletion (Fig. 4E), and splenic expression of TNF-α and IL-12β was less in KO mice (Fig. 4F and G).
Figure 4. Stat3 KO mice express reduced levels of antiviral cytokines in the spleen.
WT or KO mice were infected as described in the caption to Fig. 3. Mice of each genotype (n = 5) were sacrificed 2 dpi to collect spleen tissue. (A–H) Expression of IFN-α, USP18, IFN-β, IFN-γ, iNOS, TNF-α, and IL-12 in the spleen was determined by quantitative real-time PCR with 3 technical replicates for each mouse. All values were normalized to Gusb expression and are shown as means with 95% confidence intervals. Statistical analysis was performed with a t test. **P < 0.01, *P < 0.05, n.s., not significant.
These data show that type I IFN response in the spleens of Stat3 KO mice is also impaired during systemic HSV-1 infection, whereas expression of type II IFN, namely IFN-γ, is not dependent on STAT3. Because TNF-α signaling is important in promoting DC survival during differentiation and upon activation [43] and DC is a major producer of IL-12 in the spleen [44–46], reduced expression of TNF-α and IL-12 in KO mice prompted us to investigate the possibility of defective DC functionality in Stat3 KO mice during HSV-1 infection.
Stat3 KO mice have decreased CD8+ cDC frequency during HSV-1 infection
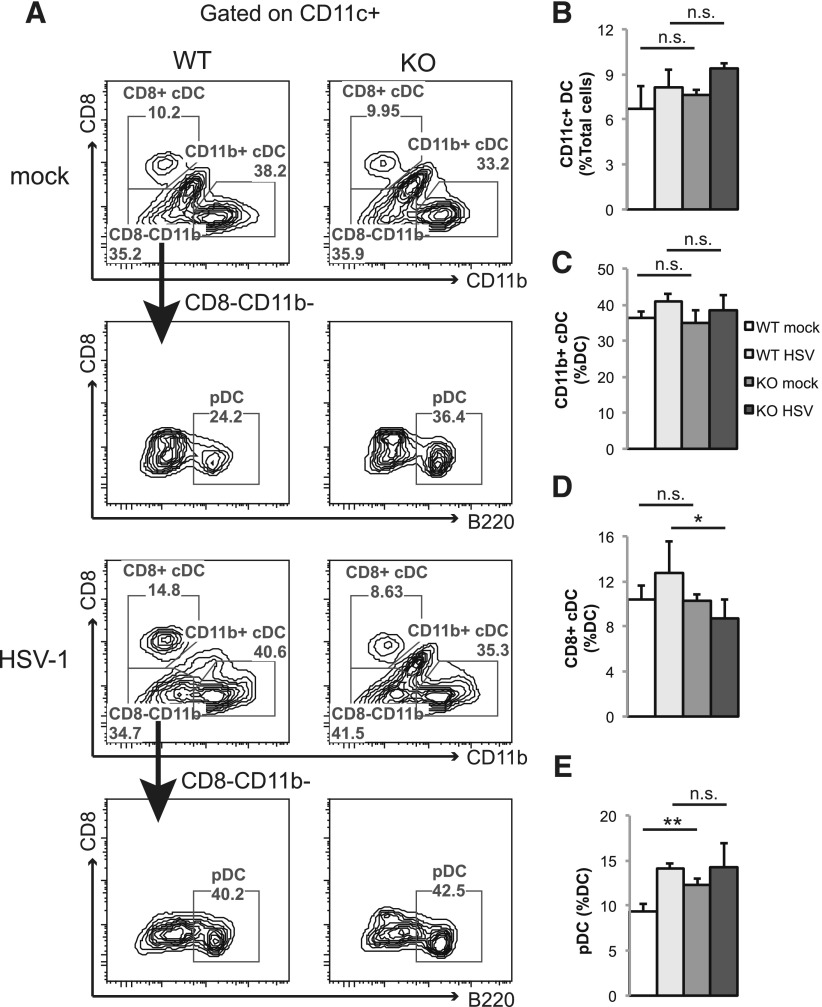
STAT3 is critical for DC differentiation from hematopoietic stem cells [47], and it has been reported that LysM-Cre can lead to partial deletion of target genes in a small fraction of splenic DCs [31]. Therefore, we asked whether LysM-Cre–mediated Stat3 deletion affected splenic DCs at steadystate and during HSV-1 infection. We found that myeloid Stat3 deletion did not affect the percentage of total splenic DCs or the CD11b+ cDCs subset at a steady state. Additionally, KO mice were able to expand the total splenic DC and CD11b+ cDC populations to a level comparable to that of WT mice in response to HSV-1 infection (Fig. 5A–C). WT and KO mice also had similar numbers of CD8+ cDCs at steadystate. However, a mild reduction in CD8+ cDC population was observed in the KO mice during HSV-1 infection (Fig. 5D). KO mice also had more steady-state pDCs, although similar levels of pDCs were observed in WT and KO mice during HSV-1 infection (Fig. 5E). These data suggest that STAT3 may have a minor role in pDC differentiation at steadystate. Importantly, the reduction of CD8+ cDC population in Stat3 KO mice during HSV-1 infection demonstrated that STAT3 contributes to CD8+ cDC differentiation, proliferation, or survival in response to HSV-1 infection.
Figure 5. Stat3 KO mice have decreased CD8+ cDC frequency during HSV-1 infection.
Sex- and age-matched WT or KO mice were infected with 1 × 107 PFU of HSV-1 by tail-vein injection. Mock-infected WT (n = 3) and KO mice (n = 3) and infected WT (n = 5) and KO mice (n = 4) were sacrificed 2 dpi, and splenic cells were collected for flow cytometry analysis. (A) Splenic cells were gated on CD11c+, followed by CD8, CD11b, and B220 analyses to determine the percentage of CD8+ cDC (CD11c+CD8+CD11b−), CD11b+ cDC (CD11c+CD8−CD11b+), and pDC (CD11c+CD8−CD11b−B220+) populations. Representative figures from 1 mouse of each group are shown. (B–E) Results summarize the percentages of total DCs, CD11b+ cDCs, CD8+ cDCs, and pDCs from flow analysis. Data shown are means ± sd. Statistical analysis was performed with a Student’s t test. n.s., not significant; *P < 0.05; **P < 0.01.
Stat3 KO mice have impaired NK and T cell activation during HSV-1 infection
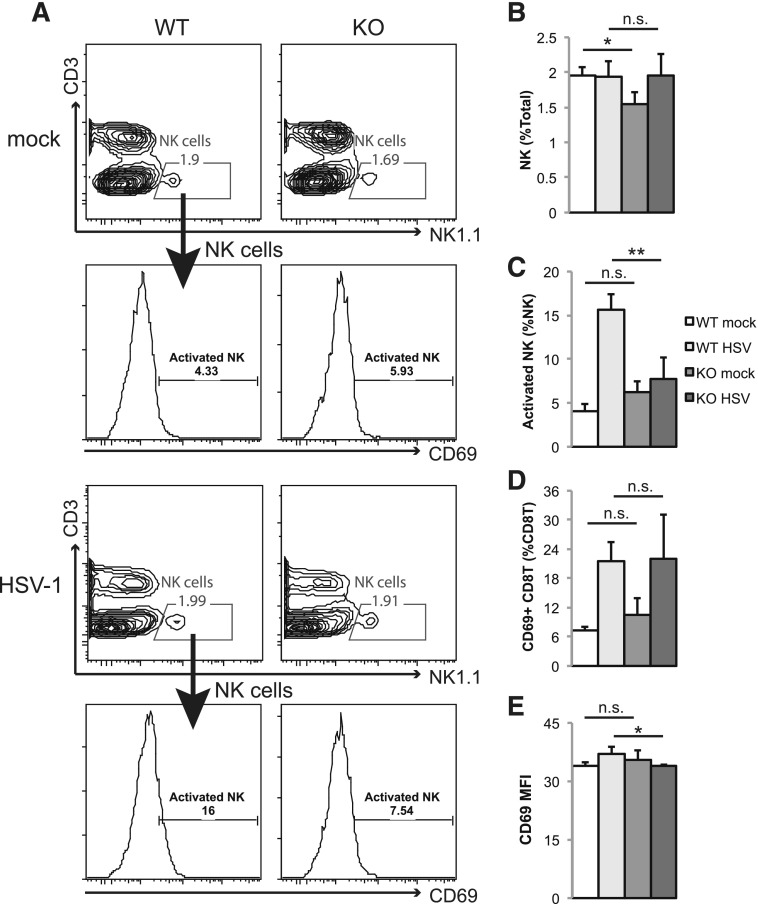
Splenic CD8+ cDCs are specialized to cross-present viral Ags to CD8 T cells to elicit virus-specific CTL immunity [20, 48]. During HSV-1 infection, DCs also have a critical role in promoting NK cell and CTL activation [15]. Given that Stat3 KO mice failed to expand CD8+ the cDC population during HSV-1 infection, we asked whether activation of NK and CD8+ T cells in Stat3 KO mice was affected. We found that mock-infected KO mice had less steady-state splenic NK cells (identified as CD3− NK1.1+), but HSV-1 infection increased the number of NK cells to a level comparable to that of WT mice (Fig. 6A and B). NK cells in WT mice up-regulated the activation marker CD69 at 2 dpi (Fig. 6C). However, NK cell activation was diminished in KO mice (Fig. 6C). Activation of CD8 T cells was mostly unaffected by Stat3 deletion (Fig. 6D), although the degree of activation was marginally lower in KO mice (Fig. 6E). These data demonstrate that myeloid Stat3 deletion leads to impaired NK cell activation and minor defects in CD8 T cell activation, and both may contribute to HSV-1 susceptibility of Stat3 KO mice.
Figure 6. Stat3 KO mice have impaired NK and T cell activation during HSV-1 infection.
WT and KO mice were infected and sacrificed for flow cytometry analysis as described in the caption to Fig. 5. (A) Representative figures of 1 mouse from each group are shown. NK cells in the spleen were defined with a CD3−NK1.1+ gate. Activated NK cells were further gated by CD69+ staining. (B and C) Results summarize the percentages of total NK and activated NK cells. (D) Results summarize the percentages of activated CD8 T cells (CD3+CD8+CD69+) in the spleen. (E) The mean fluorescence intensity (MFI) of CD69 was calculated from activated CD8 T cells defined in panel D. Data shown are means ± sd. Statistical analysis was performed with a Student’s t test. n.s., not significant; *P < 0.05; **P < 0.01.
DISCUSSION
In this study, we investigated the role of STAT3 in myeloid cells during HSV-1 infection. In vivo, Stat3 KO mice were more susceptible to HSV-1, as marked by shortened survival, higher viral loads in the brain and the spleen, and significant weight loss (Fig. 2). Myeloid Stat3 KO mice are predisposed to develop chronic colitis at later ages [26], and that may complicate our interpretation of the significant weight loss in Stat3 KO mice. In agreement with previous studies, we found that KO mice usually do not manifest symptoms of colitis until 16 wk old or older. Using healthy mice that were around 10 wk old allowed us to avoid mice with preexisting conditions, although we cannot rule out the possibility of HSV-1 infection triggering the onset of colitis and impeding the weight gain and recovery from acute HSV-1 infection. However, the fact that Stat3 KO mice also had significantly higher viral loads argues that the weight loss was associated with an aggravated disease resulting from an impaired immune system that failed to restrain the virus from replicating and spreading.
Viral infections induce production of type I IFNs, namely, IFN-α and IFN-β. Type I IFNs promote the expression of ISGs to establish an antiviral state by activating the transcription factors STAT1 and IFN-stimulated gene factor 3 (ISGF3) which comprises STAT1 STAT2 and IFN regulatory factor 9 (IRF9) [34]. STAT3 negatively regulates type I IFN-induced ISGs by inhibiting STAT1 [49, 50]. Surprisingly, our data showed that STAT3 is required for optimal IFN-α and ISG expression in the BMMs during HSV-1 infection (Fig. 1B, L, and M). Although it is possible that STAT3 directly regulates expression of ISGs positively during HSV-1 infection, it is more likely that reduced expression of ISGs is a result of diminished expression of IFN-α. Type I IFN production usually occurs in 2 phases. In the first phase, viral infection activates IRF3 to promote IFN-β expression. In the second phase, IFN-β induces expression of IRF7, which, in turn, promotes IFN-α expression and sustains IFN-β expression [34]. We speculate that STAT3 primarily affects the second phase of IFN production, such as the activation of IRF7 because the production of IFN-β was normal in Stat3 KO BMMs (Fig. 1K). Additionally, HSV-1–induced production of type I IFNs was mediated by several different cellular sensing mechanisms [45, 51–53]. It is also possible that those sensing mechanisms are impaired in the absence of STAT3 resulting in diminished IFN-α production. Apart from the impaired type I IFN response, Stat3 KO BMMs also highly expressed a subset of proinflammatory genes upon HSV-1 infection (Fig. 1D–F). Stat3 KO BMMs prolongs and heightens inflammatory responses because of defective IL-10 signaling [26]. Elevated expression of proinflammatory genes in KO BMMs is presumably driven by hyperactivation of NF-κB. It has been shown that STAT3 keeps the activity of NF-κB in check by inhibiting the expression of a positive regulator of NF-κB [54] and by promoting the expression of a microRNA that suppresses NF-κB [55]. Our data corroborate previous studies by showing that the phosphorylation of IκBα and the expression of the classic NF-κB target gene TNF-α are significantly increased in Stat3 KO BMMs (Fig. 1B and D), suggesting increased NF-κB activity.
During systemic infection of HSV-1, Stat3 KO mice expressed less IFN-α, IL-12, and TNF-α in the spleen (Fig. 4A, F, and G). The antiviral effects of IFN-α are well established [36]. TNF-α also inhibits HSV-1 replication in vitro [9]. In vivo, TNF-α protects mice from lethal herpes encephalitis [12], and IL-12 promotes the protective Th1 response against herpesvirus [11]. Therefore, dampened expression of IFN-α, IL-12, and TNF-α in the spleen may collectively contribute to the susceptibility of Stat3 KO mice. The source of these cytokines, however, remains to be determined. MMMΦ and MZMΦ are the major sources of IFN-α in the spleen during HSV-1 infection [14], but pDC, cDC, and other cell types are capable of producing ample amount of IFN-α when exposed to HSV-1 [45, 51]. Reduced expression of IL-12 in KO mice may be due to a reduced CD8+ cDC population during HSV-1 infection, because CD8+ cDCs are efficient IL-12 producers [44].
The role of neutrophils in viral infections ranges from protective to immunopathologic [56]. Early studies using Gr-1 Abs for neutrophil depletion suggested that infiltrating neutrophils suppressed HSV-1 replication in cornea [21]. However, Gr-1 is expressed by other groups of immune cells, and it was later revealed that nonneutrophil Gr-1+ cells were primarily responsible for HSV-1 inhibition [57]. Moreover, infiltrating neutrophils are a major mediator of HSV-1–induced immunopathology in the brain and the cornea [22, 23], and CXCL2 promotes neutrophil chemotaxis to the site of infection during HSV-1 infection [58]. Given that myeloid Stat3 KO mice have a significant greater neutrophil population, we originally speculated there was increased neutrophil infiltration in the brain, thereby leading to aggravated symptoms. Nonetheless, both WT and Stat3 KO mice were devoid of Ly6G+ neutrophil or CD45+ hematopoietic cell infiltrates in the brain (Fig. 3F), and CXCL2 expression level was indistinguishable between WT and KO mice (Fig. 3H), indicating that neutrophil-mediated immunopathology is not implicated in the susceptibility of Stat3 KO mice to HSV-1. The role of neutrophils in the spleen and the blood during systemic HSV-1 infection will require further investigation.
The effect of myeloid Stat3 KO on DC differentiation (Fig. 5) is minimal compared with hematopoietic Stat3 deletion, which results in a drastic reduction in the DC population [47]. Most splenic DCs are from CMPs [59, 60]. Given that LysM-Cre is only moderately expressed in CMPs [61], the minor effects of myeloid Stat3 deletion on DC differentiation can be explained by incomplete deletion of Stat3 in CMPs. The reason for the increased steady-state pDC population in our model is unclear. In fact, CD11c-Cre–mediated Stat3 KO causes a reduction in the pDC population [62]. The immediate DC progenitors express high levels of STAT3 but are CD11c−, and they up-regulate CD11c just before differentiating into distinct DC subsets [63]. Thus, CD11c-Cre–mediated Stat3 deletion would occur during the final stage of DC differentiation. To understand how STAT3 affects pDC differentiation, it will be of interest to compare how LysM-Cre and CD11c-Cre affect STAT3 expression in these immediate DC progenitors and the consequences of such perturbations. On the other hand, myeloid Stat3 KO mice have a reduction in their CD8+ cDC population during HSV-1 infection. It is possible that STAT3 facilitates the differentiation or proliferation of CD8+ cDC in response to acute HSV-1 infection. However, given that myeloid Stat3 KO does not affect the differentiation of steady-state CD8+ cDCs, we speculate that STAT3 regulates the survival of CD8+ cDCs during HSV-1 infection. Activated DCs are known to undergo apoptosis unless protected by TNF-α [43], and HSV-1 also induces apoptosis in infected DCs [64]. The reduction of splenic CD8+ cDCs in STAT3 KO mice during HSV-1 infection is likely a result of reduced splenic TNF-α and enhanced apoptosis.
DCs have a critical role in controlling HSV-1 pathogenesis by mediating NK and T cell activation, and mice with depleted DCs have higher mortality and higher viral loads in their nervous systems [15]. CD8+ cDCs are unique in their ability to cross-present viral Ags to CD8 T cells and to promote CTL response against HSV-1 infection [20, 48]. We found that activation of NK cells was dampened in Stat3 KO mice (Fig. 6C). Impaired activation of NK cells may be due to the combinatory effect of reduced CD8+ cDCs and diminished IL-12 and IFN-α expression because IL-12 secreted by CD8+ cDCs and IFN-α both mediate NK cell activation during viral infection [18, 65]. Because we did not observe a difference in NK cytotoxicity in vitro (data not shown), it will be interesting to determine whether NK cells from Stat3 KO mice are deficient in other aspects, such as cytokine production. We also noticed a minor decrease in CD8 T cell activation altitude at 2 dpi (Fig. 6E). Whether that observation translates to compromised adaptive immunity to HSV-1 at the later stage of infection awaits further investigation. Interestingly, Stat3 KO mice also showed moderate reductions in their steady-state NK cell population (Fig. 6B). It is unclear why myeloid Stat3 deletion leads to reduction in NK cells because NK cells are generally believed to arise from the lymphoid lineage [66]. However, myeloid precursors are capable of differentiating into NK cells [67, 68], which raises a possible role for myeloid STAT3 in NK cell development.
In conclusion, our data showed that myeloid-specific Stat3 deletion causes defects in multiple aspects of the innate immune system in response to HSV-1. Although individual defects may not change the outcome of infection, collectively, they result in higher susceptibility of Stat3 KO mice to HSV-1. Future studies will focus on the mechanisms of each defect, such as the mechanism by which STAT3 modulates IFN-α production. The impact of impaired innate immunity on adaptive immunity-mediated control of HSV-1 infection and reactivation is another interesting area for investigation.
AUTHORSHIP
H.-C.H., B.D., and A.S.B. conceived the study, designed the experiments, and wrote the paper. H.-C.H. performed most of the experiments and analyzed the data. C.M.S. and Z.Z. provided technical assistance, and C.M.S. performed HSV-1 infection mouse experiments. All authors have reviewed and approved the manuscript.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by U.S. National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID) Grant AI35098, awarded to A.S.B., and NIH NIAID Grants AI107810, AI109965, and NIH, National Institute of Dental and Craniofacial Research (NIDCR) DE018281, awarded to B.D. The UNC Flow Cytometry Core Facility was supported in part by P30 Grant CA016086. The UNC Translational Pathology Laboratory was supported in part by grants from the National Cancer Institute (Grant 2-P30-CA016086-40), the National Institute of Environmental Health Sciences (Grant 2-P30ES010126-15A1), the Upper Cervical Research Foundation, and the North Carolina Biotechnology Center (Grant 2015-IDG-1007). We thank members of the Baldwin Laboratory (UNC-Chapel Hill, USA) for constructive discussions and feedback on the manuscript. We are also grateful for the help of Dr. Stephanie Montgomery, the UNC Flow Cytometry Core Facility, and the UNC Translational Pathology Laboratory for expert technical assistance.
Glossary
- BMM
bone marrow–derived macrophage
- cDC
conventional dendritic cell
- CMP
common myeloid progenitor
- DC
dendritic cell
- ISG
interferon-stimulated genes
- dpi
days postinfection
- KO
knockout
- MMMΦ
marginal metallophilic macrophage
- MOI
multiplicity of infection
- MZMΦ
marginal zone macrophage
- pDC
plasmacytoid dendritic cell
- STAT3
signal transducer and activator of transcription 3
- UNC
University of North Carolina
- WT
wild type
Footnotes
The online version of this paper, found at www.jleukbio.org, includes supplemental information.
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- 1.Whitley R. J., Roizman B. (2001) Herpes simplex virus infections. Lancet 357, 1513–1518. [DOI] [PubMed] [Google Scholar]
- 2.Zuckerman R. A. (2009) The clinical spectrum of herpes simplex viremia. Clin. Infect. Dis. 49, 1302–1304. [DOI] [PubMed] [Google Scholar]
- 3.Youssef R., Shaker O., Sobeih S., Mashaly H., Mostafa W. Z. (2002) Detection of herpes simplex virus DNA in serum and oral secretions during acute recurrent herpes labialis. J. Dermatol. 29, 404–410. [DOI] [PubMed] [Google Scholar]
- 4.Howie S., Norval M., Maingay J., McBride W. H. (1986) Interactions between herpes simplex virus and murine bone marrow macrophages. Arch. Virol. 87, 229–239. [DOI] [PubMed] [Google Scholar]
- 5.Morahan P. S., Mama S., Anaraki F., Leary K. (1989) Molecular localization of abortive infection of resident peritoneal macrophages by herpes simplex virus type 1. J. Virol. 63, 2300–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zisman B., Hirsch M. S., Allison A. C. (1970) Selective effects of anti-macrophage serum, silica and anti-lymphocyte serum on pathogenesis of herpes virus infection of young adult mice. J. Immunol. 104, 1155–1159. [PubMed] [Google Scholar]
- 7.Mott K., Brick D. J., van Rooijen N., Ghiasi H. (2007) Macrophages are important determinants of acute ocular HSV-1 infection in immunized mice. Invest. Ophthalmol. Vis. Sci. 48, 5605–5615. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch M. S., Zisman B., Allison A. C. (1970) Macrophages and age-dependent resistance to herpes simplex virus in mice. J. Immunol. 104, 1160–1165. [PubMed] [Google Scholar]
- 9.Kodukula P., Liu T., Rooijen N. V., Jager M. J., Hendricks R. L. (1999) Macrophage control of herpes simplex virus type 1 replication in the peripheral nervous system. J. Immunol. 162, 2895–2905. [PubMed] [Google Scholar]
- 10.Croen K. D. (1993) Evidence for antiviral effect of nitric oxide. Inhibition of herpes simplex virus type 1 replication. J. Clin. Invest. 91, 2446–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sin J. I., Kim J. J., Arnold R. L., Shroff K. E., McCallus D., Pachuk C., McElhiney S. P., Wolf M. W., Pompa-de Bruin S. J., Higgins T. J., Ciccarelli R. B., Weiner D. B. (1999) IL-12 gene as a DNA vaccine adjuvant in a herpes mouse model: IL-12 enhances Th1-type CD4+ T cell-mediated protective immunity against herpes simplex virus-2 challenge. J. Immunol. 162, 2912–2921. [PubMed] [Google Scholar]
- 12.Lundberg P., Welander P. V., Edwards C. K. III, van Rooijen N., Cantin E. (2007) Tumor necrosis factor (TNF) protects resistant C57BL/6 mice against herpes simplex virus-induced encephalitis independently of signaling via TNF receptor 1 or 2. J. Virol. 81, 1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacLean A., Wei X. Q., Huang F. P., Al-Alem U. A., Chan W. L., Liew F. Y. (1998) Mice lacking inducible nitric-oxide synthase are more susceptible to herpes simplex virus infection despite enhanced Th1 cell responses. J. Gen. Virol. 79, 825–830. [DOI] [PubMed] [Google Scholar]
- 14.Eloranta M. L., Alm G. V. (1999) Splenic marginal metallophilic macrophages and marginal zone macrophages are the major interferon-α/β producers in mice upon intravenous challenge with herpes simplex virus. Scand. J. Immunol. 49, 391–394. [DOI] [PubMed] [Google Scholar]
- 15.Kassim S. H., Rajasagi N. K., Zhao X., Chervenak R., Jennings S. R. (2006) In vivo ablation of CD11c-positive dendritic cells increases susceptibility to herpes simplex virus type 1 infection and diminishes NK and T-cell responses. J. Virol. 80, 3985–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoneyama H., Matsuno K., Toda E., Nishiwaki T., Matsuo N., Nakano A., Narumi S., Lu B., Gerard C., Ishikawa S., Matsushima K. (2005) Plasmacytoid DCs help lymph node DCs to induce anti-HSV CTLs. J. Exp. Med. 202, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swiecki M., Wang Y., Gilfillan S., Colonna M. (2013) Plasmacytoid dendritic cells contribute to systemic but not local antiviral responses to HSV infections. PLoS Pathog. 9, e1003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews D. M., Scalzo A. A., Yokoyama W. M., Smyth M. J., Degli-Esposti M. A. (2003) Functional interactions between dendritic cells and NK cells during viral infection. Nat. Immunol. 4, 175–181. [DOI] [PubMed] [Google Scholar]
- 19.Hildner K., Edelson B. T., Purtha W. E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B. U., Unanue E. R., Diamond M. S., Schreiber R. D., Murphy T. L., Murphy K. M. (2008) Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belz G. T., Smith C. M., Eichner D., Shortman K., Karupiah G., Carbone F. R., Heath W. R. (2004) Cutting edge: conventional CD8 alpha+ dendritic cells are generally involved in priming CTL immunity to viruses. J. Immunol. 172, 1996–2000. [DOI] [PubMed] [Google Scholar]
- 21.Tumpey T. M., Chen S. H., Oakes J. E., Lausch R. N. (1996) Neutrophil-mediated suppression of virus replication after herpes simplex virus type 1 infection of the murine cornea. J. Virol. 70, 898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan X. T., Tumpey T. M., Kunkel S. L., Oakes J. E., Lausch R. N. (1998) Role of MIP-2 in neutrophil migration and tissue injury in the herpes simplex virus-1-infected cornea. Invest. Ophthalmol. Vis. Sci. 39, 1854–1862. [PubMed] [Google Scholar]
- 23.Marques C. P., Cheeran M. C.-J., Palmquist J. M., Hu S., Urban S. L., Lokensgard J. R. (2008) Prolonged microglial cell activation and lymphocyte infiltration following experimental herpes encephalitis. J. Immunol. 181, 6417–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H., Pardoll D., Jove R. (2009) STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9, 798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du T., Zhou G., Roizman B. (2013) Modulation of reactivation of latent herpes simplex virus 1 in ganglionic organ cultures by p300/CBP and STAT3. Proc. Natl. Acad. Sci. USA 110, E2621–E2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeda K., Clausen B. E., Kaisho T., Tsujimura T., Terada N., Förster I., Akira S. (1999) Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity 10, 39–49. [DOI] [PubMed] [Google Scholar]
- 27.Matsukawa A., Kudo S., Maeda T., Numata K., Watanabe H., Takeda K., Akira S., Ito T. (2005) Stat3 in resident macrophages as a repressor protein of inflammatory response. J. Immunol. 175, 3354–3359. [DOI] [PubMed] [Google Scholar]
- 28.Lee C.-K., Raz R., Gimeno R., Gertner R., Wistinghausen B., Takeshita K., DePinho R. A., Levy D. E. (2002) STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity 17, 63–72. [DOI] [PubMed] [Google Scholar]
- 29.Panopoulos A. D., Zhang L., Snow J. W., Jones D. M., Smith A. M., El Kasmi K. C., Liu F., Goldsmith M. A., Link D. C., Murray P. J., Watowich S. S. (2006) STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood 108, 3682–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsukawa A., Takeda K., Kudo S., Maeda T., Kagayama M., Akira S. (2003) Aberrant inflammation and lethality to septic peritonitis in mice lacking STAT3 in macrophages and neutrophils. J. Immunol. 171, 6198–6205. [DOI] [PubMed] [Google Scholar]
- 31.Clausen B. E., Burkhardt C., Reith W., Renkawitz R., Förster I. (1999) Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki A., Hanada T., Mitsuyama K., Yoshida T., Kamizono S., Hoshino T., Kubo M., Yamashita A., Okabe M., Takeda K., Akira S., Matsumoto S., Toyonaga A., Sata M., Yoshimura A. (2001) CIS3/SOCS3/SSI3 plays a negative regulatory role in STAT3 activation and intestinal inflammation. J. Exp. Med. 193, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen-Jackson H., Panopoulos A. D., Zhang H., Li H. S., Watowich S. S. (2010) STAT3 controls the neutrophil migratory response to CXCR2 ligands by direct activation of G-CSF-induced CXCR2 expression and via modulation of CXCR2 signal transduction. Blood 115, 3354–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivashkiv L. B., Donlin L. T. (2014) Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heise M. T., Virgin H. W. IV (1995) The T-cell-independent role of gamma interferon and tumor necrosis factor alpha in macrophage activation during murine cytomegalovirus and herpes simplex virus infections. J. Virol. 69, 904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sainz B. Jr., Halford W. P. (2002) Alpha/Beta interferon and gamma interferon synergize to inhibit the replication of herpes simplex virus type 1. J. Virol. 76, 11541–11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lokensgard J. R., Hu S., Sheng W., vanOijen M., Cox D., Cheeran M. C., Peterson P. K. (2001) Robust expression of TNF-α, IL-1β, RANTES, and IP-10 by human microglial cells during nonproductive infection with herpes simplex virus. J. Neurovirol. 7, 208–219. [DOI] [PubMed] [Google Scholar]
- 38.Aravalli R. N., Hu S., Rowen T. N., Palmquist J. M., Lokensgard J. R. (2005) Cutting edge: TLR2-mediated proinflammatory cytokine and chemokine production by microglial cells in response to herpes simplex virus. J. Immunol. 175, 4189–4193. [DOI] [PubMed] [Google Scholar]
- 39.Marques C. P., Hu S., Sheng W., Lokensgard J. R. (2006) Microglial cells initiate vigorous yet non-protective immune responses during HSV-1 brain infection. Virus Res. 121, 1–10. [DOI] [PubMed] [Google Scholar]
- 40.Cho I.-H., Hong J., Suh E. C., Kim J. H., Lee H., Lee J. E., Lee S., Kim C.-H., Kim D. W., Jo E.-K., Lee K.-E., Karin M., Lee S. J. (2008) Role of microglial IKKβ in kainic acid-induced hippocampal neuronal cell death. Brain 131, 3019–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lundberg P., Ramakrishna C., Brown J., Tyszka J. M., Hamamura M., Hinton D. R., Kovats S., Nalcioglu O., Weinberg K., Openshaw H., Cantin E. M. (2008) The immune response to herpes simplex virus type 1 infection in susceptible mice is a major cause of central nervous system pathology resulting in fatal encephalitis. J. Virol. 82, 7078–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurt-Jones E. A., Chan M., Zhou S., Wang J., Reed G., Bronson R., Arnold M. M., Knipe D. M., Finberg R. W. (2004) Herpes simplex virus 1 interaction with Toll-like receptor 2 contributes to lethal encephalitis. Proc. Natl. Acad. Sci. USA 101, 1315–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehner M., Kellert B., Proff J., Schmid M. A., Diessenbacher P., Ensser A., Dörrie J., Schaft N., Leverkus M., Kämpgen E., Holter W. (2012) Autocrine TNF is critical for the survival of human dendritic cells by regulating BAK, BCL-2, and FLIPL. J. Immunol. 188, 4810–4818. [DOI] [PubMed] [Google Scholar]
- 44.Hochrein H., Shortman K., Vremec D., Scott B., Hertzog P., O’Keeffe M. (2001) Differential production of IL-12, IFN-alpha, and IFN-γ by mouse dendritic cell subsets. J. Immunol. 166, 5448–5455. [DOI] [PubMed] [Google Scholar]
- 45.Krug A., Luker G. D., Barchet W., Leib D. A., Akira S., Colonna M. (2004) Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor 9. Blood 103, 1433–1437. [DOI] [PubMed] [Google Scholar]
- 46.Farrand K. J., Dickgreber N., Stoitzner P., Ronchese F., Petersen T. R., Hermans I. F. (2009) Langerin+ CD8α+ dendritic cells are critical for cross-priming and IL-12 production in response to systemic antigens. J. Immunol. 183, 7732–7742. [DOI] [PubMed] [Google Scholar]
- 47.Laouar Y., Welte T., Fu X.-Y., Flavell R. A. (2003) STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity 19, 903–912. [DOI] [PubMed] [Google Scholar]
- 48.Smith C. M., Belz G. T., Wilson N. S., Villadangos J. A., Shortman K., Carbone F. R., Heath W. R. (2003) Cutting edge: conventional CD8α+ dendritic cells are preferentially involved in CTL priming after footpad infection with herpes simplex virus-1. J. Immunol. 170, 4437–4440. [DOI] [PubMed] [Google Scholar]
- 49.Ho H. H., Ivashkiv L. B. (2006) Role of STAT3 in type I interferon responses: negative regulation of STAT1-dependent inflammatory gene activation. J. Biol. Chem. 281, 14111–14118. [DOI] [PubMed] [Google Scholar]
- 50.Wang W. B., Levy D. E., Lee C. K. (2011) STAT3 negatively regulates type I IFN-mediated antiviral response. J. Immunol. 187, 2578–2585. [DOI] [PubMed] [Google Scholar]
- 51.Hochrein H., Schlatter B., O’Keeffe M., Wagner C., Schmitz F., Schiemann M., Bauer S., Suter M., Wagner H. (2004) Herpes simplex virus type-1 induces IFN-α production via Toll-like receptor 9-dependent and -independent pathways. Proc. Natl. Acad. Sci. USA 101, 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson M. R., Sharma S., Atianand M., Jensen S. B., Carpenter S., Knipe D. M., Fitzgerald K. A., Kurt-Jones E. A. (2014) Interferon γ-inducible protein (IFI) 16 transcriptionally regulates type i interferons and other interferon-stimulated genes and controls the interferon response to both DNA and RNA viruses. J. Biol. Chem. 289, 23568–23581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horan K. A., Hansen K., Jakobsen M. R., Holm C. K., Søby S., Unterholzner L., Thompson M., West J. A., Iversen M. B., Rasmussen S. B., Ellermann-Eriksen S., Kurt-Jones E., Landolfo S., Damania B., Melchjorsen J., Bowie A. G., Fitzgerald K. A., Paludan S. R. (2013) Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J. Immunol. 190, 2311–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H., Hu H., Greeley N., Jin J., Matthews A. J., Ohashi E., Caetano M. S., Li H. S., Wu X., Mandal P. K., McMurray J. S., Moghaddam S. J., Sun S.-C., Watowich S. S. (2014) STAT3 restrains RANK- and TLR4-mediated signalling by suppressing expression of the E2 ubiquitin-conjugating enzyme Ubc13. Nat. Commun. 5, 5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiang M., Birkbak N. J., Vafaizadeh V., Walker S. R., Yeh J. E., Liu S., Kroll Y., Boldin M., Taganov K., Groner B., Richardson A. L., Frank D. A. (2014) STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-κB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Sci. Signal. 7, ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drescher B., Bai F. (2013) Neutrophil in viral infections, friend or foe? Virus Res. 171, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wojtasiak M., Pickett D. L., Tate M. D., Bedoui S., Job E. R., Whitney P. G., Brooks A. G., Reading P. C. (2010) Gr-1+ cells, but not neutrophils, limit virus replication and lesion development following flank infection of mice with herpes simplex virus type-1. Virology 407, 143–151. [DOI] [PubMed] [Google Scholar]
- 58.Tumpey T. M., Fenton R., Molesworth-Kenyon S., Oakes J. E., Lausch R. N. (2002) Role for macrophage inflammatory protein 2 (MIP-2), MIP-1α, and interleukin-1α in the delayed-type hypersensitivity response to viral antigen. J. Virol. 76, 8050–8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manz M. G., Traver D., Miyamoto T., Weissman I. L., Akashi K. (2001) Dendritic cell potentials of early lymphoid and myeloid progenitors. Blood 97, 3333–3341. [DOI] [PubMed] [Google Scholar]
- 60.Traver D., Akashi K., Manz M., Merad M., Miyamoto T., Engleman E. G., Weissman I. L. (2000) Development of CD8α-positive dendritic cells from a common myeloid progenitor. Science 290, 2152–2154. [DOI] [PubMed] [Google Scholar]
- 61.Ye M., Iwasaki H., Laiosa C. V., Stadtfeld M., Xie H., Heck S., Clausen B., Akashi K., Graf T. (2003) Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity 19, 689–699. [DOI] [PubMed] [Google Scholar]
- 62.Melillo J. A., Song L., Bhagat G., Blazquez A. B., Plumlee C. R., Lee C., Berin C., Reizis B., Schindler C. (2010) Dendritic cell (DC)-specific targeting reveals Stat3 as a negative regulator of DC function. J. Immunol. 184, 2638–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Onai N., Obata-Onai A., Schmid M. A., Ohteki T., Jarrossay D., Manz M. G. (2007) Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8, 1207–1216. [DOI] [PubMed] [Google Scholar]
- 64.Bosnjak L., Miranda-Saksena M., Koelle D. M., Boadle R. A., Jones C. A., Cunningham A. L. (2005) Herpes simplex virus infection of human dendritic cells induces apoptosis and allows cross-presentation via uninfected dendritic cells. J. Immunol. 174, 2220–2227. [DOI] [PubMed] [Google Scholar]
- 65.Madera S., Rapp M., Firth M. A., Beilke J. N., Lanier L. L., Sun J. C. (2016) Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J. Exp. Med. 213, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kondo M., Weissman I. L., Akashi K. (1997) Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91, 661–672. [DOI] [PubMed] [Google Scholar]
- 67.Grzywacz B., Kataria N., Kataria N., Blazar B. R., Miller J. S., Verneris M. R. (2011) Natural killer-cell differentiation by myeloid progenitors. Blood 117, 3548–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Q., Ye W., Jian Tan W., Mei Yong K. S., Liu M., Qi Tan S., Loh E., Te Chang K., Chye Tan T., Preiser P. R., Chen J. (2015) Delineation of natural killer cell differentiation from myeloid progenitors in human. Sci. Rep. 5, 15118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.