Abstract
In RNA interference (RNAi), short double-stranded RNA (known as siRNA) inhibits expression from homologous genes. Clinical or pre-clinical use of siRNAs is likely to require stabilizing modifications because of the prevalence of intracellular and extracellular nucleases. In order to examine the effect of modification on siRNA efficacy and stability, we developed a new method for synthesizing stereoregular boranophosphate siRNAs. This work demonstrates that boranophosphate siRNAs are consistently more effective than siRNAs with the widely used phosphorothioate modification. Furthermore, boranophosphate siRNAs are frequently more active than native siRNA if the center of the antisense strand is not modified. Boranophosphate modification also increases siRNA potency. The finding that boranophosphate siRNAs are at least ten times more nuclease resistant than unmodified siRNAs may explain some of the positive effects of boranophosphate modification. The biochemical properties of boranophosphate siRNAs make them promising candidates for an RNAi-based therapeutic.
INTRODUCTION
RNA interference (RNAi) is a form of post-transcriptional gene silencing in which double-stranded RNA (dsRNA) targets homologous mRNA for destruction [reviewed in (1–4)]. RNAi has been shown to occur in a wide variety of organisms from protozoa to mammals. The RNAi effector molecule, or short interfering RNA (siRNA), is double-stranded RNA about 21 bp in length with 3′ nucleotide overhangs (5). While questions remain about the precise mechanism of RNA interference, recent work has provided a clearer understanding of the process. siRNAs associate with a variety of cellular proteins to form an RNA-induced silencing complex (RISC) (6,7). A single strand from the siRNA is incorporated into the RISC, which will then target mRNA complementary to that strand (8).
There has been considerable interest in harnessing the power of RNA interference to treat human diseases (9) such as viral infections (10,11), cancer (12,13) and sepsis (14). One major problem with the pharmaceutical use of nucleic acids in general is their sensitivity to degradation by intracellular and extracellular nucleases. Work from the antisense and ribozyme fields suggests a possible solution, namely replacing the 3′–5′ phosphodiester linkage with more stable moieties to reduce susceptibility to nuclease degradation. Perhaps the best-characterized and most widely used modification of nucleic acids is the partial or complete replacement of the phosphodiester backbone with phosphorothioate linkages (in which a sulfur molecule is used in place of a non-bridging oxygen). Phosphorothioate-modified nucleic acids have several properties that have made them attractive for clinical use. They are more nuclease resistant than phosphodiester-backbone nucleic acids, have slower in vivo clearance and are recognized by DNA and RNA polymerases, and so may be synthesized enzymatically (15–17). Phosphorothioates have been the essential components of almost every successful antisense experiment in vivo (18).
Several recent studies have shown that some siRNAs with chemical modifications (including phosphorothioates) are active in RNAi assays (19–22). However, there appear to be limitations on the use of phosphorothioate siRNAs, including toxicity and impaired activity with increasing degrees of modification (19,21–23).
An alternate backbone modification that confers increased biological stability to nucleic acids is the boranophosphate linkage. In boranophosphate oligonucleotides, the non-bridging phosphodiester oxygen is replaced with an isoelectronic borane (–BH3) moiety. While boranophosphates have been less widely studied, they have many of the same advantages as phosphorothioates. Like phosphorothioates (24), boranophosphates maintain the ability to base pair with high specificity and affinity, and can be readily incorporated into DNA and RNA molecules by DNA and RNA polymerases, permitting synthesis of stereoregular boranophosphate RNA (25–29). Boranophosphates have additional properties that make them potentially more suitable for clinical use than phosphorothioate oligonucleotides (30). Boranophosphate RNA molecules are more than 300-fold more nuclease resistant than unmodified molecules and more than twice as nuclease resistant as their phosphorothioate counterparts (K. He, Z. A. Sergueeva, J. Wan and B. Ramsey Shaw, unpublished results). While each boranophosphate linkage retains a negative charge, the charge distribution of boranophosphate differs from that of normal phosphate and phosphorothioate, and thus changes the polarity and increases the hydrophobicity of the molecule (31). As a result, boranophosphate RNA may have different hydration properties and different interactions with metal ions and proteins, which could result in altered biological activity. Also, boranophosphate DNA dinucleotides are minimally toxic to rodents, and the degradation products of boranophosphate oligonucleotides (i.e. borates) are minimally toxic to humans (32,33).
In this work, we developed a new method for synthesizing stereoregular boranophosphate siRNAs. When tested in a silencing assay, the activity of boranophosphate siRNAs consistently exceeded that of phosphorothioate siRNAs, and was often greater than that of native, phosphodiester backbone siRNAs. Investigation of the structure–activity relationship in backbone-modified siRNAs revealed that a high degree of boranophosphate modification can yield very active siRNAs, particularly if the central portion of the antisense strand remains largely unmodified. Most notably, boranophosphate siRNAs are significantly more potent than unmodified siRNAs and appear to act through the standard RNAi pathway.
MATERIALS AND METHODS
Synthesis of boranophosphate and phosphorothioate siRNA
Desalted DNA oligonucleotides were purchased from Qiagen: T7 promoter primer, 5′-TAATACGACTCACTATAG-3′; EGFP1 templates: sense, 5′-AAGTTCACCTTGATGCCGTTCTATAGTGAGTCGTATTA-3′, antisense, 5′-AAGAACGGCATCAAGGTGAACTATAGTGAGTCGTATTA-3′; control EGPF1 templates: sense, 5′-AAGTTCACCGTAGTTCCGTTCTATAGTGAGTCGTATTA-3′, antisense, 5′-AAGAACGGAACTACGGTGAACTATAGTGAGTCGTATTA-3′; EGFP2 templates: sense, 5′-AAGGACTTGAAGAAGTCGTGCTATAGTGAGTCGTATTA-3′, Antisense, 5′-AAGCACGACTTCTTCAAGTCCTATAGTGAGTCGTATTA-3′; Control EGFP2 templates: sense, 5′-AAGGACTTTGAAGAAGCGTGCTATAGTGAGTCGTATTA-3′, Antisense, 5′-AAGCACGCTTCTTCAAAGTCCTATAGTGAGTCGTATTA-3′. For each in vitro transcription reaction, equimolar amounts of each template oligonucleotides were annealed with the T7 promoter primer oligonucleotide in annealing buffer (10 mM Tris–HCl pH 8.0 and 100 mM NaCl) by heating at 70°C for 5 min and cooled at room temperature to obtain partially dsDNA. This partially dsDNA was extended by Exonuclease-free Klenow fragment (Ambion) to produce fully dsDNA templates. Cytidine-5′-O-(1-thiophosphate), Sp isomer, and uridine-5′-O-(1-thiophosphate), Sp isomer, were purchased from Trilink. Ribonucleoside 5′-(α-P-borano)triphosphates (NTPαBs) were synthesized and the Rp isomer was isolated as described previously (34). For both phosphorothioate and boranophosphate oligonucleotides, sense and antisense 21-nt RNAs were generated in separate reactions from DNA templates by transcription using an AmpliScribe T7 High Yield Transcription Kit (Epicentre), and native or modified ribonucleoside 5′ triphosphates (NTPs). After incubation at 37°C for 2–4 h, 1 U RNase-free DNase (Epicentre) was added at 37°C for 15 min. Single-strand RNAs were then purified with phenol extraction and Microspin G-50 micro Columns (Amersham), and annealed in the annealing buffer (100 mM potassium acetate, 2 mM magnesium acetate, 30 mM HEPES–KOH, pH 7.4) by heating at 90°C for 1 min followed by 1 h incubation at 37°C. After purification and concentration with Microcon YM-10 centrifugal filter (Millipore), concentrations were determined by measuring A260. RNA puritywas monitored with 4% NuSieve GTG agarose gels (Cambrex). RNA concentration was determined by UV absorption. MALDI-TOF mass spectrometry with a Voyager-DE™ PRO BioSpectrometry™ Workstation was used to confirm the presence of boranophosphate and phosphorothioate linkages in single-stranded RNAs. The sample mixtures were prepared from 1 μl of ∼50 μM enzymatically synthesized and gel-purified RNA samples (normal, phosphorothioate, or boranophosphate) and 5 μl matrix [saturated solution of ATT (6-aza-thiothymine) and 0.1 M diammonium citrate]. An aliquot of 1.3 μl of this mixture was spotted in duplicate onto the MALDI-TOF gold sample carrier. The instrument settings were: linear mode, positive-ion detection with a positive 20 000 acceleration voltage, and the delayed extraction option activated at 350 ns. Calibration of the instrument was carried out with two 21-nt chemically synthesized RNAs.
Cell culture and transfection
TRex HeLa cells (Invitrogen) stably transfected with enhanced green fluorescent protein (EGFP) (Clontech) under control of a tetracycline-responsive promoter (pcDNA4/TO, Invitrogen) were cultured in MEM (Gibco) with 10% tetracycline tested fetal calf serum (FCS) (Clontech), 5 μg/ml blasticidin (Invitrogen) and 100 μg/ml Zeocin (Invitrogen). For transfections, 4.5 × 105 cells were plated per well of a 24-well plate. After 24 h, cells were transfected using Oligofectamine (Invitrogen) according to the manufacturers instructions with 1.85 μl Oligofectamine per well. The concentration of siRNA in the medium ranged from 25 to 3.1 nM, as described in figure legends. Twenty-four hours after siRNA transfection, cells were induced to express GFP with 1 μg/ml tetracycline. As the control sequence for EGFP1, we used an siRNA with the six central nucleotides inverted. As the control sequence for EGFP2, we used an siRNA with eight central nucleotides inverted because the sequence with six inverted nucleotides had an unacceptable degree of homology to cellular mRNAs.
Fluorescence activated cell sorting (FACS) analysis
After 54 h post-transfection, cells were trypsinized and fixed in 1% formaldehyde in phosphate-buffered saline (PBS). Cells were analyzed for GFP expression using a FACScan (Becton-Dickinson) flow cytometer. Data were processed using CellQuest software (Becton-Dickinson).
Cellular RNA analysis
Cells were transfected in culture medium at a final siRNA concentration of 12.5 nM. EGFP1 production was induced with 1 μg/ml tetracycline. After 22 h post-induction, cells were trypsinized and total cytoplasmic RNA isolated using the RNeasy kit (Qiagen). An aliquot of 2 μg of RNA from each group was separated by formaldehyde-agarose gel electrophoresis, transferred to a positively charged nylon membrane and cross-linked by UV irradiation. The immobilized RNA was hybridized with 32P-labeled RNA probes complementary to the EGFP coding sequence and to human β-actin (to normalize for signal). Hybridization was quantified using a Molecular Dynamics phosphorimaging system.
Nuclease stability analysis
Boranophosphate and unmodified EGFP1 siRNAs (18 pmol) were incubated with a crude mixture of RNases extracted from bovine pancreas (Roche) at 500 ng/ml in 10 μl reaction buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 75 μM NaCl). Samples were subjected to electrophoresis on a 4% Nusieve GTG agarose gel (Cambrex) with ethidium bromide. RNA was visualized by UV illumination and photographed with Nucleocam software (Nucleotech). The intensity of fluorescent bands was quantified using GelExpert software (Nucleotech).
RESULTS
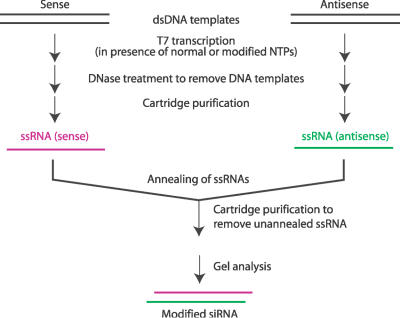
In light of the significant promise of RNAi-based therapeutics as well as the superior biological stability and potential for low toxicity of boranophosphate nucleic acids, we developed a new method for synthesis of stereoregular boranophosphate siRNAs. We used enzymatic synthesis to produce native or modified siRNAs because of the high cost and the difficulty in producing stereoregular backbone-modified RNAs with chemical synthesis. For efficient synthesis, the T7 RNA polymerase requires the initiation nucleotide to be guanosine. Thus, all siRNAs described here begin with a guanosine. When synthesizing both native and modified siRNAs, we followed the strategy outlined in Figure 1. This method is similar to one that has been described for preparation of native siRNA with T7 RNA polymerase (35), but there are several notable differences. First, single-stranded antisense or sense RNAs were synthesized and purified separately to allow incorporation of different modifications into the two strands. In addition, due to the relatively low recovery of short boranophosphate oligonucleotides after alcohol precipitation, microcentrifugal filter devices were used instead.
Figure 1.
Synthesis of modified siRNAs through T7 in vitro transcription.
For in vitro transcription of siRNAs when one type of NTPαB is substituted for one of the four native NTPs, the yield varied from 50% to 100% relative to yields of reactions with all four native NTPs. Yields were poorest when the boranophosphate modification was present at the fourth position in the nascent transcript. MALDI-TOF spectrometry was used to confirm the presence of modifications. Since boron has two natural isotopes, 11B (80.1%) and 10B (19.9%), the mass difference between a borane group (BH3) and oxygen (16O, 99.757%) is 2–3 mass units, while the mass difference between sulfur (32S, 94.93%) and oxygen is 16. To facilitate comparison of molecular mass, the antisense strand of EGFP1 siRNA which contains nine uracil residues was chosen. Upon enzymatic incorporation of UTPαB substituted for all UTP, the mass of the 21-nt RNA should decrease by 20 mass units (calculated based on the average atomic mass of boron), while the mass of the corresponding phosphorothioate RNA should have an increase of 144 mass units. Our results (Figure 2 and Table 1) clearly showed the expected differences, and proved the successful incorporation of the chemical modifications. For boranophosphate, the mass decrease of 38 instead of 20 may be due to the loss of a water molecule, or the release of BH3 as observed with small boranophosphate nucleotides under the laser desorption conditions of mass spectrometry. For all three types of RNAs, there is a 22mer RNA in addition to the 21mer transcript (Figure 2A and B). This is consistent with the well-known fact that T7 RNA polymerase usually adds one or even more additional nucleotide(s) at the end of the transcripts.
Figure 2.
(A) Gel analysis of enzymatically synthesized single-stranded RNA samples used for mass spectrometry experiments. The sequence of the RNA is 5′-pppGpUpUpCpApCpCpUpUpGpApUpGpCpCpGpUpUpCpUpU-3′. The samples were analyzed with 20% PAGE/7 M urea. The gel was stained with SYBR® Green II and visualized with a UV transilluminator. Lane Std is a chemically synthesized 21-nt RNA. Lane M is 10-bp DNA marker. Lane PO is enzymatically synthesized normal RNA. Lane BP is enzymatically synthesized boranophosphate RNA. (B) Mass spectrometry spectra of enzymatically synthesized RNA samples: PO for normal; PS for phosphorothioate; BP: boranophosphate.
Table 1. Summary of mass spectrometry results of enzymatically synthesized normal, boranophosphate and phosphorothioate RNA.
| Normal RNA | Boranophosphate RNA | Phosphorothioate RNA | Std 1 | Std 2 | ||||
|---|---|---|---|---|---|---|---|---|
| 21-nt | 22-nt | 21-nt | 22-nt | 21-nt | 22-nt | |||
| Cal. Mass | 6776 | 6756b | 6920 | 6573 | 6604 | |||
| Expt. Massc | 6777 ± 1 | 7104 ± 2 | 6739 ± 1 | 7044 ± 4 | 6922 ± 1 | 7250 ± 1 | 6574 ± 1 | 6605 ± 2 |
| ΔMass, calc. | −20 | +144 | ||||||
| ΔMass, expt. | −38 | +144 | ||||||
The sequence of the RNA is 5′-pppGpUpUpCpApCpCpUpUpGpApUpGpCpCpGpUpUpCpUpU-3′.
aAll spectra were acquired by averaging 200–300 shots acquired by rastering across the sample spot.
bFor boranophosphate RNA, the mass is calculated based on the average atomic mass of boron (10B and 11B).
cAverage values are from at least two independent experiments. Under conditions used, deviations of measurements are expected within ±0.1%.
The method described above was used to synthesize both native and modified siRNAs targeted to EGFP (Figure 3). The activity of modified siRNAs was tested in HeLa cells that had been stably transfected with the EGFP gene under control of a tetracycline-inducible promoter. Cells were transfected with EGFP-specific siRNAs, induced to express EGFP and then subjected to flow cytometry to measure the level of EGFP protein expression. The fluorescence of cells transfected with siRNA was compared to the fluorescence of mock-transfected cells to determine the percent inhibition of EGFP expression. To compare peak silencing activity of modified and unmodified siRNAs, cells were transfected at an siRNA concentration of 25 nM in the culture medium because this is the lowest concentration at which the unmodified siRNA is able to induce maximal silencing of GFP expression (78% reduction in mean cellular fluorescence). To assess nonspecific effects of short, double-stranded RNAs, some cells were transfected with control native and backbone-modified molecules in which the central nucleotide sequence was inverted (Figure 3).
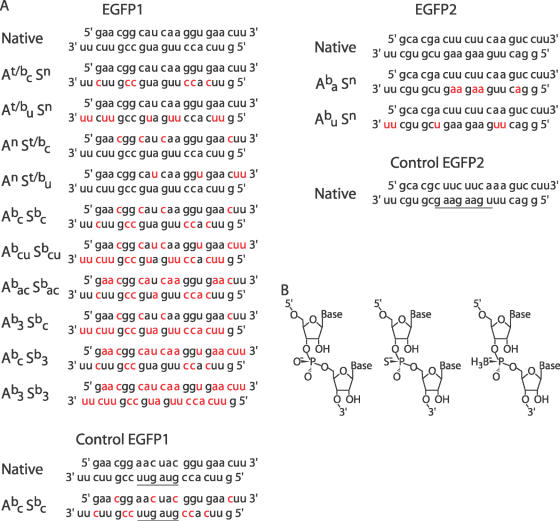
Figure 3.
(A) Native and modified siRNA species. Modified nucleotides are shown in red. For control siRNAs, the inverted sequence is underlined. A—antisense strand, S—sense strand, b—boranophosphate, t—phosphorothioate, n—native, a—adenosine, c—cytidine, u—uridine, 3—adenosine, cytidine and uridine. (B) Structure of native (left), phosphorothioate (Rp isomer, center) and boranophosphate (Sp isomer, right) ribonucleic acid backbone linkages.
siRNA activity depends on the base and strand modified
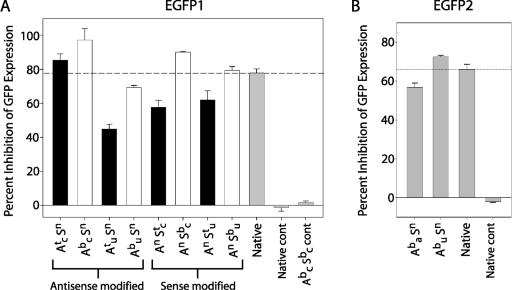
The silencing activity of molecules with modified cytidine or uridine nucleotides in either the antisense or the sense strand was examined. When the sense strand was boranophosphate-modified, the siRNAs were as active as or slightly more active than native molecules (Figure 4A). In contrast, phosphorothioate modifications on the sense strand caused moderate reductions in activity. The effect of boranophosphate or phosphorothioate modification on the sense strand was generally consistent, irrespective of the particular base that was modified. In contrast to sense strand modifications, antisense strand modifications led to striking differences in the activity of the resulting siRNAs. The effect of antisense strand modifications depended on whether the cytidine or uridine residues were modified: modification of cytidine residues on the antisense strand created highly active molecules whereas modification of uridine residues on the antisense strand caused a marked reduction in activity. This pattern was observed for both phosphorothioate and boranophosphate modification of the antisense strand. Control siRNAs with inverted sequences did not inhibit GFP expression under any of the conditions we tested (Figure 4).
Figure 4.
Position-specific effects of phosphorothioate and boranophosphate modifications on silencing activity. (A) Percent inhibition of GFP fluorescence in cells treated with native (grey bar), phosphorothioate (black bars) and boranophosphate (white bars) EGFP1 siRNA at 25 nM. To account for variations in transfection efficiency, the results of each individual experiment were normalized to results for native siRNA-treated cells in that experiment. Error bars represent the standard error. (B) Percent inhibition of GFP fluorescence in cells treated with boranophosphate modified or unmodified EGFP2 siRNA at 25 nM. Error bars represent the standard error. A—antisense strand, S—sense strand, b—boranophosphate, t—phosphorothioate, n—native, a—adenosine, c—cytidine, u—uridine, cont—inverted control sequence.
Notably, the siRNA with modified uridine residues on the antisense strand (Abu Sn) was the only boranophosphate molecule with reduced activity compared to unmodified siRNAs. There are several possible reasons why the RNAi activity of siRNAs with backbone modifications at uridine residues was impaired. One explanation is, given that Abu Sn has more modified nucleotides than any of the other species, its reduced activity is simply due to an ‘overload’ of modified nucleotides (Figure 3A). However, subsequent experiments, described below in Figure 5, demonstrated that this was not the case. Another possibility is that modified uridine residues are inherently less well tolerated in the antisense strand than are modified cytidines (perhaps because of the double uridine 3′ overhang). We also observed that Abu Sn has a relatively high concentration of uridine residues in the center of the antisense strand (Figure 3A). Thus, an alternate explanation for these results is that central modifications are less well tolerated than peripheral modifications, irrespective of the type of nucleotide that is modified. This theory is supported by previous work showing that even minor sequence changes in the middle of siRNAs can drastically reduce RNAi activity (36).
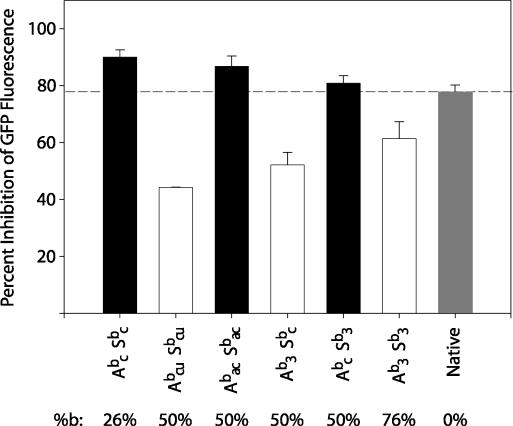
Figure 5.
Effect of increased boranophosphate modification on siRNA activity. Percent inhibition of GFP fluorescence in cells treated with native (grey bar), or boranophosphate siRNAs with the EGFP1 sequence at 25 nM. siRNAs with <3 central modifications are represented by black bars. Those with ≥3 central modifications are represented by white bars. Error bars represent the standard error. The numbers below the graph correspond to the percent of residues in the duplex region that are modified. A—antisense strand, S—sense strand, b—boranophosphate, n—native, a—adenosine, c—cytidine, u—uridine, 3—adenosine, cytidine and uridine.
Boranophosphate siRNAs are more active than phosphorothioate siRNAs
As phosphorothioate nucleotides have been extensively used and characterized in antisense and RNAi applications, we were particularly interested to compare the activity of boranophosphate siRNAs to that of phosphorothioates. The experiments described above showed that, irrespective of the base or strand modified, boranophosphate species were always more active than the corresponding phosphorothioates (Figure 4A).
Central modifications on the antisense strand inhibit siRNA activity
To test whether the position of modification or the specific base modified determines siRNA activity, we examined the silencing activity of boranophosphate and native versions of an EGFP-targeted siRNA with a different sequence (EGFP2). Like EGFP1, EGFP2 siRNA has UU 3′ overhangs but, unlike EGFP1, EGFP2 has few uridines in the center of the antisense strand. Instead, there is a high concentration of adenosine residues in this critical region (Figure 3A). Experiments showed that the EGFP2 siRNA with modified uridines in the antisense strand was actually more active than its unmodified analog, indicating that boranophosphate-modified uridines, including 3′ overhangs, are compatible with RNAi activity (Figure 4B). In contrast, the EGFP2 siRNA with boranophosphate-modified adenosine residues (including several central modifications) was significantly less active than the siRNAs synthesized with native nucleotides or boranophosphate-modified uridines. Thus, our results support the hypothesis that boranophosphate modifications placed at the center of the antisense strand reduce RNAi activity.
Highly modified boranophosphate siRNAs can retain activity
siRNAs with between 26 and 76% modified duplex nucleotides were synthesized. As shown in Figure 5, all of the siRNAs with modifications at fewer than three of five central nucleotides in the antisense strand (Abc Sbc, Abac Sbac, Abc Sb3) were as effective as, or even more effective than, the unmodified siRNAs. In contrast, the siRNAs with modified uridines (and thus, with many central modifications) were all less effective than the native siRNA. However, it is worth noting that the most highly modified siRNA species (Ab3 Sb3) was more active than the less modified Abcu Sbcu, even though both have central modifications. This finding suggests that additional peripheral modifications confer an advantage that partially compensates for the negative effects of central modifications. Thus, the activity of these highly modified siRNA species suggests that there may be no specific limit to the number of bases that can be modified, provided that relatively few bases in the central portion of the antisense strand are modified.
Boranophosphate siRNAs are more potent than native siRNAs
To compare the potency of modified and native siRNAs, we tested the activity of increasingly dilute siRNA solutions. The modified siRNAs were more potent, with greater silencing activity at low concentrations, when compared with unmodified siRNA (Figure 6A). For native molecules, reducing siRNA concentration from 25 nM to 3.1 nM reduced activity by 50%. In contrast, the activity of the most highly modified species (Ab3 Sb3) declined by <20% over the same range of concentrations. Boranophosphate siRNAs with fewer modified nucleotides than Ab3 Sb3 displayed an intermediate silencing dose–response, suggesting that there is a continuum over which a greater proportion modified bases confers increased potency.
Figure 6.
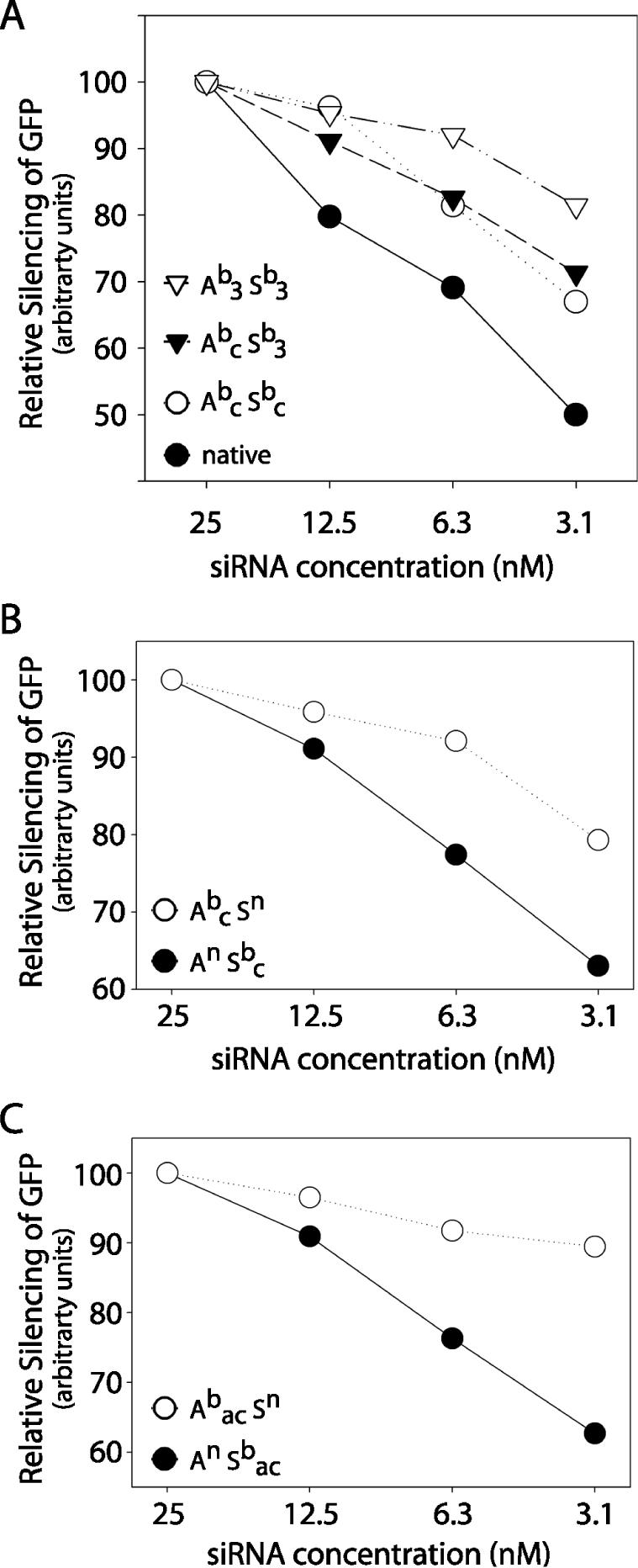
Effect of boranophosphate modification on siRNA potency. SiRNA dose–response as measured by the decrease in GFP fluorescence. The effect of each siRNA (EGFP1 sequence) at 25 nM is normalized to an arbitrary value of 100 to reveal changes in efficacy. (A) Effect of Ab3 Sb3 (open triangles), Abc Sb3 (closed triangles), Abc Sbc (open circles), and native siRNA (closed circles). A—antisense strand, S—sense strand, b—boranophosphate, n—native, a—adenosine, c—cytidine, u—uridine, 3—adenosine, cytidine and uridine. (B) Effect of boranophosphate-modified cytidines on the antisense (open circle) or sense (closed circle) strand. (C) Effect of boranophosphate-modified cytidines and adenosines on the antisense (open circle) or sense (closed circle) strand.
Modifications on the antisense strand increase siRNA potency
In order to assess the effects of modifications on the senseversus the antisense strand, siRNAs containing boranophosphate-modified cytidines on either the antisense or the sense strand were synthesized. Increasing modification of the antisense strand correlated with greater EGFP silencing potency (Figure 6B). However, as there are more cytidine residues on the antisense strand than the sense strand, it was unclear whether the difference was due to the location or the number of modified bases. To examine this issue, doubly modified siRNAs with BP-modified cytidines and adenosines on either the antisense or the sense strand were also tested. In this situation, the sense strand was more highly modified than the antisense strand (Figure 3). Nonetheless, backbone modification of the antisense strand conferred improved silencing activity at low siRNA concentrations, demonstrating that modifications on the antisense strand cause a greater increase in potency than those on the sense strand (Figure 6C).
Modified siRNAs act by reducing mRNA levels
In general, dsRNA is believed to inhibit protein expression primarily by targeting homologous mRNA for degradation. However, it has also been shown that certain RNA species, known as micro-RNAs (miRNAs,) silence genes by inhibiting translation of the target mRNA. Previous work suggests that a base mismatch or bulge in the middle of an siRNA can cause it to function through the miRNA pathway rather than by an RISC-mediated mechanism (37). Because backbone modifications generally reduce the strength of the base-pairing interaction (possibly allowing a bulge), we sought to determine whether the boranophosphate siRNAs were inhibiting translation rather than inducing degradation of target mRNA. The effect of boranophosphate-modified dsRNAs on the level of EGFP mRNA was examined by northern blot and normalized to β-actin expression. This experiment revealed that, like native siRNAs, boranophosphate-modified siRNAs caused a reduction in the level of EGFP mRNA (Figure 7). To compare the effect of the siRNAs on mRNA levels with changes in EGFP protein levels, samples of the cells used for northern analysis were analyzed by FACS. Results showed that the reduction in EGFP protein expression was essentially identical to the reduction in mRNA levels (Figure 7). Therefore, we concluded that modified siRNAs inhibit protein expression primarily by lowering EGFP mRNA levels rather than by inhibiting translation.
Figure 7.

Effect of boranophosphate-modified EGFP1 siRNA on mRNA and protein levels. Percent reduction in mRNA (white triangle) and protein (black circle) levels as determined by northern and FACS analysis when transfected with 12.5 nM siRNA. Samples of cells from the same population were used for both analyses. EGFP mRNA levels were normalized to β-actin expression. A—antisense strand, S—sense strand, b—boranophosphate, c—cytidine, 3—adenosine, cytidine and uridine, cont—native control EGFP1 sequence.
Boranophosphate siRNAs are resistant to nuclease degradation
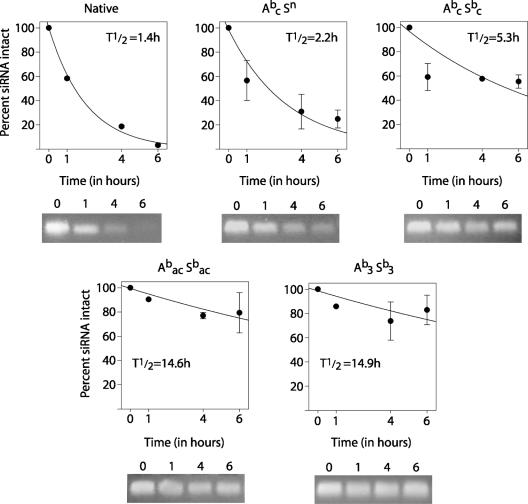
To test nuclease resistance, siRNAs were incubated with a crude preparation of RNases from bovine pancreas (roughly analogous to intracellular nucleases) with a higher RNase concentration than that found in serum. We selected assay conditions in which the unmodified siRNA had a half-life of 1.4 h, and then tested the stability of boranophosphate siRNAs under the same conditions. We found that even minimally modified samples (such as Abc Sn) showed improved stability over the unmodified siRNA, and that increasing the number of modifications yielded steadily increasing stability (Figure 8). The most highly modified species, (Abac Sbac and Ab3 Sb3), were largely intact at the longest time point of 6 h. The half-lives of these species were calculated to be >14 h—more than ten times longer than that of unmodified siRNA. Further testing revealed that over half of the Abac Sbac and Ab3 Sb3 siRNAs remain undegraded at 24 h (data not shown). It was necessary to use concentrated RNase to examine relative stability because all siRNAs tested were stable in fetal bovine serum for at least 24 h (data not shown).
Figure 8.
Stability of native and boranophosphate siRNAs. siRNAs were incubated with bovine pancreatic RNases for the times indicated, and then assessed for degradation by agarose gel electrophoresis. Above: Plot of siRNA degradation by RNase over time. Half-lives were calculated based on regression using the data shown. Error bars represent standard error. Below: photograph of siRNA samples after RNase incubation and electrophoresis. A—antisense strand, S—sense strand, b—boranophosphate, n—native, a—adenosine, c—cytidine, u—uridine, 3—adenosine, cytidine and uridine.
DISCUSSION
Structure–function relationships in modified siRNAs
Previous studies of chemically modified siRNAs suggested that, at best, some species might be as effective as native siRNAs. Therefore, we were intrigued to find that several of the boranophosphate-modified siRNAs had greater maximal silencing activity than native RNAs. In fact, we found that the most active siRNA species (Abc Sn) showed a 1.25-fold greater maximal inhibition of EGPF expression than did unmodified siRNAs (at 25 nM). There was an even greater disparity between modified and unmodified siRNA activity at concentrations that yielded less than peak silencing. At 3.1 nM, modified siRNAs were over 1.5 times more effective. We also found that boranophosphate-modified siRNAs are more active than analogous phosphorothioate siRNAs for silencing EGFP gene expression.
While boranophosphate modification of siRNAs is generally well tolerated by the RNAi pathway, three or more modifications within the central five nucleotides of the antisense strand caused a significant reduction in siRNA activity. There are several explanations for this region's particular sensitivity to modification. Because only one strand of RNA remains in the mature RISC, the antisense strand is entirely responsible for silencing activity once the RISC has formed (8). Thus, it seems reasonable that the antisense strand would have more stringent structural requirements than the sense strand, which serves essentially as a carrier for the antisense strand. Also, target mRNA cleavage occurs opposite the link between the 10th and 11th nucleotides of the antisense RNA, when counting from the 5′ end (in the center of a 21 bp siRNA) (36). It is likely that this site of enzymatic activity is especially susceptible to alterations in siRNA structure. This same region has already been shown to be particularly sensitive to sequence mismatches (36). Studies of phosphodiester-backbone siRNAs (36) have shown that sequence changes anywhere 3′ to the cleavage site also drastically reduce activity (36). In contrast, examination of siRNAs with ribose modifications showed that modification of the 5′ end of siRNAs caused the greatest inhibition of activity (38). As we did not observe either pattern with boranophosphate- or phosphorothioate-modified siRNAs, the areas of sensitivity to sequence mismatches and ribose modification appear to be more extensive than the areas sensitive to modification of the phosphodiester bond. Furthermore, while changing the identity of one of the two nucleotides opposite the expected cleavage site (i.e. in the center of the antisense strand) can abolish silencing activity, the presence of a boranophosphate nucleotide in this position does not interfere with activity (Figure 5, Abac Sbac). Therefore, we conclude that the activity of boranophosphate siRNAs is determined by the overall degree of modification in the center of the antisense strand rather than by modification at a specific position.
Because we expected that the antisense strand might be more sensitive to changes in general, we initially anticipated that the most active siRNAs would be those with many modifications on the less critical sense strand and few, if any, modifications on the antisense strand. Instead, siRNAs with peripheral modifications on the antisense strand were at least as active as those with sense strand modifications in terms of maximum efficacy (at high siRNA concentrations) (Figures 4 and 5) and had greater silencing activity at low siRNA concentrations (Figure 6B and C). Thus, we conclude that boranophosphate modification of the antisense strand is advantageous.
Possible mechanisms of enhanced activity in boranophosphate siRNAs
The observed increase in nuclease stability of boranophosphate-modified siRNAs is an attractive explanation for their increased EGFP-silencing activity. Greater stability of modified siRNAs could play a major role in determining the level of silencing when siRNA concentrations are not saturating: any reduction in siRNA levels due to nucleases would be expected to cause a corresponding reduction in activity. Thepoint in the RNAi process at which nuclease resistance is most important is unknown. In theory, modification could confer greater stability in the culture medium, in the cytoplasm, or even once the siRNA is incorporated into the RISC. We were interested to observe that boranophosphate modifications on the antisense strand caused a greater increase in potency than boranophosphate modifications on the sense strand.
While increased potency can be explained by nuclease resistance, stability alone cannot explain why modified siRNAswould have a higher maximum level of EGFP gene silencing activity. If nuclease degradation were the only factor responsible for the difference, one would expect that, at high siRNA doses, unmodified siRNAs would produce the same level of silencing activity achieved by the modified molecules. We did not find that to be the case; even at siRNA concentrations up to 200 nM, unmodified siRNAs had essentially the same level of activity as when the siRNA concentration was 25 nM (data not shown). Similarly, because the maximum efficacy of backbone-modified siRNAs exceeds that of native siRNAs, the difference in activity is unlikely to be due to increased cellular uptake of modified siRNAs. Rather, siRNA backbone modifications appear to make the RNAi process more effective, possibly by making the RISC more stable or more efficient.
How backbone modification could confer increased stability or efficiency to the RISC is unknown. It is interesting to note that boranophosphate oligonucleotides are more hydrophobic (31) than their standard phosphate analogs and that eIF2C2, one of the major RISC proteins, is believed to be associated with intracellular membranes (8,39). The increased lipophilicity of boranophosphate nucleotides could influence these RNA–protein interactions. In addition, phosphorothioate nucleotides, which have electrostatic properties similar to those of boranophosphates, have a greater affinity for serum proteins than unmodified siRNAs (16). It should also be noted that the RISC is a multiple-turnover enzyme complex (40). Since the mechanism of its activity is not yet clear, it is not known which step among target binding, target cleavage and release of cleaved RNA fragments is rate-limiting. For efficient catalysis, the product off-rate is of fundamental importance. Boranophosphate modification is known to decrease the hybridization intensity between two strands of oligonucleotides (31), which may facilitate the release of cleaved RNA products from and the turnover of the RISC/siRNA complex.
Therapeutic potential of boranophosphate siRNAs
Several properties of boranophosphate siRNAs make them attractive candidates for use in a clinical setting. Their high maximum activity and potency indicate that relatively low and infrequent doses could be used. The high activity at low concentrations is particularly important because in vivo delivery systems are unlikely to be as efficient as those used with cultured cells.
Previous work has suggested that phosphorothioate-modified siRNAs are cytotoxic, particularly when >50% of nucleotides are modified (19,22). In our system, siRNAs with up to 76% boranophosphate-modified nucleotides showed no evidence of inducing cell death (as determined by exclusion of trypan blue, data not shown). We also failed to observe toxicity with the phosphorothioate siRNAs; however, we did not test highly modified phosphorothioate siRNAs. The absence of toxicity in boranophosphate siRNAs in cell culture is encouraging in the context of possible clinical applications.
SUMMARY
We have found that boranophosphate-backbone siRNAs are highly active and potent for interference with EGFP expression in HeLa cells. Boranophosphate-modified siRNAs are more effective than their phosphorothioate counterparts and often more active than native siRNAs. siRNA backbone modifications appear to make the RNAi process more effective, perhaps by stabilizing RISC formation or enhancing the RISC's enzymatic activity.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Zinaida Sergueeva for synthesis of ribonucleoside 5′-(α-P-borano)triphosphates, and George R. Dubay for his help in MS experiments. We also wish to acknowledge Danuta Gasinski and the Duke University Cancer Center Cell Culture and Flow Cytometry facilities for excellent technical assistance. A.H.S.H. was supported by the Duke University Medical Scientist Training Program. E.E.S. was supported by an HHMI Research Training Fellowship for Medical Students. J.W. and B.R.S. were supported by R01 GM57693 to B.R.S. from the National Institutes of Health and Department of Defense Grant DAMD 17-02-1-0376, sponsored by the Department of the Army. The content of the information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred. K.A.A. was partially supported by a grant from The Children's Miracle Network and R01 CA081214 from the National Institutes of Health.
REFERENCES
- 1.Denli A.M. and Hannon,G.J. (2003) RNAi: an ever-growing puzzle. Trends Biochem. Sci., 28, 196–201. [DOI] [PubMed] [Google Scholar]
- 2.McManus M.T. and Sharp,P.A. (2002) Gene silencing in mammals by small interfering RNAs. Nature Rev. Genet., 3, 737–747. [DOI] [PubMed] [Google Scholar]
- 3.Tuschl T. (2002) Expanding small RNA interference. Nat. Biotechnol., 20, 446–448. [DOI] [PubMed] [Google Scholar]
- 4.Hutvagner G. and Zamore,P.D. (2002) RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev., 12, 225–232. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- 7.Hammond S.M., Boettcher,S., Caudy,A.A., Kobayashi,R. and Hannon,G.J. (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- 8.Martinez J., Patkaniowska,A., Urlaub,H., Luhrmann,R. and Tuschl,T. (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell., 110, 563–574. [DOI] [PubMed] [Google Scholar]
- 9.Lieberman J., Song,E., Lee,S.K. and Shankar,P. (2003) Interfering with disease: opportunities and roadblocks to harnessing RNA interference. Trends Mol. Med., 9, 397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coburn G.A. and Cullen,B.R. (2002) Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol., 76, 9225–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randall G., Grakoui,A. and Rice,C.M. (2003) Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl Acad. Sci. USA, 100, 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall A.H. and Alexander,K.A. (2003) RNA interference of human papillomavirus type 18 E6 and E7 induces senescence in HeLa cells. J. Virol., 77, 6066–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilda M., Fuchs,U., Wossmann,W. and Borkhardt,A. (2002) Killing of leukemic cells with a BCR/ABL fusion gene by RNA interference (RNAi). Oncogene, 21, 5716–5724. [DOI] [PubMed] [Google Scholar]
- 14.Sorensen D.R., Leirdal,M. and Sioud,M. (2003) Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J. Mol. Biol., 327, 761–766. [DOI] [PubMed] [Google Scholar]
- 15.Eckstein F. (1985) Nucleoside phosphorothioates. Annu. Rev. Biochem., 54, 367–402. [DOI] [PubMed] [Google Scholar]
- 16.Eckstein F. (2002) Developments in RNA chemistry, a personal view. Biochimie, 84, 841–848. [DOI] [PubMed] [Google Scholar]
- 17.Boiziau C. Dausse,E., Mishra,R., Duconge,F. and Toulme,J.J. (1997) Identification of aptamers against the DNA template for in vitro transcription of the HIV-1 TAR element. Antisense Nucleic Acid Drug Dev., 7, 369–380. [DOI] [PubMed] [Google Scholar]
- 18.Caruthers M.H. (1997) Summary. In Chadwick,D.J. and Cardew,G. (ed.) Oligonucleotides as Therapeutic Agents. Ciba Foundation Symposium 209, John Wiley, NY, pp. 235–239. [PubMed] [Google Scholar]
- 19.Amarzguioui M., Holen,T., Babaie,E. and Prydz,H. (2003) Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res., 31, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czauderna F., Fechtner,M., Dames,S., Aygun,H., Klippel,A., Pronk,G.J., Giese,K. and Kaufmann,J. (2003) Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res., 31, 2705–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braasch D.A., Jensen,S., Liu,Y., Kaur,K., Arar,K., White,M.A. and Corey,D.R. (2003) RNA interference in mammalian cells by chemically-modified RNA. Biochemistry, 42, 7967–7975. [DOI] [PubMed] [Google Scholar]
- 22.Harborth J., Elbashir,S.M., Vandenburgh,K., Manninga,H., Scaringe,S.A., Weber,K. and Tuschl,T. (2003) Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev., 13, 83–105. [DOI] [PubMed] [Google Scholar]
- 23.Parrish S., Fleenor,J., Xu,S., Mello,C. and Fire,A. (2000) Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol. Cell., 6, 1077–1087. [DOI] [PubMed] [Google Scholar]
- 24.Sanghvi Y.S., Anrade,M., Deshmukh,R.R., Holmberg,L., Scozzari,A.N. and Cole,D.L. (1999) Chemical synthesis and purification of phosphorothioate antisense oligonucleotides. In: Hartmann,G. and Endres,S. (eds) Manual of Antisense Methodology. Kluwer Academic Publisher, Boston, MA, pp. 3–24. [Google Scholar]
- 25.Porter K.W., Briley,J.D. and Ramsay Shaw,B. (1997) Direct PCR sequencing with boronated nucleotides. Nucleic Acids Res., 25, 1611–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He K. and Ramsay Shaw,B. (1999) Diastereomers of 5′-O-adenosyl 3′-O-uridyl Boranophosphate [UpBH3)A]: synthesis and nuclease resistant property. Symposium on RNA Biology III: RNA, Tool and Target in Nucleic Acids Symposium Series No. 41, Oxford University Press, Oxford, UK, pp. 99–100. [Google Scholar]
- 27.Ramsay Shaw B., Dobrikov,M., Wang,X., Wan,J., He,K., Lin,J., Li,P., Rait,V., Sergueeva,Z.A. and Sergueev,D. (2003) Reading, writing, and modulating genetic information with boranophosphate mimics of nucleotides, DNA, and RNA. Ann. N. Y. Acad. Sci., 1002, 12–39. [DOI] [PubMed] [Google Scholar]
- 28.Lato S.M., Ozerova,N.D., He,K., Sergueeva,Z., Shaw,B.R. and Burke,D.H. (2002) Boron-containing aptamers to ATP. Nucleic Acids Res. 30, 1401–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He K. (2000). Synthesis and properties of boranophosphate nucleic acids. PhD Dissertation. Duke University, Durham, NC. [Google Scholar]
- 30.Summers J.S. and Shaw,B.R. (2001) Boranophosphates as mimics of natural phosphodiesters in DNA. Curr. Med. Chem. 8, 1147–1155. [DOI] [PubMed] [Google Scholar]
- 31.Ramsay Shaw B., Sergueev,D.S., He,K., Porter,K., Summers,J., Sergueeva,Z. and Rait,V. (1999) Boranophosphate backbone: a mimic of phosphodiesters, phosphorothioates, and methylphosphonates. Meth. Enzymol., 313, 226–257. [DOI] [PubMed] [Google Scholar]
- 32.Hall I.H., Burnham,B.S., Rajendran,K.G., Chen,S.Y., Sood,A., Spielvogel,B.F. and Ramsay Shaw,B. (1993) Hypolipidemic activity of boronated nucleosides and nucleotides in rodents. Biomed. Pharmacother., 47, 79–87. [DOI] [PubMed] [Google Scholar]
- 33.Li H., Hardin,C. and Ramsay Shaw,B. (1996) Hydrolysis of thymidine boranomonophosphate and stepwise deuterium substitution of the borane hydrogens. 31P and 11B NMR studies. J. Am. Chem. Soc., 118, 6606–6614. [Google Scholar]
- 34.He K., Hasan,A., Krzyzanowska,B. and Ramsay Shaw,B. (1998) Synthesis and separation of diastereomers of ribonucleoside 5′-(alpha-P-Borano)triphosphates. J. Org. Chem., 63, 5769–5773. [DOI] [PubMed] [Google Scholar]
- 35.Donze O. and Picard,D. (2002) RNA interference in mammalian cells using siRNAs synthesized with T7 RNA polymerase. Nucleic Acids Res., 30, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elbashir S.M., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J., 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doench J.G., Petersen,C.P. and Sharp,P.A. (2003) siRNAs can function as miRNAs. Genes Dev., 17, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiu Y.L. and Rana,T.M. (2003) siRNA function in RNAi: a chemical modification analysis. RNA, 9,1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cikaluk D.E., Tahbaz,N., Hendricks,L.C., DiMattia,G.E., Hansen,D., Pilgrim,D. and Hobman,T.C. (1999) GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol. Biol. Cell., 10, 3357–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hutvagner G. and Zamore,P.D. (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297, 2056–2060. [DOI] [PubMed] [Google Scholar]