Abstract
Background:
The PIWI-interacting RNA (piRNA) pathway has an essential role in transposon silencing, meiosis progression, spermatogenesis, and germline maintenance. HIWI genes are critical for piRNA biogenesis and function. Therefore, polymorphisms in HIWI genes contribute to spermatogenesis defects and can be considered as risk factors for male infertility. The aim of the present study was to investigate the association between the HIWI2 gene rs508485 polymorphism and non-obstructive azoospermia.
Methods:
A total of 121 Iranian men with idiopathic azoospermia and 100 fertile controls were genotyped for HIWI2 rs508485 (T>C) polymorphism using Tetra-ARMS PCR. The presence of eight sequence-tagged site (STS) markers from the Y chromosome AZF region was also investigated by Multiplex PCR (M-PCR).
Results:
Thirteen (10.74%) patients showed Y chromosome microdeletions and therefore were excluded from the study. rs508485 in the 3’UTR of HIWI2 was associated with increased risk of azoospermia in our studied population with a P-value of 0.035 and odds ratio of 2.00 (CI 95%: 1.04-3.86).
Conclusions:
We provide evidence for an association between genetic variation in the HIWI2 gene involved in the piRNA pathway and idiopathic non-obstructive azoospermia in Iranian patients. Therefore, piRNA pathway gene variants can be considered as risk factors for male infertility.
Key Words: HIWI2, Non-obstructive azoospermia, PiRNA, Polymorphism
Introduction
Infertility affects 10-15% of couples worldwide, and in almost half of the cases men are responsible (1). Different types of male infertility include aspermia, azoospermia, oligozoospermia, asthenozoospermia, and teratozoospermia. Among these, azoospermia is the most common reason for male infertility by prevalence of 10-15% in infertile men (2). Studies have mentioned diverse reasons for azoospermia but in many cases the main cause is still unknown (3).
The PIWI-interacting RNA (piRNA) pathway, a newly-detected pathway essential for spermatogenesis, is expressed abundantly and exclusively in germline cells. This special class of non-coding RNAs binds to a subtype of Argonaute proteins and forms a retrotransposon-silencing complex in the germline (4, 5). Many studies showed that piRNAs are crucial for differentiation and specificity of the male germline. These small RNAs can repress transposons by degradation, histone modification, and DNA methylation (6, 7). Although piRNAs are expressed in both testis and ovaries, only male knockout mice for piRNA-associated genes become sterile, and this infertility phenotype is accompanied with overexpression of transposons (8, 9).
PIWIs, the most important proteins in this pathway, are critical for both piRNA biogenesis and function. This subtype of the Argonaute family includes HIWI, HIWI2, HIWI3, and HILI genes in humans. It has been shown that silencing of HIWI gene orthologues in mice results in meiotic arrest and sterility in males (10-13). In 2010, Gu et al. studied nine HIWI polymorphisms in a Chinese population and observed that two SNPs, rs508485 (HIWI2) and rs11703684 (HIWI3), were significantly associated with risk of oligozoospermia (14). Considering the functional and physiological importance of HIWI genes, in this study we aimed to analyze the association between the rs508485 (T>C) polymorphism and risk of non-obstructive azoospermia in Iranian infertile men.
Materials and Methods
In this case-control study 221 subjects, including 121 non-obstructive azoospermic patients and 100 proven fertile men, were considered. All patients had normal karyotypes and were aged between 21 and 60 (mean ± SD = 32.41 ± 6.43 yr). The patients were referred from the Yazd Infertility Research and Treatment Center, Kowsar Infertility Clinical Center and IVF Department of Day hospital. Semen from both cases and controls were analyzed according to WHO protocol and urological examinations were performed on all the patients for anatomical integrity of the genital system (15). Patients with anatomic disorders of genitalia, testis neoplasms, chromosomal numerical and structural abnormalities, or Y chromosome microdeletions were excluded. Ethical consent was given by all the participants. The study protocol was approved by the ethics committee of Shahid Beheshti University of Medical Sciences (SBMU). Genomic DNA was extracted from blood samples using an M&D extraction kit (Shahid Beheshti University of Medical Sciences, Iran).
To exclude the role of Y chromosome microdeletions in male infertility, a series of eight sequence-tagged site markers (STS), located on Yq11, were selected for detection of submicroscopic deletions in the AZFa, AZFb, and AZFc regions using M-PCR (16). We used the tetra primer-amplification refractory mutation system-PCR (4P-ARMS-PCR) method, which applies two pairs of primers, to analyze HWI2 rs508485 (T>C) genotypes. Primers were designed by Primer1 online software (http://primer1.soton.ac.uk/primer1.html). PCR primers were F-outer: 5' AAAAGATTGAGCTTAGTTTTCATGTCTA3', R-outer: 5' CACATGATGTTCTGAACTTTATTTTCA 3', F-inner for C allele: 5' ATAAGTGTTTGCGTGATATTTTGATTAC 3' and R-inner for T allele: 5' GTGGTGGGAATTAGACTCTGTTTATATA 3'. Each PCR contained 100 ng of DNA, 10 µl of Taq DNA Polymerase 2X Master Mix Red (Amplicon, Denmark), 10 pmol/µl of FO508, 10 pmol/µl of RO508, 5 pmol/µl of FI508, and 5 pmol/µl of RI508 in a final volume of 25 µl. Amplification was carried out on a GeneTouch (BIOER, China) with the following program: 95° C for 5' for primary denaturation, three steps of 95° C for 30", 50° C for 45", and 72° C for 45" for 32 cycles, and a final extension at 72° C for 5'. PCR products were subjected to electrophoresis on 2% agarose gels prepared in 0.5X TBE, and stained with RedSafe (iNtRON, Korea). A common 338 bp band was amplified by the outer primers in all PCRs. The T and C alleles generated 253 bp and 141 bp PCR products, respectively (Fig. 1). 10% of the samples were sequenced on an ABI 3730x1 DNA analyzer (Macrogen, Korea) to confirm the accuracy of the genotypes.
Fig. 1.
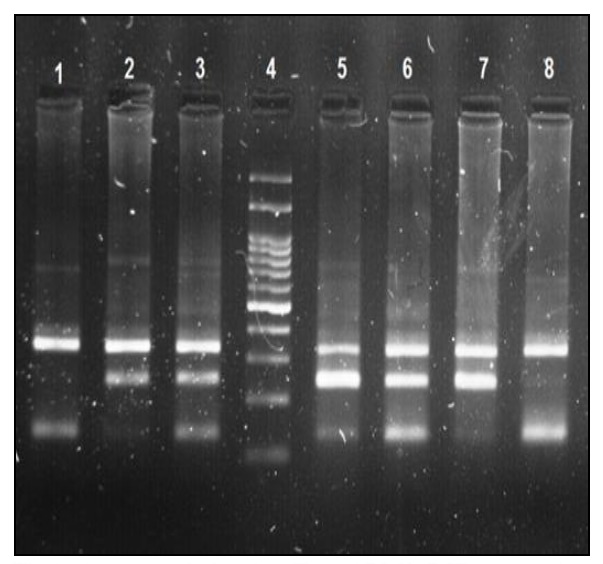
Agarose gel showing Tetra-ARMS PCR genotyping results for rs508485. Lanes 1 and 8: CC, lanes 2 and 7: TT, lanes 3, 5, and 6: TC. Lane 4: 100 bp ladder.
Statistical analyses for genotype and allele frequencies, Hardy-Weinberg equilibrium, and azoospermia association were performed by the χ2 test using MedCalc online software available from www.medcalc.org/calc/odds_ratio.php. P-Values < 0.05 were considered to be statistically significant.
Results
Thirteen (10.74%) patients showed Y chromosome microdeletions and therefore were excluded from the association study. None of the STS markers were deleted in the controls. The genotype distribution in the studied groups was all in Hardy-Weinberg equilibrium. The allele and genotype frequencies of HIWI2 rs508485 for the cases and controls are summarized in Table 1. Our study showed that the C allele for rs508485 increases the risk of azoospermia in a dominant model (CC + CT genotypes) with a P-value of 0.035 and odds ratio of 2.00 (95% CI: 1.04- 3.86).
Table 1.
Allele and genotype frequencies of rs508485 (T>C) and association with azoospermia risk.
| Genotype | Cases N=108 (%) | Controls N=100 (%) | OR (95% CI) | P Value |
|---|---|---|---|---|
| HIWI2 rs508485 (T>C) | ||||
| T/T | 19 (18%) | 30 (30%) | 1 (reference) | |
| T/C | 65 (60%) | 49 (49%) | 1.57 (0.9-2.7) | 0.10 |
| C/C | 24 (22%) | 21 (21%) | 1.07 (0.55-2.08) | 0.83 |
| Dominant C/C+C/T vs. T/T | 2.00 (1.04 -3.86) | 0.035 | ||
| Recessive C/C vs. C/T+T/T | 1.07 (0.55-2.08) | 0.83 | ||
| Allele | ||||
| T | 103 (48%) | 109 (55%) | 0.76 (0.51-1.11) | 0.16 |
| C | 113 (52%) | 91 (45%) | 1.31 (0.89-1.93) | 0.16 |
Discussion
Spermatogenesis is a complicated process with the cooperation of thousands of genes and proteins (17). Although the prevalence of male infertility is currently increasing, only a few genes are known as main reasons for this condition in humans. Gene knockout studies and genome-wide association studies (GWAS) are useful to find new genes involved in male infertility (18, 19). Proteins involved in the piRNA pathway including PIWIs and TDRDs are essential for spermatogenesis and recently considered as new candidates for male infertility (20).
We analyzed the association of the rs508485 (T>C) polymorphism in HIWI2 with risk of azoospermia in Iranian men. Single nucleotide variations could hypothetically influence gene expression and/or protein structure by altering cis-acting elements, RNA transcript stability, or RNA splicing. The studied SNP locates in the 3’UTR of HIWI2, and due to its position, may affect mRNA stability or alter the binding affinity of regulatory miRNAs. According to HaploReg v4, it was suggested that the mentioned polymorphism may also influence the binding affinities of several transcription factors including CEBPG, Fox, Foxd1, Hoxa9, and GATA (21). Despite a previous study, which reported that rs508485 was associated only with oligozoospermia (P-Value: 0.04, OR: 1.49, and 95% CI: 1.02-2.18) in a Chinese population (14), an association was observed between this SNP and increased risk of azoospermia in an Iranian population (P-Value: 0.035, OR: 2.00, and 95% CI: 1.04 - 3.86). Several factors including ethnicity, sample size, and the male infertility type may explain the observed discrepancies between studies.
Overall, this study showed that genetic variants in piRNA pathway genes may predispose to spermatogenesis defects. To better understand the relationship between the piRNA pathway and male infertility, studying a larger sample, as well as other genes of this pathway in other types of male infertility, should be considered.
Acknowledgments
The authors are grateful to Yazd Infertility Research and Treatment Center, Kowsar Infertility Clinical Center and IVF Department of Day hospital for their kind collaboration. Authors declare there are no conflicts of interest.
References
- 1.Massart A, Lissens W, Tournaye H, Stouffs K. Genetic causes of spermatogenic failure. Asian J Androl. 2012;14(1):40–8. doi: 10.1038/aja.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cocuzza M, Alvarenga C, Pagani R. The epidemiology and etiology of azoospermia. Clinics. 2013;68:15–26. doi: 10.6061/clinics/2013(Sup01)03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boettger-Tong H. Genes Causing Azoospermia and Oligozoospermia. Glob Libr Women's Med. 2008 [Google Scholar]
- 4.Hartig JV, Tomari Y, Förstemann K. piRNAs—the ancient hunters of genome invaders. Gene Dev. 2007;21(14):1707–13. doi: 10.1101/gad.1567007. [DOI] [PubMed] [Google Scholar]
- 5.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135(1):3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 6.Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annual Rev Genet. 2011;45 doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moshkovich N, Lei EP. HP1 recruitment in the absence of argonaute proteins in Drosophila. Plos Genet. 2010;6(3):e1000880-e. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Z, Kokkinaki M, Pant D, Gallicano GI, Dym M. Small RNA molecules in the regulation of spermatogenesis. Reproduction. 2009;137(6):901–11. doi: 10.1530/REP-08-0494. [DOI] [PubMed] [Google Scholar]
- 9.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761–4. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 10.Thomson T, Lin H. The biogenesis and function PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng W, Lin H. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2(6):819–30. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 12.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131(4):839–49. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 13.Carmel MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12(4):503–14. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Gu A, Ji G, Shi X, Long Y, Xia Y, Song L, et al. Genetic variants in Piwi-interacting RNA pathway genes confer susceptibility to spermatogenic failure in a Chinese population. Hum Reprod. 2010 doi: 10.1093/humrep/deq274. deq274. [DOI] [PubMed] [Google Scholar]
- 15.WHO laboratory manual for the examination and processing of human semen. 5. Geneva, Switzerland: WHO; 2012. World Health Organisation. [Google Scholar]
- 16.Mirfakhraie R, Mirzajani F, Kalantar SM, Montazeri M, Salsabili N, Pourmand GR, et al. High prevalence of AZFb microdeletion in Iranian patients with idiopathic non-obstructive azoospermia. Indian J Med Res. 2010;132(3):265–70. [PubMed] [Google Scholar]
- 17.Krausz C, Giachini C. Genetic risk factors in male infertility. . Arch Androl. 2007;53(3):125–33. doi: 10.1080/01485010701271786. [DOI] [PubMed] [Google Scholar]
- 18.Yan W. Male infertility caused by spermiogenic defects: lessons from gene knockouts. Mol Cell Endocrinol. 2009;306(1):24–32. doi: 10.1016/j.mce.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosova G, Scott NM, Niederberger C, Prins GS, Ober C. Genome-wide association study identifies candidate genes for male fertility traits in humans. Am J Hum Genet. 2012;90(6):950–61. doi: 10.1016/j.ajhg.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Gene & Dev. 2009;23(15):1749–62. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr917. (D1):D930-D934. [DOI] [PMC free article] [PubMed] [Google Scholar]


