Systemic inflammation induces innate activation of effector-like CD8+ T cells to infiltrate the liver and produce IL-10.
Keywords: cytokine, hemophagocytic syndrome, hepatitis
Abstract
Immune-mediated liver injury is a central feature of hyperinflammatory diseases, such as hemophagocytic syndromes, yet the immunologic mechanisms underlying those processes are incompletely understood. In this study, we used the toll-like receptor 9 (TLR9)–mediated model of a hemophagocytic syndrome known as macrophage activation syndrome (MAS) to dissect the predominant immune cell populations infiltrating the liver during inflammation. We identified CD8+ T cells that unexpectedly produce interleukin-10 (IL-10) in addition to interferon-γ (IFN-γ) as a major hepatic population induced by TLR9 stimulation. Despite their ability to produce this anti-inflammatory cytokine, IL-10+ hepatic CD8+ T cells in TLR9–MAS mice did not resemble CD8+ T suppressor cells. Instead, the induction of these cells occurred independently of antigen stimulation and was partially dependent on IFN-γ. IL-10+ hepatic CD8+ T cells demonstrated an activated phenotype and high turnover rate, consistent with an effector-like identity. Transcriptional analysis of this population confirmed a gene signature of effector CD8+ T cells yet suggested responsiveness to liver injury–associated growth factors. Together, these findings suggest that IL-10+ CD8+ T cells induced by systemic inflammation to infiltrate the liver have initiated an inflammatory, rather than regulatory, program and may thus have a pathogenic role in severe, acute hepatitis.
Introduction
PALF is a serious, rapidly progressive syndrome, frequently requiring liver transplantation and often resulting in patient death [1, 2]. Many PALF cases result from known causes, such as acetaminophen toxicity, inherited metabolic disease, autoimmune hepatitis, or acute viral hepatitis; however, the etiology eludes specific diagnosis in nearly one-half of cases (designated iPALF) [3]. Mounting evidence suggests that dysregulated, inflammatory processes may underlie a large portion of iPALF cases. Elevated serum levels of sIL-2Rα, a marker of T cell activation, are found in ∼one-half of patients with iPALF [4]. There are also striking similarities in clinical presentation between iPALF and hemophagocytic syndromes, both of which can include fever, hepatomegaly, abnormal liver function tests, BM suppression, and elevated sIL-2Rα [5]. Liver failure in these “cytokine storm” syndromes, which include FHL and MAS, is a well-recognized complication of the disease and can sometimes dominate the clinical presentation. These hemophagocytic syndromes are caused by a failure of healthy immune regulation during a reactionary inflammatory process, resulting in hypercytokinemia, hepatosplenomegaly, cytopenias, inflammatory cell infiltration in multiple organs (particularly spleen and liver), and can rapidly lead to multiorgan failure and death [6, 7]. Murine models of numerous forms of hemophagocytic syndrome, particularly variants of FHL, have implicated activated CD8+ T cells that overproduce cytokines as having a major pathogenic role [8, 9]. Interestingly, recent histologic studies of FHL and iPALF identified a predominant hepatic CD8+ T cell infiltrate in most cases [10]. Together, these findings suggest the possibility of a common immunologic mechanism responsible for liver injury in both hemophagocytic syndrome and iPALF. Inflammatory liver injury is, therefore, likely important in many scenarios.
To better understand the nature of hepatic inflammation in hemophagocytic syndrome and other immune-mediated liver diseases, we used the murine model of MAS induced by TLR9 stimulation (TLR9-MAS) to investigate liver-infiltrating immune cells [11]. We identified a large population of hepatic CD8+ T cells that surprisingly produces both IL-10 and IFN-γ. This subset bears little resemblance to previously described IL-10+ CD8+ suppressor T cells. Instead, these cells arise independent of Ag stimulation and rely indirectly on IFN-γ for their induction. Using extensive phenotypic and transcriptional analysis, we determined that these IL-10+ hepatic CD8+ T cells resemble activated, early effector CD8+ T cells, similar to those elicited by viral or intracellular bacterial pathogens. However, they also exhibit a particular signature of responsiveness to growth factors induced by hepatic damage. Thus, our study provides evidence that IL-10+CD8+ T cells are a prominent immune cell subset at the site of liver injury in the context of systemic inflammatory disease. Future studies investigating inflammatory infiltrates in iPALF and other systemic inflammatory illnesses with liver dysfunction should include analysis of IL-10+ hepatic CD8+ T cells.
MATERIALS AND METHODS
Mice
Male and female C57BL/6, B6.SJL (B6.SJL-PtprcaPepcb/BoyJ), tiger (B6.129S6-Il10tm1Flv/J, referred to as IL-10 reporter) [12], OT-I (C57BL/6-Tg[TcraTcrb]1100Mjb/J), perforin-deficient (C57BL/6-Prf1tm1Sdz/J, referred to as Prf1−/−), and IFNGR-deficient (B6.129S7-Ifngr1tm1Agt/J, referred to as Ifngr−/−) mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA). Unless otherwise stated, WT refers to IL-10 reporter mice. All experiments were performed with institutional animal care and use committee approval of the University of Pennsylvania (Philadelphia, PA, USA) and Children’s Hospital of Philadelphia (Philadelphia, PA, USA).
In vivo treatment
To induce TLR9–MAS, mice were injected i.p. with 50 μg CpG oligodeoxynucleotides 1826 (Integrated DNA Technologies, Coralville, IA, USA) on d 0, 2, 4, 7, and 9; control mice were injected i.p. with PBS on the same dosing schedule, and all mice were analyzed on d 10. To induce FHL, Prf1−/− IL-10 reporter mice were infected with 1 × 105 PFU LCMV-Armstrong i.p.
Flow cytometry
Hepatic leukocytes were isolated by Percoll density gradient centrifugation (GE Healthcare Life Sciences, Little Chalfont, United Kingdom). Cells were stained with LIVE/DEAD fixable viability dye (Thermo Fisher Scientific, Waltham, MA, USA) and Fc blocked (clone 2.4G2) before surface Ab staining. Cells were fixed and permeabilized using the Foxp3/Transcription Factor Fixation/Permeabilization kit (eBioscience, San Diego, CA, USA) or Cytofix/Cytoperm (BD Bioscience, San Jose, CA, USA) before staining for intracellular markers or cytokines, respectively.
BM chimeras
Irradiated B6.SJL (CD45.1+) hosts received BM from CD45.1/2+ IL-10 reporter donors (WT), CD45.2+ Ifngr−/− IL-10 reporter donors (Ifngr−/−), or both. Mixed BM chimeras received a 9:1 mix of WT and Ifngr−/− cells to ensure an IFN-γ response sufficient to induce TLR9–MAS.
Viability assays
Live (LIVE/DEAD−) cells were FACS purified and cultured with BMDCs, 10 μg/ml CpG, plate-bound α-CD3/α-CD28, 50 U/ml IL-2, 0.5 ng/ml IL-12, or 10-fold, diluted TLR9–MAS serum before restaining with LIVE/DEAD and viability analysis.
Microarrays and transcriptional analysis
Paired IL-10− and IL-10+ populations were sorted from among the live CD44+ hepatic CD8+ T cell pools from 2 male and 2 female TLR9–MAS IL-10 reporter mice into Buffer RLT (Qiagen, Hilden, Germany), and RNA was isolated using the RNeasy Micro Kit (Qiagen). Amplification, hybridization to GeneChip Mouse Gene 1.0 ST arrays (Affymetrix, Santa Clara, CA, USA), and data collection was performed by the Children’s Hospital of Philadelphia Nucleic Acid/Protein Research Core. Probe sets lacking gene symbol annotation or with log2 intensity means of <5 among the IL-10+ samples were filtered out. Hierarchical clustering of IL-10− and IL-10+ samples was performed with GenePattern software (Broad Institute, Cambridge, MA, USA). Differentially expressed genes were defined as those with at least 1.5-fold difference in expression of IL-10+ as compared with IL-10− cells and statistical significance using paired Student’s t test, with a false-discovery rate <0.2. Pathway analysis was performed using IPA (Qiagen). Putative upstream regulators with an activation z-score of >2 or < −2 and a P value of <5 × 10−5 were considered significant. Comparison to ImmGen data sets ([13] and Gene Expression Omnibus Accession GSE15907 [National Center for Biotechnology Information, Bethesda, MD, USA]) was performed on RMA normalized raw data, and corrected for batch effect using ComBat software [14].
Statistics
Data were analyzed in Prism 5 software (GraphPad, La Jolla, CA, USA) using statistical tests indicated in the Figure legends. Unless otherwise specified, P values are represented in Figures by the number of asterisks (e.g., *P < 0.05, **P < 0.01, ***P < 0.001).
RESULTS AND DISCUSSION
IL-10–producing CD8+ T cells are prominent among hepatic inflammatory infiltrates in murine hemophagocytic syndrome
The TLR9–MAS murine model of hemophagocytic syndrome results in severe liver damage, as evidenced by hepatomegaly, marked lymphohistiocytic inflammatory infiltration, and lobular necrosis [11]. To investigate this hepatotoxic effect of systemic inflammation, we first surveyed the principal immune cell populations induced by inflammation in the liver. CpG-treated TLR9–MAS mice demonstrated a mixed hepatic infiltrate with CD8+ T cell predominance (Fig. 1A). Given the known pathogenic role of CD8+ T cells in the perforin-deficient (Prf1−/−)/LCMV model of FHL [8, 15] and their conspicuous presence in the livers of patients with iPALF [10], we proceeded to characterize this intrahepatic CD8+ T cell pool.
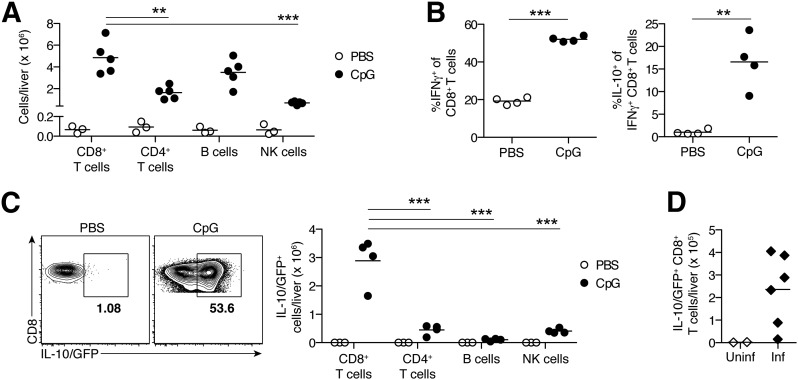
Figure 1. IL-10–producing CD8+ T cells are prominent among hepatic inflammatory infiltrates in murine hemophagocytic syndrome.
(A) Total liver lymphocytes in control (PBS-treated) or TLR9–MAS (CpG-treated) WT mice. n = 3–5 mice/treatment group; data were pooled from 2 independent experiments. (B) Cytokine production capacity of hepatic CD8+ T cells from PBS- or CpG-injected WT mice. n = 4 biologic replicates/group; each obtained from individual TLR9–MAS mice or by pooling cells from 4 PBS-injected mice. (C) Representative flow plots of liver leukocytes isolated from PBS- or CpG-injected IL-10 reporter mice, gated on TCRβ+CD8+CD4− cells. Numbers indicate frequency of IL-10/GFP+ cells among CD8+ T cells. Summary data for total numbers of IL-10/GFP+ cells in livers of PBS- and CpG-injected mice are shown. n = 3–4 mice/treatment group; data were pooled from 2 independent experiments. (D) Numbers of IL-10/GFP+ hepatic CD8+ T cells in uninfected (Uninf) or LCMV-infected (Inf) Prf1−/− IL-10 reporter mice. n = 2–6 mice/group. (A and C) Cell populations within TLR9–MAS mice were analyzed by 1-way ANOVA; significance of Dunnett’s posttests comparing CD8+ T cells to all other groups are indicated. **P < 0.01, ***P < 0.001.
Most hepatic CD8+ T cells in these mice were capable of producing IFN-γ, the primary cytokine driving inflammation in TLR9–MAS [11] (Fig. 1B). Surprisingly, many of these IFN-γ+ hepatic CD8+ T cells also produced IL-10 (Fig. 1B), the main negative regulator of inflammation in TLR9–MAS mice [11]. This IFN-γ+IL-10+ functional profile was not observed in the absence of inflammation because hepatic CD8+ T cells obtained from PBS-treated mice showed negligible IL-10 production capacity among IFN-γ+CD8+ T cells (Fig. 1B). Having confirmed that IL-10 was produced at the protein level, we then made use of IL-10 reporter mice, which report IL-10 transcription through the production of GFP and show excellent correlation between IL-10 protein and GFP expression (Supplemental Fig. 1) [12]. CpG induced substantial IL-10 production by CD8+ T cells, making this subset the largest population of IL-10 producers in the livers of TLR9–MAS mice (Fig. 1C). Numerous IL-10+CD8+ T cells were also present in the livers of LCMV-infected Prf1−/− mice, representing the murine model of FHL (Fig. 1D). These data suggest that IL-10 production marks an atypical population of hepatic CD8+ T cells, and their localization to the liver in multiple models of hemophagocytic syndrome suggests a connection between IL-10+CD8+ T cells and severe hepatic inflammation. Thus, we decided to investigate the IL-10+ hepatic CD8+ T cells further.
IL-10+ hepatic CD8+ T cells in TLR9–MAS mice are a unique population induced in an IFN-γ–dependent, Ag-independent manner
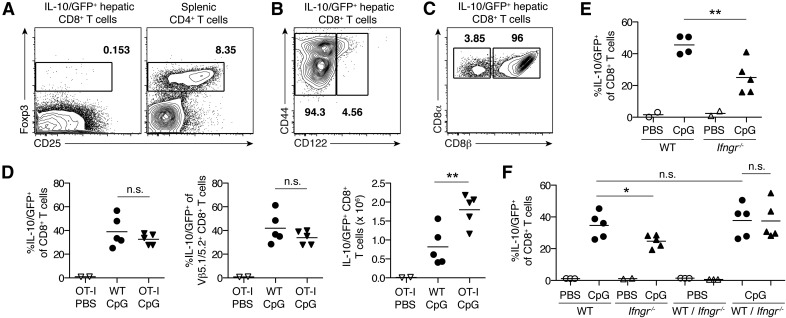
Several populations of IL-10+CD8+ T cells with suppressive function have been described, including natural CD8+ Treg cells (expressing Foxp3, CD122, or CD8αα) and induced, Ag-dependent CD8+ Treg cells [16–22]. However, IL-10/GFP+CD8+ T cells in TLR9–MAS mice lacked expression of Foxp3 and were nearly all CD122− conventional CD8αβ+ T cells (Fig. 2A–C). To determine whether IL-10+ hepatic CD8+ T cells rely on Ag in a manner similar to inducible CD8+ Treg cells, we induced TLR9–MAS in OT-I IL-10 reporter mice, whose T cells express a transgenic Vβ5.1/5.2 TCR specific for the MHC class I–restricted SIINFEKL peptide of ovalbumin. Notably, CpG induced similar frequencies of both total and Vβ5.1/5.2+ hepatic CD8+ T cells expressing IL-10/GFP in OT-I IL-10 reporter (OT-I) mice as in IL-10 reporter (WT) mice, despite the absence of ovalbumin (Fig. 2D). The total number of IL-10/GFP+ CD8+ T cells in the livers of OT-I mice was at least as high as it was in WT mice (Fig. 2D), demonstrating that substantial numbers of IL-10+ hepatic CD8+ T cells were induced independent of Ag. Thus, IL-10+ hepatic CD8+ T cells in TLR9–MAS mice are distinct from other IL-10–expressing CD8+ Treg populations.
Figure 2. IL-10+ hepatic CD8+ T cells in TLR9–MAS mice are a unique population induced in an IFN-γ–dependent, Ag-independent manner.
(A–C) Representative flow plots of live hepatic CD8+ T cells from TLR9–MAS IL-10 reporter mice. Numbers indicate frequency of cells within the gate. (A) FACS-purified IL-10/GFP+ cells stained for surface CD25 and intracellular Foxp3 expression. Splenic CD4+ T cells from an untreated C57BL/6 mouse are shown for comparison. n = 2 mice/group; data are representative of 2 independent experiments. (B) Flow plot gated on IL-10/GFP+ hepatic CD8+ T cells, showing surface staining of CD122. Representative of n = 4 mice. (C) Flow plot gated on IL-10/GFP+ hepatic CD8+ T cells, showing surface staining of CD8α and CD8β. Representative of n = 2 mice. (D) IL-10 reporter mice with OT-I transgenic TCR (OT-I) or without (WT) received either PBS or CpG to induce TLR9–MAS, without exposure to ovalbumin. Frequency and number of total and Vβ5.1/5.2+ IL-10/GFP+ hepatic CD8+ T cells are shown. n = 2–5 mice/group; data are representative of 2 independent experiments. Analyzed by unpaired Student’s t test. (E) Frequency of IL-10/GFP-expressing hepatic CD8+ T cells in IL-10 reporter mice either sufficient for IFNGR (WT) or deficient (Ifngr−/−). n = 2–5 mice/group; data were pooled from 2 independent experiments. Analyzed by unpaired Student’s t test. (F) BM chimeras reconstituted with cells from IL-10 reporter donors (WT), Ifngr−/− IL-10 reporter donors (Ifngr−/−), or a mixture of both (WT/Ifngr−/−) received either PBS or CpG to induce TLR9-MAS. Frequencies of IL-10/GFP+ hepatic CD8+ T cells among WT cells (circles) and Ifngr−/− cells (triangles) are shown. N = 2-5 mice/group, data are representative of 4 independent experiments. Analyzed by unpaired Student’s t test (WT CpG vs. Ifngr−/− CpG; WT CpG vs. mixWT CpG) or paired Student’s t test (mixWT CpG vs. mixIfngr−/− CpG). n.s., not significant. *P < 0.05, **P < 0.01.
We hypothesized that IL-10+ hepatic CD8+ T cells may instead rely on cytokine-mediated activation, similar to populations of effector and memory CD8+ T cells that have been described [23–26]. Because IFN-γ has a central role in the pathogenesis of TLR9–MAS [11, 27], we examined the livers of CpG-injected mice deficient in IFNGR. Ifngr−/− IL-10 reporter mice demonstrated a lower frequency of IL-10–producing cells among hepatic CD8+ T cells than did IL-10 reporter mice (mean 25.0% vs. 45.6%, respectively), resulting in a decrement of 50% in total frequency (Fig. 2E). To determine whether that partial dependence on IFN-γ was CD8+ T cell intrinsic or extrinsic, we generated BM chimeras using donor cells from IL-10 reporter (WT) mice, Ifngr−/− IL-10 reporter (Ifngr−/−) mice, or both. Consistent with data in the global IFNGR knockout (Fig. 2E), Ifngr−/− BM recipients showed a lower frequency of IL-10/GFP+ hepatic CD8+ T cells induced by CpG than did WT BM recipients (Fig. 2F). However, equivalent frequencies of WT and Ifngr−/− hepatic CD8+ T cells produced IL-10 in response to CpG stimulation in mixed BM chimeras (Fig. 2F), demonstrating that the partial dependence of IL-10+ hepatic CD8+ T cells on IFN-γ is CD8+ T cell-extrinsic. Thus, IL-10+ CD8+ T cells in the livers of TLR9–MAS mice rely largely on indirect effects of IFN-γ signaling, rather than on Ags, for their induction.
Together, these data suggest that TLR9-induced IL-10+ hepatic CD8+ T cells do not resemble previously described populations of IL-10–producing CD8+ T cells and may, therefore, represent a unique subset of CD8+ T cells. This study provides, to our knowledge, the first description of the induction of IL-10 production by CD8+ T cells in an Ag-independent manner. These results, therefore, suggest that Ag-independent CD8+ T cell activation, leading to the production of both pro- and anti-inflammatory cytokines, is an underappreciated part of the innate immune response and may have important physiologic implications for bystander cells in the vicinity.
IL-10+ hepatic CD8+ T cells have the activated phenotype and a high turnover rate of terminally differentiated effectors
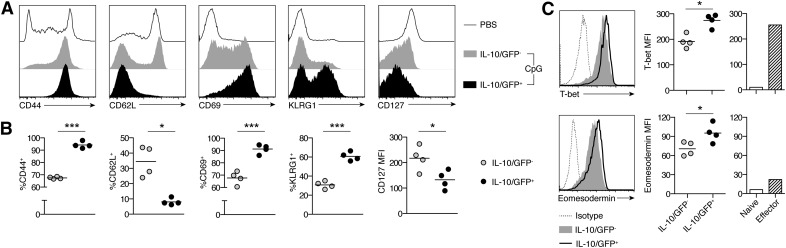
To further characterize this unique population, we sorted IL-10/GFP+ and IL-10/GFP − CD8+ T cells from the livers of TLR9–MAS IL-10 reporter mice and examined their surface phenotype. IL-10/GFP+ hepatic CD8+ T cells were uniformly CD44+CD62L−CD69+, suggesting they were recently activated effector cells (Fig. 3A and B). Furthermore, these cells had low expression of CD127 and a high proportion of them expressed KLRG1, consistent with terminally differentiated effectors (Fig. 3A and B). In contrast, IL-10/GFP−CD8+ T cells were a more heterogeneous population, composed of CD44− naïve cells, CD44+CD62L+ memory cells, CD44+CD62L−CD127+KLRG1− memory precursors, and a few terminally differentiated effectors (Fig. 3A and B). Among Ag-experienced CD44+ hepatic CD8+ T cells, those producing IL-10 had greater expression of T bet, a key transcription factor driving terminal effector differentiation, compared with their IL-10− counterparts (Fig. 3C). IL-10/GFP+CD8+ T cells also demonstrated relative up-regulation of eomesodermin, which is induced shortly after T cell activation (Fig. 3C) [28]. Interestingly, IL-10/GFP+ hepatic CD8+ T cells tended to have greater expression of both T bet and eomesodermin than control bona fide effector CD8+ T cells from LCMV-infected mice (Fig. 3C). This difference may be a reflection of the different organs and inflammatory inducers used in these 2 systems but may also identify a characteristic distinguishing IL-10+ hepatic CD8+ T cells from conventional effector CD8+ T cell populations.
Figure 3. IL-10+ hepatic CD8+ T cells have an activated phenotype consistent with terminally differentiated effectors.
Phenotype of hepatic CD8+ T cells from TLR9–MAS IL-10 reporter mice. (A) Representative histograms showing expression of surface markers differentiating naïve, effector, and memory populations. Plots are gated on CD8+ T cells, either in total (for PBS conditions) or IL-10/GFP− and IL-10/GFP+ subsets (for CpG conditions). (B) Summary of data as shown in (A). n = 4 mice/group, either pooled from 2 independent experiments (CD44, CD62L) or representative of 2 independent experiments (CD69, KLRG1, CD127). (C) Representative histograms and summary data for T bet and Eomesodermin expression. Plots are gated on CD44+CD8+ T cells. Naïve and effector splenic CD8+ T cells are included for comparison. N = 4 paired samples per group. Data are representative of 2 independent experiments. Analyzed by paired Student’s t test. MFI, mean fluorescent intensity. *P < 0.05, *** P < 0.001.
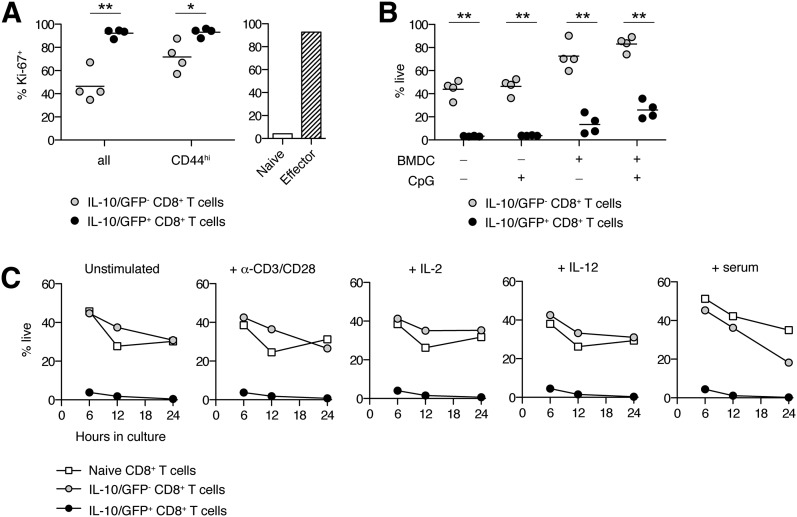
Consistent with their effector phenotype, >90% of CD44+ IL-10/GFP+ hepatic CD8+ T cells were Ki-67+, suggesting a high degree of proliferation, similar to control effector CD8+ T cells (Fig. 4A). Most IL-10/GFP− hepatic CD8+ T cells were Ki-67−, and even among those expressing CD44, the frequency of proliferating cells was persistently less than among their IL-10+ counterparts (Fig. 4A). Similar to highly proliferative, terminally differentiated effectors, IL-10/GFP+ hepatic CD8+ T cells also displayed markedly enhanced susceptibility to cell death. Regardless of culture conditions, the in vitro viability of IL-10/GFP+ hepatic CD8+ T cells was 40.9–59.1% lower than that of their IL-10/GFP− counterparts (Fig. 4B), suggesting intrinsically differential survival. In contrast to naïve splenic CD8+ T cells and IL-10/GFP− hepatic CD8+ T cells, IL-10/GFP+ hepatic CD8+ T cells showed reduced viability after as little as 6 h in culture (Fig. 4C). The addition of BMDCs activated with CpG, TCR stimulation, exogenous cytokines, or serum from a TLR9–MAS mouse failed to rescue the viability of IL-10/GFP+CD8+ T cells (Fig. 4B and C). It is possible that the viability of IL-10+ hepatic CD8+ T cells may be better preserved in situ, where supportive anatomic compartments within the liver, such as those formed by intrahepatic myeloid-cell aggregates for T cell population expansion [29], are induced by administration of CpG or other inflammatory stimuli. However, our findings are also consistent with work by the Crispe laboratory (University of Washington, Seattle, WA, USA) demonstrating selective retention of activated, apoptotic CD8+ T cells in the liver [30]. Together, these findings show that IL-10+ hepatic CD8+ T cells in TLR9–MAS mice display a phenotype and high degree of turnover, resembling that of short-lived effector cells.
Figure 4. IL-10+ hepatic CD8+ T cells demonstrate a high turnover rate.
(A) Frequency of Ki-67–expressing hepatic CD8+ T cells among IL-10/GFP− and IL-10/GFP+ subsets, isolated from TLR9–MAS IL-10 reporter mice. Naïve and effector splenic CD8+ T cells are included for comparison. n = 4 paired samples/group. Data are representative of 2 independent experiments. Analyzed by paired Student’s t test. (B) Viability of live-sorted IL-10/GFP+ or IL-10/GFP− hepatic CD8+ T cells after in vitro culture with or without BMDCs and CpG. n = 4 paired samples/group; data were pooled from 3 independent experiments. (C) Viability time course of live-sorted IL-10/GFP+ or IL-10/GFP− hepatic CD8+ T cells or naïve splenic CD8+ T cells. *P < 0.05, ** P < 0.01.
IL-10+ hepatic CD8+ T cells possess a distinct transcriptional signature most similar to d-6 effector CD8+ T cells but showing responsiveness to liver growth factors
Because the poor viability of IL-10+ hepatic CD8+ T cells precluded in vitro functional studies, we conducted ex vivo transcriptional analysis to further define those cells. We performed genome-wide microarray studies on biologic pairs of IL-10+ and IL-10− hepatic CD44+CD8+ T cells isolated from TLR9–MAS IL-10 reporter mice. Unsupervised clustering analysis grouped all IL-10+ samples separate from the IL-10− samples, confirming the distinctiveness of IL-10+CD8+ T cells at a transcriptional level (Supplemental Fig. 2). Indeed, 178 genes were significantly increased and 204 genes were significantly decreased in IL-10+ cells compared with IL-10− cells. Importantly, Il10 was the gene most up-regulated in the IL-10+ population as compared with the IL-10− cells, thereby validating our sorting purity and analysis methods (Fig. 5A).
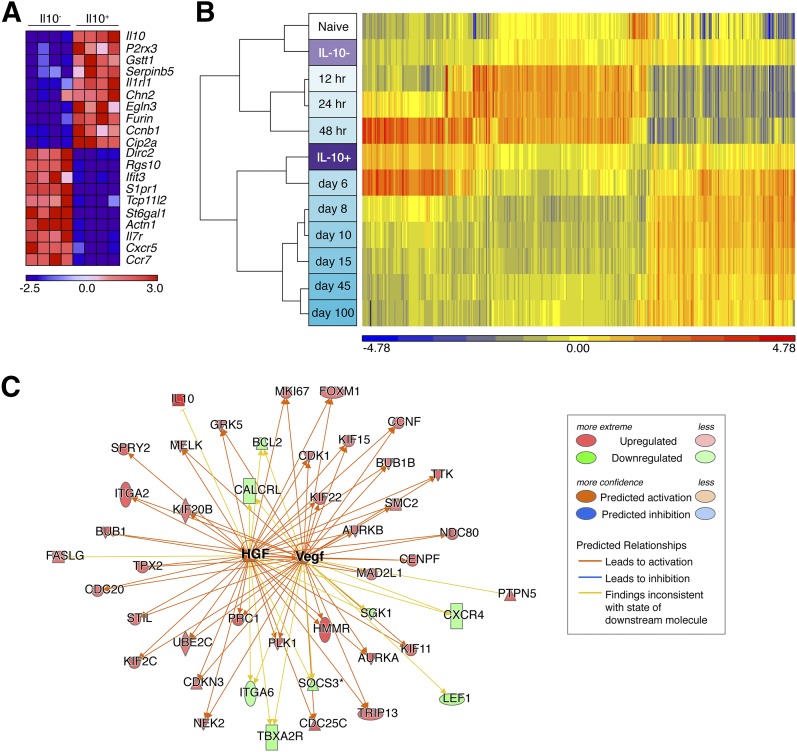
Figure 5. IL-10+ hepatic CD8+ T cells possess a distinct transcriptional signature most similar to d-6 effector CD8+ T cells but showing responsiveness to liver growth factors.
(A) Heat map depicting the top 10 genes most differentially up-regulated or down-regulated in IL-10+ vs. IL-10− cells. Colors indicate the number of standard deviations from the mean intensity for a given gene. Paired samples correspond to the same columns within each group. (B) Hierarchical clustering analysis comparing IL-10+ and IL-10− hepatic CD8+ T cell subsets (rows labeled in purple) to ImmGen CD8+ T cell subsets (rows labeled in white/turquoise). (C) Representation of HGF and VEGF networks, showing differential expression of downstream targets in IL-10+ cells and predicted activation state of upstream regulators.
Analysis of differentially expressed genes revealed hallmarks characteristic of effector CD8+ T cells among the IL-10+ population. Expression of Il1rl1 and Furin, both of which are induced upon TCR stimulation [31, 32], were highly up-regulated in IL-10+ hepatic CD8+ T cells relative to their IL-10− counterparts (Fig. 5A). The most down-regulated genes were notable for several chemokine receptors (S1pr1, Cxcr5, Ccr7) and other genes typically expressed at low levels in effector CD8+ T cells (Il7r, Actn1) (Fig. 5A) [33, 34]. These data suggest that IL-10+ hepatic CD8+ T cells are programmed to circulate through nonlymphoid tissues, consistent with their accumulation in the liver. A number of up-regulated genes were regulators of cell cycle (e.g., Ccnb1; Fig. 5A), similar to more putative upstream regulators identified by IPA, including FOXM1, MYC, S100A6, CCND1, MAPK1 (predicted activation; Table 1) and TP53, CDKN2A, CDKN1A, RBL1 (predicted inhibition; Table 1). These findings indicated that IL-10+ cells are highly proliferative and, thus, corroborated our results in Fig. 4A.
TABLE 1.
Upstream pathway analysis and predicted regulators
| Upstream regulator | Molecule type | Activation z score | P value of overlap |
|---|---|---|---|
| ESR1 | Ligand-dependent nuclear receptor | 5.435 | 6.87 × 10−12 |
| PTGER2 | G-protein coupled receptor | 5.222 | 2.52 × 10−37 |
| CSF2 | Cytokine | 4.083 | 4.99 × 10−31 |
| FOXM1 | Transcription regulator | 3.794 | 4.92 × 10−13 |
| TBX2 | Transcription regulator | 3.769 | 7.00 × 10−15 |
| HGF | Growth factor | 3.535 | 5.80 × 10−17 |
| MYC | Transcription regulator | 3.529 | 1.06 × 10−6 |
| Vegf | Group (growth factor) | 3.139 | 1.78 × 10−18 |
| ACKR2 | G-protein coupled receptor | 3.000 | 1.95 × 10−8 |
| TRIM24 | Transcription regulator | 2.985 | 1.41 × 10−5 |
| MED1 | Transcription regulator | 2.945 | 7.99 × 10−6 |
| MITF | Transcription regulator | 2.889 | 6.30 × 10−12 |
| S100A6 | Transporter | 2.828 | 4.59 × 10−8 |
| RARA | Ligand-dependent nuclear receptor | 2.820 | 6.56 × 10−13 |
| AR | Ligand-dependent nuclear receptor | 2.589 | 5.91 × 10−12 |
| CCND1 | Transcription regulator | 2.496 | 2.16 × 10−24 |
| AHR | Ligand-dependent nuclear receptor | 2.440 | 2.87 × 10−5 |
| NKX2-3 | Transcription regulator | 2.324 | 6.66 × 10−6 |
| E2f | Group (transcription regulator) | 2.204 | 4.90 × 10−10 |
| MAPK1 | Kinase | 2.144 | 3.98 × 10−6 |
| TRAF2 | Enzyme | 2.105 | 2.30 × 10−7 |
| NUPR1 | Transcription regulator | −5.864 | 4.50 × 10−17 |
| TCF3 | Transcription regulator | −4.359 | 5.46 × 10−13 |
| TP53 | Transcription regulator | −3.764 | 2.30 × 10−25 |
| KDM5B | Transcription regulator | −3.733 | 2.86 × 10−7 |
| IRF7 | Transcription regulator | −3.533 | 4.59 × 10−9 |
| IRF3 | Transcription regulator | −3.412 | 3.79 × 10−8 |
| CDKN2A | Transcription regulator | −3.300 | 4.56 × 10−7 |
| IFNA2 | Cytokine | −3.193 | 3.85 × 10−12 |
| IFNAR1 | Transmembrane receptor | −3.090 | 9.00 × 10−10 |
| CDKN1A | Kinase | −2.715 | 3.89 × 10−23 |
| TGM2 | Enzyme | −2.688 | 3.52 × 10−7 |
| DDX58 | Enzyme | −2.608 | 2.46 × 10−5 |
| BACH2 | Transcription regulator | −2.607 | 7.22 × 10−6 |
| IRF5 | Transcription regulator | −2.353 | 9.86 × 10−7 |
| SMARCE1 | Transcription regulator | −2.236 | 3.24 × 10−5 |
| DYRK1A | Kinase | −2.224 | 4.25 × 10−5 |
| RBL1 | Transcription regulator | −2.200 | 4.21 × 10−5 |
| TLR9 | Transmembrane receptor | −2.193 | 5.46 × 10−9 |
| STAT5A | Transcription regulator | −2.081 | 2.32 × 10−5 |
| IFNL1 | Cytokine | −2.030 | 6.96 × 10−7 |
Consistent with the reduced viability of IL-10+ cells (Fig. 4B and C), downstream analysis of cell death pathways demonstrated a significant number of differentially expressed apoptosis-related genes in IL-10+ cells, including Fasl (up-regulated 1.535-fold) and Bcl2 (down-regulated 1.923-fold). IPA predicted increased T cell apoptosis with a P value of 4.6 × 10−5 and a z-score of 1.98, suggesting apoptosis was likely enhanced in IL-10+ cells. Furthermore, FasL expression by IL-10+ hepatic CD8+ T cells has hepatotoxic potential because hepatocytes constitutively express Fas and are thus highly susceptible to apoptosis [35, 36]. Fas/FasL signaling has also been implicated in the liver damage observed in murine FHL [37], raising the possibility of a similar mechanism in TLR9–MAS.
As an alternative analytic approach, we entered our differentially expressed gene list into the Database for Annotation, Visualization and Integrated Discovery (National Institute of Allergy and Infectious Diseases, Bethesda, MD, USA) and performed a clustering analysis. The results suggested that genes differentially expressed in IL-10+CD8+ T cells represent altered cell cycle, cell death, and T cell activation status, consistent with our IPA analysis (see Supplemental File).
These results point to an effector-like classification for IL-10+ hepatic CD8+ T cells, but we wanted to compare them to a known standard. The Immunologic Genome Project (ImmGen) has generated data on CD8+ T cell subtypes by isolating adoptively transferred OT-I cells from Listeria–OVA-infected mice at different time points postinfection [13]. We performed clustering analysis to compare the transcriptional profiles of our IL-10+ and IL-10− CD8+ T cells to those ImmGen CD8+ T cell subsets, using the top 500 genes able to differentiate among ImmGen subsets. Remarkably, the IL-10+ hepatic CD8+ T cells clustered closest to the d-6 ImmGen subset, whereas the IL-10− hepatic CD8+ T cells were most similar to naïve CD8+ T cells (Fig. 5B). These data provide further evidence that IL-10 marks a distinct CD8+ T cell population in the liver which best resembles early effectors, and suggest that not all IL-10–producing CD8+ T cells may be suppressive.
However, IL-10+ hepatic CD8+ T cells were not merely identical to conventional effector cells. Strikingly, their transcriptional profile showed hallmarks of activation by HGF and VEGF (Fig. 5C and Table 1), 2 growth factors highly induced upon hepatic injury [38]. HGF enhances T cell adhesion and promotes leukocyte migration to the liver [39]. VEGF is important for the maintenance of liver sinusoidal endothelial cells, which are specially equipped to recruit lymphocytes to the liver [40]. Thus, the predicted responsiveness of IL-10+ hepatic CD8+ T cells to HGF and VEGF provides evidence that these cells may specifically home to injured liver and undergo transcriptional reprogramming there.
Taken together, these data demonstrate that IL-10+ hepatic CD8+ T cells represent a unique subset of highly activated, Ag-independent, early effector cells induced by systemic inflammation to accumulate within damaged liver, with the potential to contribute to pathogenesis. Further study will not only be important for understanding the role of this particular IL-10+ hepatic CD8+ T cell subset, but may also offer insights that will enable us to dissect mechanisms of inflammatory liver damage in iPALF and other diseases.
AUTHORSHIP
J.E.R., S.W.C., and L.K.W. designed, conducted, and analyzed experiments. J.W.T. and E.M.B. performed bioinformatics analyses. J.E.R. prepared the manuscript figures and wrote the manuscript. E.M.B. edited the manuscript and supervised the overall research.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the U.S. National Institutes of Health (Grants R01 HL112836-A1 from National Heart, Lung, and Blood Institute [E.M.B.], T32 AR007442-27 from National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) [J.E.R.], T32 HD043021-09 from Eunice Kennedy Shriver National Institute of Child Health and Human Development and T32 AR007442-26A1 from NIAMS [L.K.W.], and F32-AI-98337 from National Institute of Allergy and Infectious Diseases [S.W.C.]), the Rheumatology Research Foundation (Scientist Development Award to S.W.C.), and the Nancy Taylor Foundation (E.M.B.). We thank Katharine Slade and Julia Wrobel for technical assistance, Eric Rappaport and the Nucleic Acid/Protein Research Core Facility at Children’s Hospital of Philadelphia for performing microarrays, and Gary Koretzky, Taku Kambayashi, Martha Jordan, and E. John Wherry for thoughtful input.
Glossary
- BM
bone marrow
- BMDC
bone marrow–derived dendritic cell
- FHL
familial hemophagocytic lymphohistiocytosis
- HGF
hepatocyte growth factor
- IFNGR
IFN-γ receptor
- IPA
Ingenuity Pathway Analysis (Qiagen)
- iPALF
indeterminate pediatric acute liver failure
- LCMV
lymphocytic choriomeningitis virus
- MAS
macrophage activation syndrome
- PALF
pediatric acute liver failure
- sIL
soluble interleukin
- VEGF
vascular endothelial growth factor
- WT
wild-type
Footnotes
The online version of this paper, found at www.jleukbio.org, includes supplemental information.
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- 1.Squires R. H., Jr (2008) Acute liver failure in children. Semin. Liver Dis. 28, 153–166. [DOI] [PubMed] [Google Scholar]
- 2.Baliga P., Alvarez S., Lindblad A., Zeng L.; Studies of Pediatric Liver Transplantation Research Group (2004) Posttransplant survival in pediatric fulminant hepatic failure: the SPLIT experience. Liver Transpl. 10, 1364–1371. [DOI] [PubMed] [Google Scholar]
- 3.Squires R. H. Jr., Shneider B. L., Bucuvalas J., Alonso E., Sokol R. J., Narkewicz M. R., Dhawan A., Rosenthal P., Rodriguez-Baez N., Murray K. F., Horslen S., Martin M. G., Lopez M. J., Soriano H., McGuire B. M., Jonas M. M., Yazigi N., Shepherd R. W., Schwarz K., Lobritto S., Thomas D. W., Lavine J. E., Karpen S., Ng V., Kelly D., Simonds N., Hynan L. S. (2006) Acute liver failure in children: the first 348 patients in the pediatric acute liver failure study group. J. Pediatr. 148, 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucuvalas J., Filipovich L., Yazigi N., Narkewicz M. R., Ng V., Belle S. H., Zhang S., Squires R. H. (2013) Immunophenotype predicts outcome in pediatric acute liver failure. J. Pediatr. Gastroenterol. Nutr. 56, 311–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiPaola F., Grimley M., Bucuvalas J. (2014) Pediatric acute liver failure and immune dysregulation. J. Pediatr. 164, 407–409. [DOI] [PubMed] [Google Scholar]
- 6.Canna S. W., Behrens E. M. (2012) Making sense of the cytokine storm: a conceptual framework for understanding, diagnosing, and treating hemophagocytic syndromes. Pediatr. Clin. North Am. 59, 329–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henter J. I., Horne A., Aricó M., Egeler R. M., Filipovich A. H., Imashuku S., Ladisch S., McClain K., Webb D., Winiarski J., Janka G. (2007) HLH-2004: Diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr. Blood Cancer 48, 124–131. [DOI] [PubMed] [Google Scholar]
- 8.Jordan M. B., Hildeman D., Kappler J., Marrack P. (2004) An animal model of hemophagocytic lymphohistiocytosis (HLH): CD8+ T cells and interferon gamma are essential for the disorder. Blood 104, 735–743. [DOI] [PubMed] [Google Scholar]
- 9.Brisse E., Wouters C. H., Matthys P. (2015) Hemophagocytic lymphohistiocytosis (HLH): a heterogeneous spectrum of cytokine-driven immune disorders. Cytokine Growth Factor Rev. 26, 263–280. [DOI] [PubMed] [Google Scholar]
- 10.McKenzie R. B., Berquist W. E., Nadeau K. C., Louie C. Y., Chen S. F., Sibley R. K., Glader B. E., Wong W. B., Hofmann L. V., Esquivel C. O., Cox K. L. (2014) Novel protocol including liver biopsy to identify and treat CD8+ T-cell predominant acute hepatitis and liver failure. Pediatr. Transplant. 18, 503–509. [DOI] [PubMed] [Google Scholar]
- 11.Behrens E. M., Canna S. W., Slade K., Rao S., Kreiger P. A., Paessler M., Kambayashi T., Koretzky G. A. (2011) Repeated TLR9 stimulation results in macrophage activation syndrome-like disease in mice. J. Clin. Invest. 121, 2264–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamanaka M., Kim S. T., Wan Y. Y., Sutterwala F. S., Lara-Tejero M., Galán J. E., Harhaj E., Flavell R. A. (2006) Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity 25, 941–952. [DOI] [PubMed] [Google Scholar]
- 13.Best J. A., Blair D. A., Knell J., Yang E., Mayya V., Doedens A., Dustin M. L., Goldrath A. W.; Immunological Genome Project Consortium (2013) Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat. Immunol. 14, 404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson W. E., Li C., Rabinovic A. (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127. [DOI] [PubMed] [Google Scholar]
- 15.Terrell C. E., Jordan M. B. (2013) Perforin deficiency impairs a critical immunoregulatory loop involving murine CD8(+) T cells and dendritic cells. Blood 121, 5184–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rifa’i M., Kawamoto Y., Nakashima I., Suzuki H. (2004) Essential roles of CD8+CD122+ regulatory T cells in the maintenance of T cell homeostasis. J. Exp. Med. 200, 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endharti A. T., Rifa’I M., Shi Z., Fukuoka Y., Nakahara Y., Kawamoto Y., Takeda K., Isobe K., Suzuki H. (2005) Cutting edge: CD8+CD122+ regulatory T cells produce IL-10 to suppress IFN-γ production and proliferation of CD8+ T cells. J. Immunol. 175, 7093–7097. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y., Lan Q., Lu L., Chen M., Xia Z., Ma J., Wang J., Fan H., Shen Y., Ryffel B., Brand D., Quismorio F., Liu Z., Horwitz D. A., Xu A., Zheng S. G. (2014) Phenotypic and functional characteristic of a newly identified CD8+Foxp3−CD103+ regulatory T cells. J. Mol. Cell Biol. 6, 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xystrakis E., Dejean A. S., Bernard I., Druet P., Liblau R., Gonzalez-Dunia D., Saoudi A. (2004) Identification of a novel natural regulatory CD8 T-cell subset and analysis of its mechanism of regulation. Blood 104, 3294–3301. [DOI] [PubMed] [Google Scholar]
- 20.Bienvenu B., Martin B., Auffray C., Cordier C., Bécourt C., Lucas B. (2005) Peripheral CD8+CD25+ T lymphocytes from MHC class II-deficient mice exhibit regulatory activity. J. Immunol. 175, 246–253. [DOI] [PubMed] [Google Scholar]
- 21.Tang X., Maricic I., Purohit N., Bakamjian B., Reed-Loisel L. M., Beeston T., Jensen P., Kumar V. (2006) Regulation of immunity by a novel population of Qa-1-restricted CD8αα+TCRαβ+ T cells. J. Immunol. 177, 7645–7655. [DOI] [PubMed] [Google Scholar]
- 22.Vinay D. S., Kim C. H., Choi B. K., Kwon B. S. (2009) Origins and functional basis of regulatory CD11c+CD8+ T cells. Eur. J. Immunol. 39, 1552–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Sun S., Hwang I., Tough D. F., Sprent J. (1998) Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity 8, 591–599. [DOI] [PubMed] [Google Scholar]
- 24.Berg R. E., Crossley E., Murray S., Forman J. (2003) Memory CD8+ T cells provide innate immune protection against Listeria monocytogenes in the absence of cognate antigen. J. Exp. Med. 198, 1583–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kambayashi T., Assarsson E., Lukacher A. E., Ljunggren H.-G., Jensen P. E. (2003) Memory CD8+ T cells provide an early source of IFN-γ. J. Immunol. 170, 2399–2408. [DOI] [PubMed] [Google Scholar]
- 26.Kohlmeier J. E., Cookenham T., Roberts A. D., Miller S. C., Woodland D. L. (2010) Type I interferons regulate cytolytic activity of memory CD8+ T cells in the lung airways during respiratory virus challenge. Immunity 33, 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canna S. W., Wrobel J., Chu N., Kreiger P. A., Paessler M., Behrens E. M. (2013) Interferon-γ mediates anemia but is dispensable for fulminant toll-like receptor 9-induced macrophage activation syndrome and hemophagocytosis in mice. Arthritis Rheum. 65, 1764–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearce E. L., Mullen A. C., Martins G. A., Krawczyk C. M., Hutchins A. S., Zediak V. P., Banica M., DiCioccio C. B., Gross D. A., Mao C. A., Shen H., Cereb N., Yang S. Y., Lindsten T., Rossant J., Hunter C. A., Reiner S. L. (2003) Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 302, 1041–1043. [DOI] [PubMed] [Google Scholar]
- 29.Huang L. R., Wohlleber D., Reisinger F., Jenne C. N., Cheng R. L., Abdullah Z., Schildberg F. A., Odenthal M., Dienes H. P., van Rooijen N., Schmitt E., Garbi N., Croft M., Kurts C., Kubes P., Protzer U., Heikenwalder M., Knolle P. A. (2013) Intrahepatic myeloid-cell aggregates enable local proliferation of CD8+ T cells and successful immunotherapy against chronic viral liver infection. Nat. Immunol. 14, 574–583. [DOI] [PubMed] [Google Scholar]
- 30.Mehal W. Z., Juedes A. E., Crispe I. N. (1999) Selective retention of activated CD8+ T cells by the normal liver. J. Immunol. 163, 3202–3210. [PubMed] [Google Scholar]
- 31.Yang Q., Li G., Zhu Y., Liu L., Chen E., Turnquist H., Zhang X., Finn O. J., Chen X., Lu B. (2011) IL-33 synergizes with TCR and IL-12 signaling to promote the effector function of CD8+ T cells. Eur. J. Immunol. 41, 3351–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pesu M., Muul L., Kanno Y., O’Shea J. J. (2006) Proprotein convertase furin is preferentially expressed in T helper 1 cells and regulates interferon gamma. Blood 108, 983–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung Y. W., Rutishauser R. L., Joshi N. S., Haberman A. M., Kaech S. M. (2010) Differential localization of effector and memory CD8 T cell subsets in lymphoid organs during acute viral infection. J. Immunol. 185, 5315–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benechet A. P., Menon M., Xu D., Samji T., Maher L., Murooka T. T., Mempel T. R., Sheridan B. S., Lemoine F. M., Khanna K. M. (2016) T cell-intrinsic S1PR1 regulates endogenous effector T-cell egress dynamics from lymph nodes during infection. Proc. Natl. Acad. Sci. USA 113, 2182–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malhi H., Guicciardi M. E., Gores G. J. (2010) Hepatocyte death: a clear and present danger. Physiol. Rev. 90, 1165–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wesche-Soldato D. E., Chung C.-S., Gregory S. H., Salazar-Mather T. P., Ayala C. A., Ayala A. (2007) CD8+ T cells promote inflammation and apoptosis in the liver after sepsis: role of Fas-FasL. Am. J. Pathol. 171, 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiossone L., Audonnet S., Chetaille B., Chasson L., Farnarier C., Berda-Haddad Y., Jordan S., Koszinowski U. H., Dalod M., Mazodier K., Novick D., Dinarello C. A., Vivier E., Kaplanski G. (2012) Protection from inflammatory organ damage in a murine model of hemophagocytic lymphohistiocytosis using treatment with IL-18 binding protein. Front. Immunol. 3, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLeve L. D. (2013) Liver sinusoidal endothelial cells and liver regeneration. J. Clin. Invest. 123, 1861–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams D. H., Harvath L., Bottaro D. P., Interrante R., Catalano G., Tanaka Y., Strain A., Hubscher S. G., Shaw S. (1994) Hepatocyte growth factor and macrophage inflammatory protein 1 beta: structurally distinct cytokines that induce rapid cytoskeletal changes and subset-preferential migration in T cells. Proc. Natl. Acad. Sci. USA 91, 7144–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lalor P. F., Lai W. K., Curbishley S. M., Shetty S., Adams D. H. (2006) Human hepatic sinusoidal endothelial cells can be distinguished by expression of phenotypic markers related to their specialised functions in vivo. World J. Gastroenterol. 12, 5429–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.