Abstract
Site-specific modification of nucleosomal histones plays a central role in the formation of transcriptionally active and inactive chromatin structures. These modifications may serve as specific recognition motifs for chromatin proteins, which act as a signal for the adoption of the appropriate regulatory responses. Here, we show that the orphan nuclear receptor SHP (small heterodimer partner), a coregulator that inhibits the activity of several nuclear receptors, can associate with unmodified and lysine 9-methylated histone-3, but not with the acetylated protein. The naturally occurring SHP mutant (R213C), which exhibits decreased transrepression potential, interacts less avidly with K9-methylated histone 3. We demonstrate that SHP can functionally interact with histone deacetylase-1 and the G9a methyltransferase and that it is localized exclusively in nuclease-sensitive euchromatin. The results point to the involvement of a multistep mechanism in SHP-dependent transcriptional repression, which includes histone deacetylation, followed by H3-K9 methylation and stable association of SHP itself with chromatin.
INTRODUCTION
Post-translational modification of histones is a key regulatory signal in eukaryotic gene expression (1–3). Current evidence suggests that H3 lysine 9 (H3-K9) methylation is a mark for transcriptionally silent chromatin (4–6). In mammalian cells, K9-methylated H3 produced by Suv39h1 interacts with the chromodomain of HP1 proteins to generate a stable repressive structure, which could be propagated to the neighboring nucleosomes via the recruitment of additional Suv39h1–HP1 protein complexes (7,8). This mechanism has been shown to play a crucial role in the formation of pericentric heterochromatin (9). However, the broad methylation pattern of H3-K9 in the euchromatic regions of the nucleus suggests that the function of this modification is not restricted to heterochromatin silencing (9–11).
G9a is a newly identified H3-K9 methyltransferase, which is localized exclusively in euchromatic regions (12). The dominant role of this enzyme in euchromatic histone methylation is substantiated by the observation that the broad euchromatic H3-K9 methylation pattern was largely abolished in G9a-deficient cells (11). To further explore the role of G9a and H3-K9 methylation mark in euchromatic silencing, it is important to identify target genes regulated by the enzyme, the factors responsible for its recruitment and to decipher the mechanism by which the created histone modification is ‘translated’ into a repressive function.
In this report, we show that SHP (small heterodimer partner, NROB2) is a potential factor that may target G9a to promoters, as it can physically and functionally interact with G9a both in vivo and in vitro. SHP is an atypical orphan nuclear receptor, which contains an N-terminal receptor dimerization domain, but lacks a DNA-binding domain (13). SHP, which functions as a specific repressor of transcription, is a key regulator of genes involved in cholesterol–bile acid homeostasis (14,15). Transcriptional regulation of the genes coding for cholesterol-catabolizing enzymes, such as the Cyp7A1 and Cyp8B genes, involves a negative feedback loop induced by bile acids. Bile acids are ligands for the nuclear receptor FXR, which activates the expression of SHP. SHP is then recruited to the promoters of Cyp7A1 and Cyp8B genes by CPF (also known as LRH), or HNF-4, two orphan nuclear receptors implicated in the activation of the above genes (14,15). In humans, mutations in SHP gene associate with insulin resistance and mild obesity (16). SHP inhibits the activity of several hormone receptors by a mechanism that is believed to involve competition with coactivators for the common AF2-binding surface of nuclear receptors and a less well-understood direct repression mechanism mediated by its C-terminal autonomous repressor domain (17–21). Here, we present supporting evidence for the role of histone deacetylase-1 (HDAC-1) and G9a in the latter mechanism. Our results suggest that SHP may not only control the recruitment of G9a to the promoters but can also associate with underacetylated and/or K9-methylated histone 3, which may also play a role in its repressive function.
MATERIALS AND METHODS
Plasmid constructions
The full-length SHP cDNA fragment was isolated by RT–PCR from total HepG2 RNA, and the open reading frame was subcloned into pGEX-2TK (APB Biotech) and pMT3-HA vectors. Deletion mutant derivatives were constructed by standard methods and verified by DNA sequencing. pBXG1-HNF-4 FL and pCMV-myc-HDAC were described previously (22,23). pCMV-T7-G9a containing the long form of G9a cDNA (Ng36/G9a) fused to the T7-tag was a gift from C. M. Sanderson (24). SHP-R213C and G9a-R1109H mutants were generated by the GeneEditor site-directed mutagenesis kit (Promega).
Protein–protein interactions
In vitro glutathione S-transferase (GST) pull-down experiments were performed either by using in vitro translated proteins, or nuclear extracts from HepG2 cells prepared as described previously (23). 35S-labeled recombinant proteins were synthesized in vitro using the TNT coupled reticulocyte lysate system (Promega). An aliquot of 2 μg of GST-fusion proteins coupled to glutathione–Sepharose column (Pharmacia) were incubated with the in vitro translated proteins in a buffer containing 20 mM HEPES, pH 7.9, 200 mM NaCl, 5 mM MgCl2, 0.1% NP-40, 0.2% BSA, 10% glycerol, 1 mM PMSF and 10 μg/ml aprotinin at 4°C with constant agitation. In salt titration experiments, the buffer composition was the same except for the concentration of NaCl, which ranged between 150 and 500 mM. After excessive washing with the same buffer lacking BSA, the beads were resuspended in SDS sample buffer, and the proteins were separated by electrophoresis in SDS–polyacrylamide gels and visualized by autoradiography or western blot analysis. For peptide pull-down assays, biotinylated histone 3 peptides (1–21 amino acids) (Upstate Biotechnology) were immobilized in streptavidin–agarose and incubated with in vitro translated 35S-labeled SHP as above.
For far-western analysis, core histones were purified from HeLa cells by acid extraction and separated in either 12% SDS–polyacrylamide gels as described previously (25). After electrotransfer, the membranes were blocked and hybridized with 500 000 c.p.m./ml 32P-GST-SHP probe in a buffer containing phosphate-buffered saline (PBS), 0.3% BSA, 1% fetal calf serum (FCS), supplemented with protease inhibitor cocktail (Roche). After extensive washings with the same buffer, interacting proteins were visualized by autoradiography.
Co-immunoprecipitation and western blot assays were performed as described previously (23). The following antibodies were used in this study: the polyclonal antibody for SHP was raised in New Zealand white rabbits against a KLH-conjugated peptide corresponding to the 234–248 amino acid region of human SHP. The αHNF-4 antibody has been described previously (22). α-acH3, α-K9 dimethyl-H3 and αG9a were from Upstate Biotechnology; αHDAC, α-K9 mono and trimethyl-H3 were from Abcam; αHP1 was from Euromedex; and αCBP, αHA-tag and α-myc-tag antibodies were from Santa Cruz Biotechnology.
Cell culture, transfections and chromatin fractionation
Caco-2, HepG2, HeLa and Cos-1 cells were grown in Dulbecco's Modified Eagle Medium supplemented with 10 or 20% FCS. Caco-2 cells were used in chromatin fractionation and immunofluorescence experiments, because they constitutively express high levels of SHP. HepG2 cells were used in experiments where endogenous factors were analyzed, while HeLa cells were used to obtain highly purified histones. In order to avoid complications with endogenous factors, all transfection and reporter assays were performed with Cos-1 cells. RT–PCR analysis, transfections and luciferase reporter assays were performed as described previously (26,27). Intact nuclei were prepared as described previously (28). Briefly, the cells were lysed in a buffer containing 0.32 M sucrose, 15 mM HEPES, pH 7.9, 60 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5% BSA, 0.5 mM spermidine, 0.15 mM spermine and protease inhibitor cocktail (Roche). After Dounce homogenization, the nuclei were layered over 5 ml of a 30% sucrose-containing buffer with the same composition, but without BSA. After centrifugation at 3000 r.p.m. for 15 min, the nuclei were recovered in a buffer containing 15 mM HEPES, pH 7.5, 60 mM KCl, 15 mM NaCl, 0.34 mM sucrose, 0.15 mM mercaptoethanol. Chromatin fractionation was performed as described previously (29,30). Briefly, after the addition of CaCl2 to a final concentration of 3 mM, the nuclei were digested with 2, 5 and 10 U of micrococcal nuclease (MNase) for 2 min at 37°C. Digested nuclei were cooled on ice and centrifuged at 12 800 g for 5 min. The supernatant (S1 fraction) was removed and the pellet was resuspended in 2 mM EDTA. After a 10 min incubation on ice, the samples were centrifuged as above and the supernatant (S2 fraction) was saved. The pellet (P) was resuspended in 2 mM EDTA. Equal amounts of samples, in terms of the initial number of nuclei, were then analyzed either by western blot assays for SHP and HP1 protein detection, or by agarose gel electrophoresis for the estimation of DNA fragmentation after deproteinization.
Confocal microscopy
The cells were seeded on glass coverslips and fixed with methanol. After blocking with 1% BSA in PBS for 30 min, the cells were stained with the polyclonal rabbit antibody against SHP, followed by the secondary antibodies anti-rabbit Alexa Fluor 568 (Molecular Probes). The coverslips were then counterstained with DAPI, mounted into glass slides with Mowiol (Sigma) and observed in a Leica SP confocal microscope.
RESULTS
Functional interactions of SHP with HDAC-1 and G9a methyltransferase
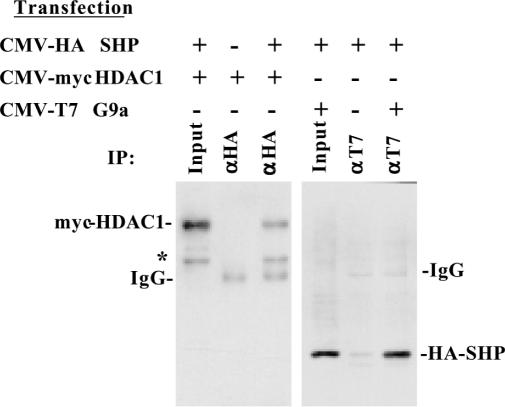
In an effort to decipher the potential role of chromatin modifying factors in the direct repression mechanism, in vivo protein–protein interactions between SHP and HDAC-1 or G9a were investigated by co-immunoprecipitation assays. Cos-1 cells were cotransfected with CMV-HA-SHP and CMV-Myc-HDAC-1 or CMV-T7-G9a expression vectors. Lysates from the transfected cells were immunoprecipitated with either αHA or αT7 antibodies, and the presence of HDAC-1 or SHP in the immunoprecipitates was assayed by western blots using αMyc or αHA antibody, respectively. The results shown in Figure 1 indicate that both HDAC-1 and G9a can specifically interact with SHP in vivo.
Figure 1.
Physical interaction of SHP with HDAC-1 and G9a in vivo. Nuclear extracts of Cos-1 cells transfected with the indicated expression vectors were immunoprecipitated with either αHA or αT7 antibodies. The immunoprecipitated materials were then analyzed in western blots with α-Myc antibody to detect myc-tagged HDAC-1 (left panel), or with αHA antibody to detect HA-tagged SHP proteins (right panel). The asterisk at left depicts an unidentified band, which probably corresponds to HDAC-1 degradation product.
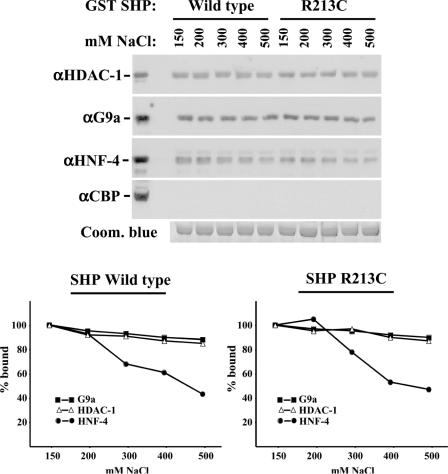
To exclude the possibility of potential artifacts generated by overexpressing the proteins, we performed interaction assays using untransfected HepG2 nuclear extracts as a source of endogenous G9a and HDAC-1 proteins and GST-SHP fusion proteins immobilized on glutathione–Sepharose beads. In order to gain insight into the binding affinities, we performed titration experiments by increasing the salt concentration of the interaction buffer. As expected, no interaction could be observed with CBP/p300, which was used as a negative control (Figure 2). As a positive control, we examined SHP-HNF-4 interaction, where we observed a standard binding curve, with ∼40% of the bound protein observed at 150 mM NaCl concentration, surviving high stringency (500 mM NaCl) conditions. At NaCl concentrations higher than 500 mM, part of the GST fusion proteins dissociated from the beads and thus were non-informative (data not shown). When the interactions between SHP and G9a, or HDAC-1 were examined, we could detect a much higher affinity of binding (80–90% of bound protein survived the high stringency conditions) (Figure 2). We also examined the interactions of HDAC-1 and G9a with a naturally occurring mutant (R213C) of SHP, which has previously been shown to exhibit reduced repression potential (21). Surprisingly, the interaction of SHP-R213C with both G9a and HDAC-1 was indistinguishable from that of wild-type SHP (Figure 2).
Figure 2.
SHP interacts with HDAC-1 and G9a with high affinity. GST pull-down experiments were performed with bacterially expressed GST-SHP fusion proteins and HepG2 nuclear extracts. The binding and wash buffers contained the indicated NaCl concentrations. Equal GST-fusion protein absorption to the beads was verified by Coomassie blue staining of a gel from a parallel experiment (bottom panel). The interacting proteins were identified by western blot analysis using the indicated antibodies. Serial exposures of enhanced chemiluminescence images were quantitated by the NIH-Image 1.63 software or by a Fujifilm LAS-1000 Luminescent Image analyzer and plotted as a percentage of the values obtained using buffers containing 150 mM NaCl.
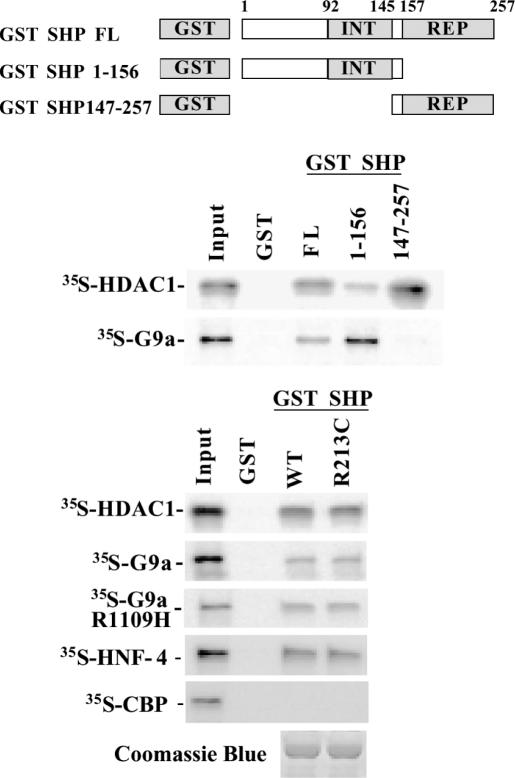
In order to determine whether these interactions are direct and not mediated by other intermediary factors, in vitro pull-down experiments were performed. Both HDAC-1 and G9a efficiently interacted in vitro with the full-length protein, but with preference for distinct SHP domains (Figure 3). SHP-G9a association was observed mainly with the receptor interaction domain (1–156 amino acids) of SHP, while HDAC-1 associated with the active repression domain (147–257 amino acids) of the protein (Figure 3). Furthermore, the in vitro translated wild-type G9a, methylase activity-deficient mutant form of G9a (G9aR1109H) and HDAC-1 proteins interacted with the SHP-R213C mutant as efficiently as with the wild-type SHP protein (Figure 3). To validate the results of the above direct interactions, we performed control experiments analyzing the interaction between SHP and HNF-4 (positive control), or CBP/p300 (negative control). As shown in the bottom panel of Figure 3, only HNF-4 binding could be observed.
Figure 3.
Direct interaction of SHP with HDAC-1 and G9a in vitro. In vitro GST pull-down experiments were performed with 35S-labeled in vitro translated HDAC-1 or with G9a and bacterially expressed GST-SHP fusion proteins containing the indicated regions or the R213C mutation of the protein. INT depicts the receptor interaction domain of SHP (92–145 amino acids), while REP corresponds to the autonomous repression domain of SHP (157–257 amino acids). Equal coupling of wild-type and the R213C mutant GST-SHP proteins were verified by Coomassie blue staining.
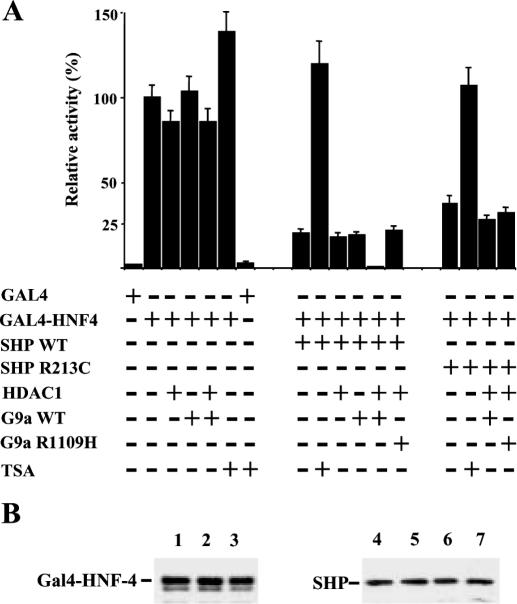
In order to examine the functional interplay of HDAC-1 and G9a on SHP-mediated repression, we coexpressed SHP together with HDAC-1 and G9a in Cos-1 cells and evaluated their effects on Gal-4-HNF-4 mediated transcription. HNF-4, a factor playing crucial roles in hepatocyte and pancreatic β-cell function (22,31–33), is a known target for SHP (20). As expected, overexpression of SHP inhibited Gal4-HNF-4 activity by a factor of ∼5-fold, while the inhibitory effect of the R213C mutant of SHP was compromised (∼3-fold) (Figure 4). Treatment of the cells with the HDAC inhibitor trichostatin A (TSA) had a small stimulatory effect (∼1.3-fold) on Gal4-HNF-4-driven transcription. TSA treatment, however, completely reversed wtSHP and R213C mutant SHP-mediated repression. Because the basal promoter activity observed with Gal-4 control plasmid was only marginally affected by TSA treatment (∼20%), the results suggest that SHP is required for targeting HDAC to the promoter and that histone deacetylation is involved in the repression mechanism. Overexpression of HDAC-1, or G9a alone, had no significant inhibitory effect, either on Gal4-HNF-4 activity, or its SHP-repressed activity. This indicates that increasing the endogenous levels of either protein is not sufficient to reduce further SHP-driven repression. On the other hand, when both HDAC-1 and G9a were coexpressed with SHP, reporter activity at background levels (∼0.4%) was detected (Figure 4). The significance of this finding is substantiated by the fact that no such repressive effect was observed when the R213C mutant of SHP was analyzed. Importantly, the methylase activity-deficient mutant of G9a (R1109H), which can interact with SHP as efficiently as wild-type G9a (Figure 3), failed to further inhibit SHP-induced repression, suggesting that histone methylation may also play a role in the repression mechanism. Although, transiently transfected reporter DNA may not necessarily adopt the same chromatin configuration as the endogenous gene, in most cases they represent a valid model for the study of the interplay of activators and repressors. The results obtained with TSA and the methylase-deficient G9a mutant point to effects mediated by histone modifications, which suggest that a significant portion of the reporter was assembled into chromatin.
Figure 4.
Functional roles of HDAC-1 and G9a in SHP-mediated transcriptional repression. (A) Cos-1 cells were transfected with 0.5 μg 4xGal4-E1B-luc reporter together with 10 ng of SHP expression vectors and 100 ng of the other indicated expression vectors. Bars represent means ± SE of normalized luciferase activities, expressed as a percentage of the activity obtained by Gal4-HNF-4 alone (100%). Where indicated, the cells were treated with 0.1 μM trichostatin A (TSA) 12 h before harvest. (B) Whole cell extracts from Cos-1 cells were transfected with the above amounts of expression vectors and were analyzed in western blots using αHNF4 and αSHP antibodies, as indicated. The cells were transfected with vectors for Gal4-HNF-4 alone (lane 1), Gal-4-HNF-4 and SHPwt (lane 2), Gal-4-HNF-4, SHPwt, HDAC-1 and G9aWT (lane 3), SHPwt alone (lane 4), SHP R213C alone (lane 5), SHPwt, Gal4-HNF-4, HDAC-1 and G9aWT (lane 6), SHP R213C, Gal4-HNF-4, HDAC-1 and G9aWT (lane 7). The results show that under the conditions used, overexpression of the different proteins does not significantly alter the expression of Gal4-HNF4, SHPwt and SHP R213C.
Together, these results suggest that HDAC-1 and G9a can both be recruited to promoters via direct physical interactions with distinct domains of SHP. The enzymatic activity and the synergistic action of HDAC-1 and G9a are required to achieve high-level repression.
SHP can associate with underacetylated and K-9 methylated histone 3
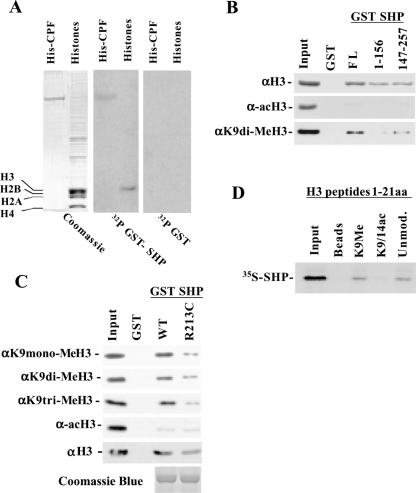
As described above, the R213C mutant of SHP can interact with both HDAC-1 and G9a. However, unlike what was observed with wtSHP, the repression activity of the R213C mutant was not further potentiated by overexpression of the two enzymes together. This indicates that, besides the recruitment of enzymatically active HDAC-1 and G9a to the promoters, other events may also play important roles in the repression mechanism. The function of active chromatin modifying enzymes in SHP-mediated repression raised the possibility that the resulting histone modifications may influence the formation of a repressor complex. To this end, we first investigated whether SHP itself can associate with chromatin. Far-western blot analysis using 32P-labeled GST-SHP recombinant protein as a probe revealed that SHP could selectively interact with histone 3 (Figure 5A). As a positive control, we included recombinant CPF/LRH-1 protein in the blot, which is known to interact with SHP (21). This interaction was further verified by GST pull-down experiments. Immobilized fusion proteins containing different domains of SHP were incubated with purified histones and the presence of SHP-bound histone 3 was detected in western blots using an αH3 antibody that can recognize both modified and unmodified histone 3, or an α-acH3 that detects hyperacetylated H3, respectively. Only negligible amounts of hyperacetylated H3 could be observed in the bound material, while SHP-histone 3 interaction was clearly evident using this assay as well (Figure 5B). Interestingly, however, SHP-bound histone 3 fractions gave a strong signal in western blots using α-diMeK9-H3, an antibody that reacts specifically with K9-dimethylated histone 3. This interaction was constrained to the C-terminal autonomous repression domain of SHP (Figure 5B). When the same assay was performed with antibodies recognizing K9-mono and trimethylated histone 3, we observed no difference in SHP binding. Interestingly however, the R213C mutant SHP protein, interacted much less efficiently with all forms of methylated histone 3 (Figure 5C). The total H3 bound to SHP-R213C was somewhat less than that bound to wtSHP (Figure 5C, bottom panel). We, however, note that the difference observed between wild-type and mutant SHP in binding K9-methylated H3, especially with trimetylated H3, was much more pronounced. We also performed in vitro pull-down experiments using modified and unmodified histone 3 peptides immobilized to streptavidin–agarose beads. SHP specifically interacted with the unmodified and K9-methylated histone N-terminal peptides but not with the acetylated one (Figure 5D). Although SHP may possess an intrinsic ability to interact with unmodified histone 3 tails, the finding that a methylase activity-deficient G9a mutant failed to repress transcription (Figure 4) suggests that K9-methylation of histone 3 is functionally relevant. In addition, the R213C mutant of SHP, which has a decreased transrepression potential, interacted less efficiently with K9-methylated histone 3. This points to the functional importance of SHP-histone 3 association, as this mutation did not affect the interactions of SHP with HDAC-1 and G9a (Figures 2 and 3).
Figure 5.
Association of SHP with K9-methylated histone 3. (A) Core histones from HeLa cells were separated by SDS–PAGE, transferred to PVDF membranes and hybridized with 32P-GST as control (right panel), or 32P-GST-SHP (middle panel). Coomassie blue staining of the gel is shown at left. As a positive control for interaction, recombinant His-tagged CPF was run in parallel with histones (His-CPF lane). (B and C) An aliquot of 2 μg of the indicated GST-SHP fusion proteins linked to glutathione–sepharose beads were used to pull-down purified core histones. After washings, the retained material was subjected to western blot analysis with the indicated antibodies that detect total histone 3 (αH3), hyperacetylated histone 3 (α-acH3) and K9-mono, di or trimethylated histone 3 (αK9 mono-MeH3, αK9 di-MeH3, αK9 tri-MeH3). (D) Unmodified, K9-dimethylated (K9Me), or K9/K14-acetylated, biotin-conjugated histone 3 peptides (1–21 amino acids) were immobilized on streptavidin–agarose beads and used to pull-down 35S-labeled in vitro-translated SHP.
SHP is localized in nuclease-sensitive euchromatin
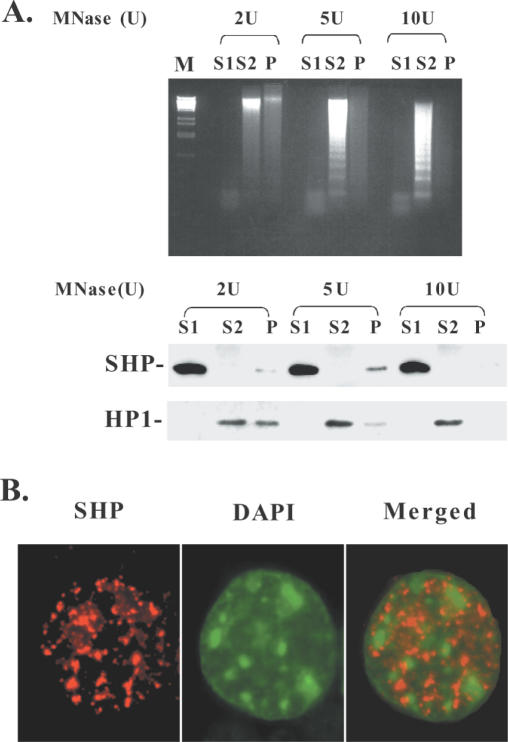
Since K9 methylation of histone 3 has been considered as an epigenetic mark that drives heterochromatin formation when recognized by HP-1 (9), we investigated the localization of SHP in nuclear territories corresponding to euchromatic and heterochromatic regions. We performed biochemical fractionation and in situ immunofluorescence analysis with differentiated Caco-2 cells, which constitutively express SHP at high levels. According to an established fractionation procedure (29,30), mild treatment of nuclei with MNase in a low salt buffer leads to the initial release of mono- and oligonucleosomes, which are enriched in non-histone chromosomal proteins. This fraction, called S1, lacks histone H1 and represents the potentially ‘active’ chromatin (euchromatin). Hypotonic lysis of the resulting pellet leads to the recovery of the S2 fraction, which corresponds to transcriptionally inactive heterochromatin that contains polynucleosomes and is enriched for histone H1. Finally, the remaining insoluble pellet (fraction P) represents transcriptionally competent, matrix-associated chromatin. Western blot analysis with an antibody against SHP showed that almost all the proteins were fractionated in the euchromatin-enriched S1 fraction (Figure 6A). On the other hand, HP-1, used as a control for heterochromatin, was distributed mainly in the S2 and to a lesser extent in the P fraction (Figure 6A).
Figure 6.
SHP is localized in euchromatic nuclear territories. (A) Caco-2 cell nuclei were treated with the indicated amounts of MNase and fractionated to obtain euchromatin-enriched (S1), heterochromatin-enriched (S2) and insoluble nuclear matrix (P) containing fractions. DNA from each fraction was purified and separated on 1.5% agarose gels (upper panel). Western blot analysis of the same fractions was performed with antibodies against SHP and HP1 (bottom panel). (B) In situ immunofluorescence analysis of Caco-2 cells was performed to detect SHP (left panel) and DAPI (middle panel). The merged image of the two stainings is shown at right.
In agreement with the results of the chromatin fractionation experiment, immunohistochemical analysis of CaCo-2 cells revealed that SHP shows a broad localization in the interphase nuclei, but is completely excluded from DAPI-dense heterochromatin (Figure 6B). This distribution suggests that SHP can be involved in gene silencing at euchromatic regions and that the mechanism by which SHP represses target gene expression is unlikely to involve transitions from euchromatic to heterochromatic states.
DISCUSSION
SHP is an unusual member of the nuclear receptor superfamily, containing the putative ligand-binding domain, but lacking the conserved DNA-binding domain (13). SHP was originally isolated as a cofactor that interacts with and inhibits the activity of other nuclear receptors. Recent studies exploring the molecular mechanism involved in SHP-mediated repression revealed that besides the N-terminal nuclear receptor interaction domain, which targets the AF-2 regions of nuclear receptors and thus competes with coactivators, the C-terminal domain of SHP is also important for repression (20,21). The C-terminal domain possesses an autonomous repression activity, which is poorly characterized. The relevance of the proposed dual mechanism, i.e. coactivator competition and ‘direct transcriptional repression’, is supported by the fact that naturally occurring mutations in humans with clinical phenotype have been identified at both the N-terminal and the C-terminal parts of SHP (16).
The main findings of this work addressing the mechanism of ‘direct repression’ can be summarized as follows: (i) SHP is localized exclusively in nuclease-sensitive euchromatin regions and can physically interact with HDAC-1 and the euchromatic histone 3 methylase G9a. (ii) SHP can interact with unmodified and K9-methylated, but not with acetylated histone 3 N-terminal tails. (iii) Association with HDAC-1, G9a and histone 3 is functionally important for SHP-mediated transcriptional silencing.
The observations extend previous models by pointing to the involvement of a multistep mechanism in SHP-dependent transcriptional repression. In the first step, as suggested before, SHP may compete with coactivators for a common interaction surface on nuclear receptors (20,21). In the second step, the C-terminal domain of SHP, which has been shown to function as an autonomous repressor domain (21), can recruit HDAC-containing complexes to the promoters, whose activity may erase the acetylation marks on the neighboring nucleosomes. Next, SHP-recruited G9a can methylate the deacetylated histone 3 tails at the lysine 9 residue. Underacetylated histone 3 may serve as a new surface with which SHP itself can interact and create a stable repressive complex. This association is prevented by acetylation, but not by K9 methylation of histone 3. Therefore, SHP may be responsible for both the recruitment of chromatin modifiers to the promoters and for the interpretation of histone code in a feed-forward mechanism that promotes and maintains histone deacetylation and/or H3K9 methylation at genomic sites of SHP recruitment. In certain promoter contexts, the recently identified potential SHP partner E1D (34) may also contribute to the last step of the mechanism. Hence, SHP may serve as a common molecular platform to recruit different chromatin modifying activities to promoters, which may act in consecutive steps to achieve transcriptional repression of target genes.
In principle, several of the above individual events should result in efficient transcriptional repression. For example, coactivator dissociation or histone deacetylation should be sufficient to repress the promoters. The molecular features of SHP interaction with G9a and histone 3 may provide additional means for long-term repression on certain promoter contexts. Thus, we speculate that on different genes SHP-mediated repression may involve one or more of the above steps. In line with this is the recent finding pointing to an SHP-dependent recruitment of an HDAC-containing mSin3A-Swi/Snf complex onto the human Cyp7A1 promoter during bile acid-mediated repression (35).
G9a is the major mammalian methylase, responsible for euchromatic H3-K9 methylation (11,12), and SHP is identified as an interaction partner that can target this enzyme to promoters. Since SHP can inhibit the activity of various nuclear receptors (13,19–21), this finding predicts an extensive inventory of potential target genes regulated by G9a.
Acknowledgments
ACKNOWLEDGEMENTS
We thank C.M. Saunderson for the G9a cDNA plasmid, N. Katrakili for expert technical assistance, M. Denaxa for help with confocal microscopy and P. Hatzis for comments on the manuscript. This work was supported by grants from EU (QLRT-2000 01513, HPRN-CT 2000-00087 and LSHG-CT-2004-502950) and from HFSP (RGP-0024).
REFERENCES
- 1.Turner B.M. (2000) Histone acetylation and an epigenetic code. BioEssays, 2, 836–845. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 93, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 3.Turner B.M. (2002) Cellular memory and the histone code. Cell, 111, 285–291. [DOI] [PubMed] [Google Scholar]
- 4.Rea S., Eisenhaber,F., O'Carroll,D., Strahl,B.D., Sun,Z.W., Schmid,M., Opravil,S., Mechtler,K., Ponting,C.P., Allis,C.D. and Jenuwein,T. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406, 593–599. [DOI] [PubMed] [Google Scholar]
- 5.Melcher M., Schmid,M., Aagaard,L., Selenko,P., Laible,G. and Jenuwein,T. (2000) Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell. Biol., 20, 3728–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kouzarides T. (2002) Histone methylation in transcriptional control. Curr. Opin. Genet. Dev., 12, 198–209. [DOI] [PubMed] [Google Scholar]
- 7.Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- 8.Lachner M., O'Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- 9.Jenuwein T. (2001) Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol., 11, 266–273. [DOI] [PubMed] [Google Scholar]
- 10.Schultz D.C., Ayyanathan,K., Negorev,D., Maul,G.G. and Rauscher,F.J.,III (2002) SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev., 16, 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tachibana M., Sugimoto,K., Nozaki,M., Ueda,J., Ohta,T., Ohki,M., Fukuda,M., Takeda,N., Niida,H., Kato,H. and Shinkai,Y. (2002) G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev., 16, 1779–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tachibana M., Sugimoto,K., Fukushima,T. and Shinkai,Y. (2001) Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem., 276, 25309–25317. [DOI] [PubMed] [Google Scholar]
- 13.Seol W., Choi,H.S. and Moore,D.D. (1996) An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science, 272, 1336–1339. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin B., Jones,S.A., Price,R.R., Watson,M.A., McKee,D.D., Moore,L.B., Galardi,C., Wilson,J.G., Lewis,M.C., Roth,M.E. et al. (2000) A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell, 6, 517–526. [DOI] [PubMed] [Google Scholar]
- 15.Lu T.T., Makishima,M., Repa,J.J., Schoonjans,K., Kerr,T.A., Auwerx,J. and Mangelsdorf,D.J. (2000) Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell, 6, 507–515. [DOI] [PubMed] [Google Scholar]
- 16.Nishigori H., Tomura,H., Tonooka,N., Kanamori,M., Yamada,S., Sho,K., Inoue,I., Kikuchi,N., Onigata,K., Kojima,I. et al. (2001) Mutations in the small heterodimer partner gene are associated with mild obesity in Japanese subjects. Proc. Natl Acad. Sci. USA, 98, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seol W., Chung,M. and Moore,D.D. (1997) Novel receptor interaction and repression domains in the orphan receptor SHP. Mol. Cell. Biol., 17, 7126–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson L., Bavner,A., Thomsen,J.S., Farnegardh,M., Gustafsson,J.A. and Treuter,E. (2000) The orphan nuclear receptor SHP utilizes conserved LXXLL-related motifs for interactions with ligand-activated estrogen receptors. Mol. Cell. Biol., 20, 1124–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson L., Thomsen,J.S., Damdimopoulos,A.E., Spyrou,G., Gustafsson,J.A. and Treuter,E. (1999) The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERalpha and ERbeta. J. Biol. Chem., 274, 345–353. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y.K., Dell,H., Dowhan,D.H., Hadzopoulou-Cladaras,M. and Moore,D.D. (2000) The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol. Cell. Biol., 20, 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y.K. and Moore,D.D. (2002) Dual mechanisms for repression of the monomeric orphan receptor liver receptor homologous protein-1 by the orphan small heterodimer partner. J. Biol. Chem., 277, 2463–2467. [DOI] [PubMed] [Google Scholar]
- 22.Ktistaki E. and Talianidis,I. (1997) Modulation of hepatic gene expression by hepatocyte nuclear factor 1. Science, 277, 109–112. [DOI] [PubMed] [Google Scholar]
- 23.Soutoglou E., Viollet,B., Vaxillaire,M., Yaniv,M., Pontoglio,M. and Talianidis,I. (2001) Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J., 20, 1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown S.E., Campbell,R.D. and Sanderson,C.M. (2001) Novel NG36/G9a gene products encoded within the human and mouse MHC class III regions. Mamm. Genome, 12, 916–924. [DOI] [PubMed] [Google Scholar]
- 25.Edmondson D.G. and Roth,S.Y. (1998) Interactions of transcriptional regulators with histones. Methods, 15, 355–364. [DOI] [PubMed] [Google Scholar]
- 26.Hatzis P. and Talianidis,I. (2001) Regulatory mechanisms controlling human hepatocyte nuclear factor 4alpha gene expression. Mol. Cell. Biol., 21, 7320–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soutoglou E. and Talianidis,I. (2002) Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science, 295, 1901–1904. [DOI] [PubMed] [Google Scholar]
- 28.Hatzis P. and Talianidis,I. (2002) Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell, 10, 1467–1477. [DOI] [PubMed] [Google Scholar]
- 29.Rose S.M. and Garrard,W.T. (1984) Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J. Biol. Chem., 259, 8534–8544. [PubMed] [Google Scholar]
- 30.Huang S.Y. and Garrard,W.T. (1989) Electrophoretic analyses of nucleosomes and other protein–DNA complexes. Meth. Enzymol., 170, 116–142. [DOI] [PubMed] [Google Scholar]
- 31.Ferrer J. (2002) A genetic switch in pancreatic beta-cells: implications for differentiation and haploinsufficiency. Diabetes, 51, 2355–2362. [DOI] [PubMed] [Google Scholar]
- 32.Hayhurst G.P., Lee,Y.H., Lambert,G., Ward,J.M. and Gonzalez,F.J. (2001) Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol., 21, 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., Ning,G. and Duncan,S.A. (2000) Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev., 14, 464–474. [PMC free article] [PubMed] [Google Scholar]
- 34.Bavner A., Johansson,L., Toresson,G., Gustafsson,J.A. and Treuter,E. (2002) A transcriptional inhibitor targeted by the atypical orphan nuclear receptor SHP. EMBO Rep., 3, 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kemper J.K., Kim,H., Miao,J., Bhalla,S. and Bae,Y. (2004) Role of an mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid synthesis by SHP. Mol. Cell. Biol., 24, 7707–7719. [DOI] [PMC free article] [PubMed] [Google Scholar]