Abstract
Bacteria growing in different conditions experience a broad range of demand on the rate of protein synthesis which profoundly affects cellular resource allocation. During fast growth, protein synthesis is long known to be modulated by adjusting the ribosome content, with the vast majority of ribosomes engaged at a near-maximal rate of elongation. Here we characterized protein synthesis by E. coli systematically, focusing on slow growth conditions. We establish that the translational elongation rate decreases as growth slows down, exhibiting a Michaelis-Menten dependence on the abundance of the cellular translational apparatus. However, an appreciable elongation rate is maintained even towards zero growth including the stationary phase. This maintenance, critical for timely protein synthesis in harsh environments, is accompanied by a drastic reduction in the fraction of active ribosomes. Interestingly, well-known antibiotics such as chloramphenicol also cause substantial reduction in the pool of active ribosomes, instead of slowing down translational elongation as commonly thought.
Recent studies have shown that proteome allocation constraint plays a major role in shaping the global gene expression pattern of bacteria in response to both genetic and environmental perturbations during exponential growth1–4. Resolving this constraint has also been established as the physiological origin of such widely known microbial responses as catabolite repression and metabolic overflow5,6. In a nutshell, proteome allocation constraint reflects the limitation of ribosomes in protein synthesis – the vast majority of ribosomes are engaged in translation such that under growth limiting conditions that require the synthesis of certain proteins, the synthesis of others must decrease. Conversely, under favorable conditions that permit fast growth, ribosomal proteins are preferentially synthesized at the expense of all other proteins4.
The notion of a limited translational capacity in bacteria was suggested by Maaloe and colleagues long ago7, in attempt to rationalize earlier finding that the cellular ribosome content exhibited a linear dependence on the growth rate for a broad range of nutrient conditions8. This linear dependence, referred to as a bacterial “growth law”, was theorized to result from the ribosomes elongating at their maximal rate regardless of nutrient conditions, such that the rate of protein synthesis (reflected by the growth rate) is proportional to the number of translating ribosomes. However, the elongation rate was later found to decrease for E. coli growing in poor nutrient conditions9–11. In a recent theoretical study, Klumpp et al12 noted that the ternary complexes (TC, comprising of aminoacyl-tRNA, elongation factor Tu, and GTP), which are the “substrates” of ribosomes and which diffuse slowly in the crowded cytoplasm, present another significant bottleneck to the translational capacity that may be the cause of slow down in translational elongation at slow growth. They further showed that this slow down in translational elongation could still give rise to a linear relation between the ribosome content and growth rate for fast to moderate growth, assuming a Michaelis-Menten dependence of the elongation rate on TC abundances, and a proportionality between TC and ribosome abundances due to co-regulation in their synthesis13; see Supplementary Note 1 for a review. This theory therefore provides a plausible reconciliation between the linear growth law and the known data on translational elongation. However for slow growing cells in nutrient-limited conditions, the same theory would predict an abrupt drop in the ribosome content and a vanishing elongation rate, contrary to the known fact that protein synthesis proceeds effectively even in the stationary phase14 and can moreover jump to a high rate immediately after nutrient upshift15.
In this study, we systematically characterized the translational elongation rate of E. coli in vivo by varying nutrients and applying translation-inhibiting antibiotics. A very broad range of growth conditions was studied, particularly for slow growth (up to 20-hour doubling time) and including stationary phase. Our results quantitatively established the Michaelis-Menten dependence of the elongation rate on components of the ternary complexes, the key assumption of Klumpp et al’s theory, for all growth conditions studied. Nevertheless, the elongation rate is maintained at a significant level (>50% of that in rich medium) even in extremely slow growth. Our results suggest that this maintenance, important physiologically for the effective synthesis of proteins when nutrients run out, is accomplished by limiting the pool of active ribosomes. Also, a number of translation-inhibiting antibiotics are found to dramatically reduce the pool of active ribosomes, without slowing down the rate of translational elongation as is commonly thought.
RESULTS AND ANALYSIS
Translation under nutrient limitation
We first characterized the translational elongation rate (ER, denoted mathematically by k) of wild type E. coli K-12 cells growing exponentially in a variety of nutrient conditions. A broad range of growth rates (GR, denoted by λ), corresponding to doubling time from 20 minutes to 20 hours, was probed by varying both the carbon and nitrogen sources in saturating minimal medium (Supplementary Fig. 1AB). In each case, ER was determined by the classical β-galactosidase (LacZ) induction assay (Supplementary Fig. 2)9,16, with correction of the initiation time, ~10 sec across all the growth conditions probed, by repeating the induction assay using the short LacZα fragment (Supplementary Fig. 3). This method was further validated by measuring ER of a previously characterized slow translational mutant (Supplementary Fig. 4).
As shown in Fig. 1A (black open circle), ER was nearly constant (k = 16~17 aa/s) under good growth conditions (λ > 1 h−1), but steadily decreased in poorer growth conditions. The decreases in ER found here are in agreement with those obtained from sporadic previous studies determined by several different methods9,10,11 (Supplementary Fig. 5). Notably, ER remained at significant values (>9 aa/sec) even close to zero growth (doubling time of 20 hours). We also determined ER after cells entered stationary phase (Supplementary Fig. 6). The value obtained, ~8 aa/s (red open circle in Fig. 1A, Fig. 1B), is in line with ER values obtained from slow steady state growth and also with the known demand of protein synthesis in stationary phase14,17.
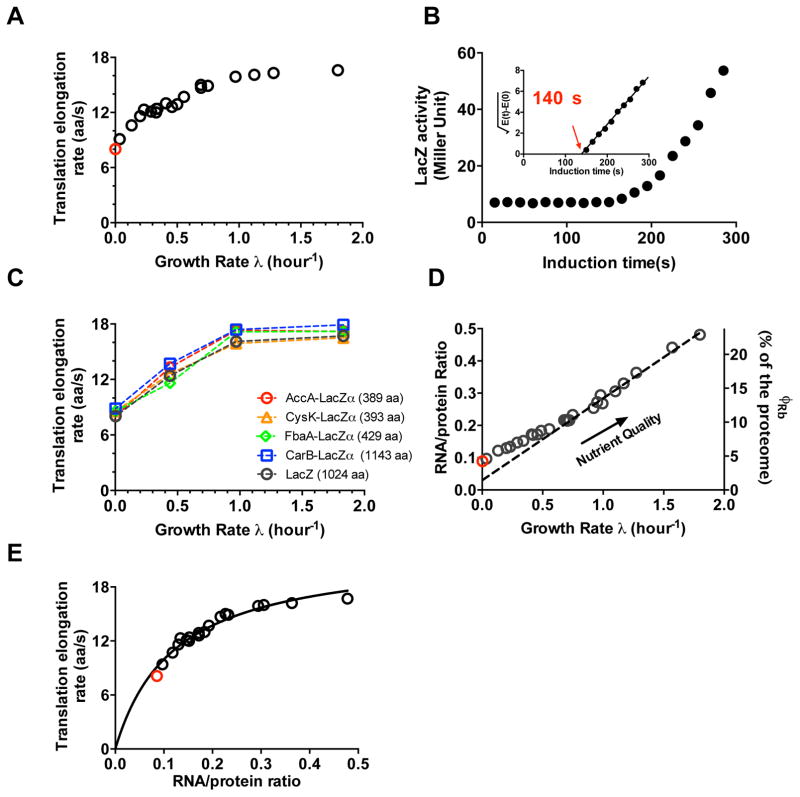
Figure 1. Characteristics of protein synthesis capacity under nutrient limitation.
(A) Growth-rate dependence of the in vivo translational elongation rate (ER) was obtained by the LacZ induction assay from exponentially growing cultures with various nutrient sources. These data are shown as black circles (Supplementary Table 1). The red circle shows the ER obtained from a culture in early stage of stationary phase (panel B). (B) LacZ induction kinetics of E. coli in early stationary phase (Supplementary Fig. 6), with the corresponding Schleif plot shown as the inset. LacZ expression appeared ~140 s after induction, giving an ER of ~8 aa/s after taking into account of initiation time via the LacZα assay (Supplementary Fig. 3). (C) ERs of four other LacZα-fused proteins under nutrient limited growth were obtained using the LacZα complementation assay (Supplementary Table 2, Supplementary Fig. 7). ER values obtained under the same conditions by the LacZ induction assay are shown in black. (D) Growth-rate dependence of the RNA-protein ratio (R/P) obtained for the same growth medium as in panel A. Original data are given in Supplementary Table 3. Right y-axis gives the ribosomal fraction, ϕRb; see Supplementary Note 1 for conversion. The dashed line is a linear fit to the data in the fast-growth regime (λ > 0.7 h−1). Red circle corresponds to stationary phase data. (E) ER from panel A is plotted against R/P from panel D. The black line is a fit to the Michaelis-Menten relation (with KM ≈ 0.11 and kelong ≈ 22 aa/s). Data shown in panel A to D are averages over three biological replicates. Standard deviations are within 5–10% (~ size of the symbols).
To investigate whether our results may be specific to the translation of LacZ, we developed a variant of LacZ induction assay, translationally fusing a target gene of choice to LacZα (Supplementary Fig. 7A)18. We then measured the ER of four such LacZα hybrid proteins of different lengths for different growth conditions (Supplementary Fig, 7B-E). The ERs obtained for these proteins, shown as colored symbols in Fig. 1C, are quantitatively consistent with those obtained from LacZ for all conditions tested. Together, these results suggest that the growth-rate dependence of ER shown in Fig. 1A are generic to the translation of E. coli proteins.
In order to understand how cells manage to maintain an appreciable ER at slow growth, we characterized the abundance of the cellular ribosome content ϕRb for a number of slow growth conditions using quantitative mass spectrometry3. The results are seen to correlate well (Supplementary Fig. 8) with the RNA/protein ratio (R/P)1,19, which we use below as a proxy of the cellular ribosome content; see Supplementary Note 1A. R/P values were obtained for wild-type cells grown in the same medium as those shown in Fig. 1A. They are seen to exhibit a linear correlation with the growth rate at moderate to fast growth (Fig. 1D) as is well known1,4,5,20. However, this linear correlation (dashed line in Fig. 1D), referred to as the linear ‘growth law’, is broken at slow growth (λ ≲0.7/h) by a slight but notable upward bend. Such deviations from linear correlation of the ribosome content with GR at slow growth have been reported previously21.
Could this deviation from linear relation be the source of finite ER at slow growth? We next made a scatter plot of ER against R/P obtained under the same growth condition. The data is well described by a Michaelis-Menten (MM) relation (black line, Fig. 1E), predicted12 based on an assumed underlying MM relation between ER and the concentration of ternary complexes (TC, the substrates of translation) and co-regulation in the expression of ribosomes and components of TC. We will quantitatively examine the relation between ribosome and TC shortly below. Here we remark that the existence of such a relation would indeed tie the maintenance of ER at slow growth to the upward bend of the linear growth law: A slight upward bend would increase significantly the y-intercept of R/P-vs-GR curve, which would significantly increase the TC content and hence significantly increase ER at slow growth (Supplementary Fig. 9).
Translation under drug inhibition
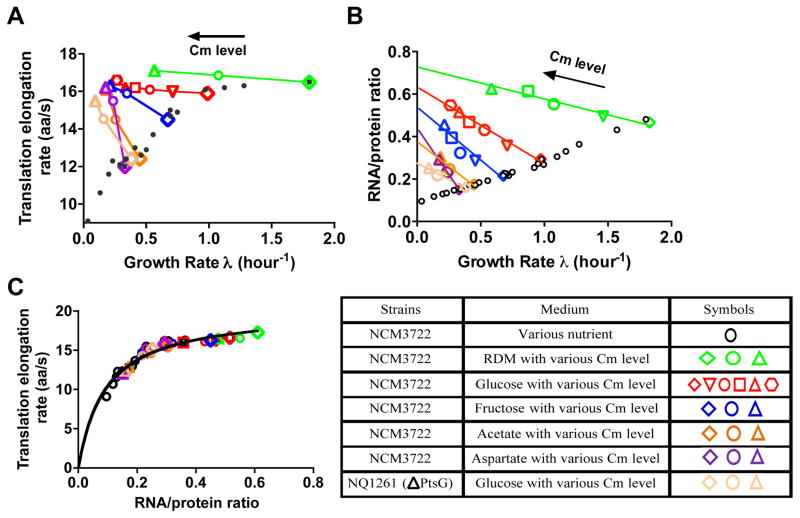
Translational inhibition is an alternative way to vary the ribosomal and TC content1, thus offering a different perspective on the determinants of the in vivo translational elongation rate. Addition of sub-lethal doses of chloramphenicol (Cm), a well-known translational elongation blocker, inhibits overall protein synthesis in a graded manner and slows down cell growth1,22. We characterized the ER of exponentially growing cells for a range of Cm concentrations in glucose minimal medium. Although the overall LacZ production rate was substantially reduced by the addition of Cm, the synthesis time of the first LacZ remained nearly constant for the different Cm concentrations applied (Supplementary. Fig. 2D), implying a nearly constant ER as plotted in Fig. 2A (red symbols). This seemingly surprising result is not specific to the synthesis of LacZ, as the same finding is recapitulated for the translation of other proteins using the pulse-chase labeling method11 (Supplementary Fig. 10). Similar result was obtained for Cm addition to rich nutrient conditions (RDM + Glucose, green symbols in Fig. 2A). Even more remarkably, clear increases in ER were obtained by the addition of Cm in poor nutrient conditions (minimal medium with fructose, acetate, or aspartate as the sole carbon source), or for a ΔptsG strain (NQ1261, deleted of the main glucose transporter), which grew slowly in glucose medium and hence mimics carbon-limited growth without changing the substrate (Fig. 2A). This result is further verified by applying the hybrid LacZα method (Supplementary Fig. 11).
Figure 2. Characteristics of protein synthesis capacity under translation limitation by chloramphenicol (Cm) inhibition.
(A) Translational elongation rate (ER) plotted against the growth rate for steady state culture treated with sub-lethal doses of chloramphenicol (Cm). Each color represents a fixed nutrient source as indicated in the legend table, except for the orange symbols, representing a mutant strain (NQ1261, ΔptsG) which grows slowly in glucose medium due to the lack of the major glucose transporter. (It is another way to generate a “poor carbon” condition.) Different symbols indicate varied Cm levels in the medium. The small black dots represent the ER obtained under nutrient limited growth as shown in Fig. 1A. Original data are given in Supplementary Table 4. (B) RNA-protein ratio (R/P) under Cm inhibition. R/P has a negative correlation with growth rate as previously reported1. The lines are straight-line fits to guide the eyes. Original data are given in Supplementary Table 5. The small black open circles are the data of nutrient limitation as shown at Fig. 1D. (C) ER in panel A is plotted against R/P in Panel B (same color code). The data of Fig. 1E are shown as black symbols. ER exhibits a Michaelis-Menten dependence on R/P under both nutrient limitation and Cm inhibition. The black line is the best-fit to the Michaelis-Menten relation with kM ≈ 0.11 and kelong ≈ 22 aa/s. Data shown in panel A and B are averages over three biological replicates. Standard deviations are within 5–10% (~ size of the symbols).
Cm is known to block the translational elongation process by binding to the 50S ribosome subunit22. The binding rate is slow (kon = 0.034 μM/min), but when it does bind, it binds tightly with a half-life exceeding 7 min (koff = 0.074 min−1)22. Our finding of a constant ER in good growth medium can be rationalized if the observed ER was derived from a fraction of leading ribosomes that escaped Cm binding at the sub-lethal doses applied, while the majority of ribosomes stalled when “hit” by Cm, resulting in aborted translation. This scheme is illustrated in Supplementary Fig. 12 and elaborated in Supplementary Note 2.
If the above scenario is correct, then we expect to see similar effect on ER for other drugs that bind tightly to ribosomes, but not for those that exhibit rapid reversible binding. Towards this end, we characterized the effect of four other protein synthesis inhibitors, tetracycline (Tet), erythromycin (Ery), fusidic acid (FA), and Mupirocin (Mup). Tet inhibits translational elongation by binding tightly to 30S ribosome subunit23,24. Erythromycin binds to the 50S subunit of ribosome with a comparably low koff as Cm does (0.15 min−1)25. As expected, the effect of Tet and Ery on ER is similar to that of Cm: ER remained nearly constant for cells grown in glucose and increased for cells grown in poor growth medium (aspartate) upon the addition of sub-lethal doses of these two drugs (Supplementary Fig. 13AB). On the other hand, FA, which prevents the rapid turnover of elongation factor G (EF-G) from the ribosome via binding to the EF-G-ribosome complex26,27, affects the ribosome movement more reversibly28. In this case, we found ER to decrease gradually with sub-lethal doses of FA treatment (Supplementary Fig. 13C). Finally, we characterized Mup, which inhibits the charging of isoleucine tRNA29. Here, we observed a reduced ER similar to the case of nutrient limitation (Supplementary Fig. 13D).
Model of translational elongation
The above picture of tight drug-ribosome binding addresses the effect of these drugs on ER in good growth medium. To understand why ER in poor growth medium increased upon drug treatment, we again looked into the ribosome content in these cultures by characterizing R/P, which again reflects the ribosomal protein content as shown in Supplementary. Fig. 8. As found previously, R/P is negatively correlated with growth rate upon Cm inhibition (Fig. 2B). Similar effects are seen for Tet and Ery treatment (Supplementary Fig. 14AB). The opposing pattern of the GR-dependence of R/P results from the differential regulation of ribosome synthesis mediated by guaninetetraphosphate (ppGpp)20,30. Making the scatter plot of ER against R/P again, we find remarkably that the two diverging datasets for nutrient limitation and Cm inhibition (Fig. 1AD and 2AB) collapsed onto a single curve (Fig. 2C), which is well-described by the Michaelis-Menten relation (solid line). Thus, the exact same Michaelis-Menten relation between ER and the ribosomal content holds despite the very different physiological conditions between nutrient-limited and translation-limited growth. Graphically, the rise in ER is simply illustrated in Supplementary Fig. 9(cyan symbols).
To appreciate the origin of this MM relation, we developed a coarse-grained model of translation12,31, in which tRNA ternary complexes (TC) are treated as a single species of substrates for the ribosomes, and referred to as TCeff. The time to translate one codon, which is the inverse of the elongation rate (k), is divided into two coarse-grained time scales: (i) the binding of the ternary complex to the ribosome, which depends inversely on the effective TC concentration [TCeff] due to limitation by TC diffusion12,32, and (ii) other enzymatic processes (e.g., translocation) whose rate does not depend on TC concentration. Let these two time scales be 1/(kon[TCeff]) and 1/kelong, respectively. Then we have
| [1] |
which leads to a Michaelis-Menten relation between the elongation rate k and the effective TC concentration, with kelong interpreted as the maximal rate of peptide elongation, and kelong/kon being the binding constant of the effective TC with the ribosome. The precise mathematical definition of [TCeff] and its effective binding constant in terms of the corresponding quantities for individual tRNA species are given in Supplementary Note 3A and 3B. The Michaelis-Menten relation of ER versus R/P then follows if [TCeff] and R/P are proportional, i.e.,
| [2] |
This proportionality, suggested previously12 based on the known co-regulation between rRNA, tRNA, and EF-Tu, is directly verified (Supplementary Fig. 15, 16) for nutrient limitation and Cm inhibition. We further established the constancy of the tRNA charging ratio, for a number of major tRNA species (Supplementary Fig. 17) for both types of growth perturbations. Taken together, these results strongly support the validity of the relation proposed in Eq. [2].
Based on the quantitative relations obtained between the abundances of EF-Tu, total tRNA, and ribosomes, and based on the known composition of individual tRNA species33 assuming the constancy of the relative composition of the individual tRNA species and the tRNA charging ratio, we obtained the concentration of TCeff for each of our conditions, summarized by the constant of proportionality C≈31 μM in Eq. [2]; see Supplementary Note 3B for details. Then from the Michaelis-Menten fit of Fig. 2C, we obtained kelong = 22 aa/s, and kon = 6.4 μM−1s−1. We note that the value of kelong is consistent with previous estimation31,34 and the value of kon is in agreement with prediction made based on the assumption that kon is diffusion-limited in the crowded cytoplasmic environment12,32. The same analysis can be extended to include the effect of other substrates of translational elongation such as EF-G (which is also proportional to R/P as seen in Supplementary Fig. 16CD). However, our calculations show that EF-G diffusion is unlikely to be the limiting step because of its much higher concentration compared to individual tRNA species in cells (Supplementary Note 3C).
The coarse-grained model described here gives a simple, quantitative explanation to the counterintuitive pattern of increased ER under Cm inhibition in poor growth medium: The increased R/P under Cm inhibition (Fig. 2B) is accompanied by increased levels of TCs (Supplementary Figs 15–17). For cells growing in good nutrient conditions, the higher TC levels do not noticeably increase ER since it is already saturated. However, in poor nutrient condition where ER is limited by the substrate TC levels in the absence of drugs, the observed increase in ER under Cm treatment is quantitatively accounted for by the increase of TC levels. Thus, we conclude that the observed dependences of ER on growth rate upon nutrient limitation and Cm inhibition arise largely from the changing TC levels. The same conclusion can also be applied to other drugs like Tet and Ery, which exhibited similar correlation between ER and R/P as Cm (Supplementary Fig. 14CD).
Fraction of active ribosome equivalent
For cells under Cm inhibition (also Ery, Tet), the reduced rate of protein synthesis cannot be accounted for by either the measured ER (which either increases or remains constant) or the observed ribosomal content (which increases). These findings point to a necessary reduction in the number of productively translating ribosomes, due to a variety of possible reasons including ribosome mis-assembly35, inactivation36, and drop-off37 as will be discussed below. However direct quantitative characterization of the state of ribosome activity by polysome profiling have yielded inconclusive and conflicting results in the past38,39, due to experimental challenges such as the fragility of polysomes or polysome run off which convert a fraction of polysomes to ribosome subunits or 70S monosomes during the sample collection process10,40. Here, we introduce a related quantity, the “active ribosome equivalent”, which can be deduced based on mass balance in steady state.
For exponentially growing cells with negligible rate of protein turnover41, mass balance implies
| [3] |
where Naa is the number of amino acids contained in all cellular proteins in a standard culture volume, and is the total number of “ribosome equivalent” needed to synthesize all the cellular proteins, assuming that they are all translating at the observed rate k. Since Naa, λ, and k are all measured quantities, Eq. [3] can be used to compute , or more meaningfully, the fraction of active ribosome equivalent, , with NRb being the total number of ribosomes per standard culture volume; see details in Supplementary Note 1A.
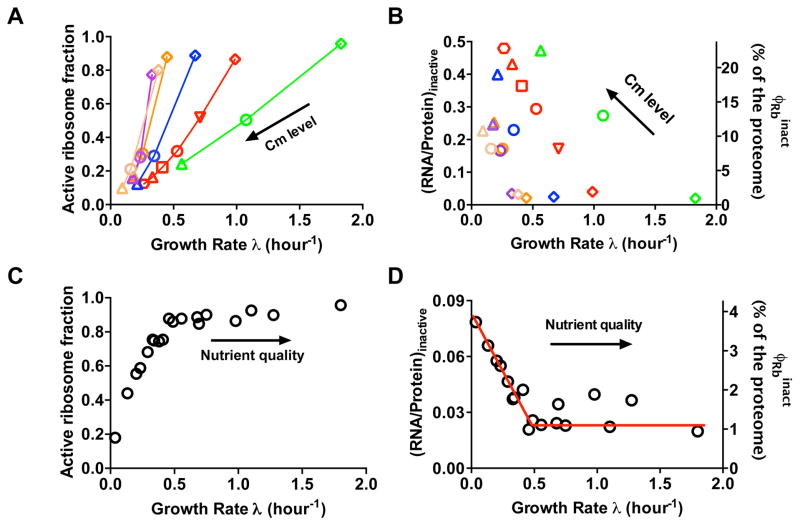
The result obtained for Cm inhibition is shown in Fig. 3A: The fraction of active ribosome equivalent, factive so-defined, decreases steeply with increasing Cm level (decreasing GR) for each of the nutrient condition characterized (colors same as those defined in Fig. 2). Similar patterns were observed for Tet and Ery inhibition as well (Supplementary Fig. 18). It is also convenient to introduce the inactive ribosome equivalent, whose absolute abundance (1 − factive)×ϕRb is shown in Fig. 3B. We see that this quantity reaches over 20% of the proteome for some of the conditions. Note that our definition of active ribosome equivalent based on Eq. [3] would classify those that drop-off during the translation process and produce unstable partial protein products (Supplementary Fig. 12) as inactive.
Figure 3. Growth-rate dependence of the active ribosome fraction.
(A) Growth-rate dependent fraction of active ribosome equivalent (factive) for cultures under sub-lethal doses of Cm, computed according to Eq. [N1.5] in Supplementary Note 1A. Symbol shapes and colors are the same as those shown in Fig. 2. Original data are given in Supplementary Table 6. (B) The absolute abundance of inactive ribosomes upon Cm inhibition. Left y-axis shows the portion of RNA-protein ratio (R/P) attributed to inactive ribosomes, given by (1 − factive) ×R/P, with R/P taken from Fig. 2B. Right y-axis shows the protein mass fraction of inactive ribosomes, as defined in Supplementary Note 1A. (C) Growth-rate dependent fraction of active ribosome equivalent (factive) under nutrient limitation. Original data are given in Supplementary Table 7. (D) The absolute abundance of inactive ribosomes upon nutrient limitation. The left and right y-axis are the same as those defined in Panel B, with R/P taken from Fig. 1D. The red line, showing a constant from moderate to fast growth range (λ>0.5/h) together with a linear relation between with growth rate at slow growth range ((λ<0.5/h), is shown to guide the eyes. Note that the error in the estimates, of factive arising from errors in ER and R/P values, is ~10%. Data shown in panel A to D are calculated based on the ER and R/P data (each with at least three replicates) shown in Fig. 1 and Fig. 2, respectively.
We applied the same analysis to deduce the fraction of active ribosome equivalent and the abundance of inactive ribosome equivalent under nutrient limitation alone; the results are shown in Fig. 3C, 3D. From moderate to fast growth (λ > 0.5 h−1), factive in Fig. 3C remained nearly constant at a high level (>80%), which is consistent with previous estimates over the same growth range20,38. But at slower growth, the fraction of active ribosome equivalent steeply decreased to below 20%. Note that the upward bend in R/P occurred in the same slow growth range (Fig. 1D). From Fig. 3D, we see that the abundance of inactive ribosome equivalent, kept at ~1% of the total protein mass for λ > 0.5 h−1, increased in the corresponding slow growth range to 4%.
DISCUSSION
In this study, we firmly established a single Michaelis-Menten relation between the elongation rate and the ribosome content for E. coli cells subjected to both nutrient limitation and translational inhibition (Fig. 2C). This observation reflects an underlying Michaelis-Menten relation between ER and the concentrations of ternary complexes (TC), whose components are proportional to the ribosome content (Supplementary Figs 15–17). Different ERs observed in vivo are seen as manifestations of changes in substrate availability. The maintenance of ER in poor nutrient conditions including the stationary phase is then understood as a consequence of cells upholding their TC levels, reflected in an upward-bend of the ribosome-GR relation at slow growth (Fig. 1D and Supplementary Fig. 9). The appearance of the latter is in turn related to the rise of inactive ribosomes at slow growth (Fig. 3D). In fact, the upward bend of the ribosome-GR relation as well as the maintenance of elongation at slow growth can be quantitatively captured by incorporating the observed fraction of inactive ribosome (Fig. 3D) into the model of Klumpp et al12 without introducing any ad-hoc fitting parameters (Supplementary Fig. 19 and Supplementary Note 4). This maintenance of a significant elongation rate is of direct physiological importance at slow growth, as timely synthesis of proteins is needed in poor nutrient conditions including the stationary phase14,42.
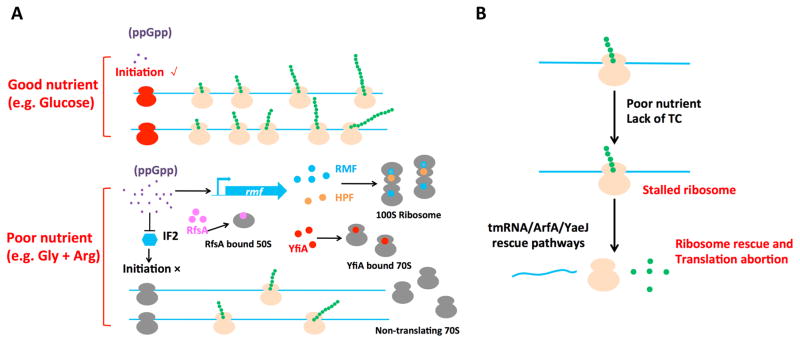
To gain insights into the underlying mechanisms of reduction in active ribosome fraction during slow growth, we performed polysome profiling analysis for cells grown in poor conditions. There is a clear shift from polysome to 70S monosome for cells grown in poor nutrient compared to glucose (Supplementary Fig. 20A). This shift indicates a programmed reduction in translational initiation at slow growth, probably through inhibition of IF2-mediated translation initiation by ppGpp, whose concentration increases in poor nutrient conditions (Fig. 4A)20,43. This reduction can also be implemented by various regulators that titrate ribosomes away from translation, including RMF, HPF, YfiA, and RsfA36,44,45 (Fig. 4A). Alternatively, ribosomal inactivation may arise passively due to increased occurrences of abortive translation, with ribosome stalling triggered by low levels of ternary complexes at slow growth; see Supplementary Note 5. The rescue of stalled ribosomes (through such factors as tmRNA-SmpB, ArfA, YaeJ)46,47 would also elevate the 70S monosome fraction (Fig. 4B). Both the programmed and passive scenarios of Fig. 4 would be consistent with early findings that polyribosomes were converted to 70S monosomes with the loss of protein synthesizing capacity for E. coli under carbon starvation48.
Figure 4. Models for the reduction of the active ribosome fraction at slow growth.
(A) Examples of regulated mechanisms of ribosome inactivation: Under good nutrient conditions such as glucose medium (upper case), the intracellular ppGpp level (small dots) is low. Translation initiates frequently (with initiating ribosomes in red), giving a high fraction of polysomes (the actively translating ribosome, in orange color). However, in poor nutrient conditions (lower case), the intracellular ppGpp level is increased. Ribosomes may be inactivated by ribosome modulation factor (RMF) (positively regulated by ppGpp)45 and dimerize by hibernation promotion factor (HPF) to form 100S ribosome36. Alternatively, YfiA can inactivate the 70S ribosome36, while RsfA can bind to the 50S subunit, thereby reducing the available ribosomes for translation. The reduced active ribosome fraction can also result from inhibition of translation initiation: Increased ppGpp is reported to inhibit the IF2-dependent translation initiation process43. All these mechanisms lead to decrease in polysome number, with some increasing the number of non-translating 70S monosomes (in gray). (B) Possible passive mechanism of ribosome inactivation: Under poor nutrient condition, ternary complexes (TC) which are the substrates of ribosomes become limiting, causing the ribosomes to stall more frequently. Prolonged stalling triggers abortion of the translation process, with the stalled ribosome rescued by the tmRNA/ArfA/YaeJ pathway46. Partially translated proteins resulting from the abortive process are usually not well-folded and are further degraded by proteases such as ClpX50. The ribosomes involved in abortive translation are “futile” because they do not produce stable products. They are classified as inactive ribosomes according to the way active ribosomes are defined (Eq. [3] and Supplementary Note 1).
The growth-rate dependence of the active ribosome fraction may be at the core of a long-standing puzzle regarding the low efficiency of bacterial protein synthesis in poor nutrient conditions: In a classical nutrient-upshift assay15,49 where a very slow-growing chemostat culture was shifted to a rich nutrient broth, the rate of protein synthesis increased 4–5× within minutes, well before significant number of new ribosomes and ternary complexes could be synthesized15. Our results on the reduction of the active ribosome fraction, from ~90% at fast growth to <20% in slow growth (Fig. 3C), provides a natural resolution to this puzzle, if the inactive ribosomes resulting from regulated sequestration or inhibition (Fig. 4A) can quickly engage in protein synthesis upon nutrient upshift.
In summary, our results reveal that E. coli maintains the speed of elongation by reducing the fraction of active ribosomes which in turn elevates the expression of translational machineries including components of the ternary complex. One might argue that TC synthesis could in principle be separated from that of ribosomal synthesis, so that a separate control can be used to maintain the TC levels without also increasing the inactive ribosomes at slow growth. However, given the additional benefit of preparing cells for rapid nutrient upshift as just discussed, co-regulation of these two quantities conveniently couples the potential advantage of growth recovery and the functional necessity of maintaining the elongation rate at slow growth.
The other major finding of this study is that sub-lethal doses of translational elongation blockers (Cm, Tet, Ery) inhibit protein synthesis through decreasing the active ribosome fraction instead of reducing the rate of translational elongation in vivo (Fig. 3AB). There may occur via several possible routes: First, it is known that translational inhibitors such as Cm lead to ribosome misassembly, forming large amounts of inactive precursors22,30 (Supplementary Fig. 12). This is manifested in our polysome profiling data where a substantial subunit fraction is obtained, with an extra peak corresponding to ribosome intermediates being clearly visible for cells grown in Cm (Supplementary Fig. 20B). In addition, there is again a significant shift of the polysome distribution to 70S monosomes. This may result from the rescue of the stalled Cm-bound ribosomes and the trailing ribosomes (Supplementary Fig. 12). The rescued monosomes would be rendered “inactive” by our classification, since the partial protein products they produce before stalling would be rapidly degraded50. This expectation is supported by previous report22 showing that under Cm inhibition, the 70S monosomes contributed little to the overall protein synthesis compared to the polysome fraction. In the future, we expect global methods such as ribosome profiling to reveal more directly the in vivo action of these drugs; particularly, regarding ribosome stalling and traffic jam as depicted in Supplementary Fig. 123,47.
Methods
Strains
The strains used in this study are either wild type E. coli NCM3722 strain51,52 or its derivatives: NQ1261 (ΔptsG, deleted of the gene encoding the main glucose transporter PtsG), which grows slowly in glucose medium53, NQ1468 used for measuring LacZα induction kinetics, and the WE2015 series of strains used for measuring the translational elongation rate of various proteins using the hybrid LacZα approach described below. E. coli MG1655 strain used in the ribo-seq study of Li et al3 was also obtained directly from G. W. Li.
Strain NQ1468 contains chromosome expression of the LacZα fragment (the first 90 codons of the LacZ protein) driven by its native lac promoter, and also a plasmid, which constitutively expresses the complementary LacZω fragment (wild type LacZ with 11-41 aa deleted)54 by the synthetic Ptet promoter.
To construct the strain: The lacZ coding region corresponding to 91-1024 aa residues of NCM3722 was first replaced with selective maker kanr genes (kanamycin resistant gene) using λ Red system55. The kanr marker was further flipped out by plasmid pCP20. This generates the chromosome-expressed LacZα strain, NQ1441. NQ1441 strain is white when grown at X-gal plate (50 μg/mL) due to its lack of LacZ activity. The LacZω fragment coding sequence was amplified from E. coli DH5α strain with upstream primer and downstream primer containing KpnI site and BamHI site, respectively. The PCR product was gel purified and inserted between the KpnI site and BamHI site of a pZA31-luc derived plasmid pZA31*-luc (a BamHI site was introduced between the luciferase gene and T1 terminator of pZA31-luc). This yields pZA31*Ptet-lacω, with the lacZω fragment driven by the constitutive Ptet promoter of the pZA31 plasmid56. Since for measuring LacZα induction kinetics, chloramphenicol needs to be added to modulate protein synthesis (see translation rate measurement below), the cmr gene in pZA31* needs to be replaced by another antibiotic marker. To do this, the ampr gene fragment of pZE12-luc was cut off with XhoI and SpeI, gel purified, and inserted into the same site of pZA31*-Ptet-lacω to replace cmr. The resultant plasmid pZA31**-Ptet-lacω was then transformed into strain NQ1441, yielding strain NQ1468. NQ1468 colony is blue in a LB plate supplemented with 50 μg/mL X-gal and 100 μg/mL ampicillin, indicating that LacZα is successfully complemented in vivo.
To measure the translational elongation rate of several other proteins (AccA, CarB, CysK, and FbaA) using the LacZα complementation system, we amplified the native Plac promoter through PCR and inserted it into the HindIII/KpnI site of pZS24*-MCS plasmid. The coding sequence of the lacZα fragment (for this time, we use a smaller version of lacZα fragment coding for N-terminal 1-60 aa of LacZ57 together with an upstream sequence coding for a 10 aa (GGGGS)2 linker) was further inserted to the KpnI/MluI site of the above vector, yielding pKU1500 vector. The fragment containing Ptet-lacZω-T1 (T1 terminator) was further PCR amplified from the above pZA31*Ptet-lacZω, and cloned into the SalI/HindIII sites of pKU1500 vector. A pair of XhoI/NotI site was introduced into the pKU1500 vector between Plac promoter and the lacZα fragment. The coding sequence of accA, carB, cysK, and fbaA was inserted individually into XhoI and NotI site to be fused with lacZα fragment to yield the final constructs. Each final construct was further transferred into a lacZ deficient NCM3722 strain constructed using λ Red system3 system to yield strains WE2015-accA, WE2015-carB, WE2015-cysK and WE2015-fbaA, respectively.
Growth medium
All the growth media used in this study are MOPS buffered media as described in Cayley et al58 except in the case of growing the MG1655 strain in Li et al3 (see methods of mass spectroscopy) where we followed Li et al3 and used MOPS buffered media as described in Neidhardt et al59. The medium contains 40 mM MOPS and 4 mM Tricine (adjusted to pH 7.4 with NaOH), 0.1 mM FeSO4, 0.276 mM Na2SO4, 0.5 μM CaCl2, 0.523 mM MgCl2, and also micronutrients mixtures used in Neidhardt et al59. Carbons sources and nitrogen sources were varied for creating different extent of nutrient limitations. In addition, rich defined medium (RDM) + glucose medium contains 0.2% (w/v) glucose, micronutrients, various amino acids, nucleotides, and vitamins. Glucose + cAA medium contains 0.2% (w/v) glucose and 0.2% (w/v) casamino acids. Chloramphenicol (Cm), Tetracycline (Tet), Mupirocin (Mup), fusidic acid (FA) concentrations were varied for different extent of translation inhibition. All the MOPS minimal media contain 0.1 M NaCl.
Cell growth
Exponentially cell growth was always performed in a 37°C water bath shaker shaking at 240 rpm. A standard cell growth round always follows three steps: seed culture, pre-culture, and experimental culture. Typically, cells were first grown as seed cultures in LB broth for several hours, then as pre-cultures overnight in MOPS medium that is identical in composition to the experimental conditions. Finally, the experimental cultures were started by diluting the pre-cultures to OD600 around 0.01 to 0.02. For calculating growth rates, 7–9 OD600 points within the range of OD600 0.05 ~ 0.5 (spanning three generations) were measured.
Total RNA quantification
Total RNA quantification method is the same as used by You et al5.
Total Protein quantification
Total Protein quantification method is the same as used by You et al5.
Measurement of Translational elongation rate by LacZ or LacZα induction assay
Translational elongation rates were measured using LacZ induction as described by Andersson et al60 with modifications. NCM3722 was grown to OD600 ~ 0.4 followed by induction of the lac operon with 5 mM Isopropyl β-D-Thiogalactoside (IPTG). Immediately after induction, at 10 or 15-second intervals, aliquots of either 200 μL or 400 μL culture were transferred into pre-chilled microfuge tubes containing 5 μL chloramphenicol (34 mg/mL) for a total of 16–18 time points. Samples were frozen using dry ice and stored at −80 °C prior to LacZ assay.
LacZ assay was mainly performed as per the traditional Miller’s colorimetric method using o-Nitrophenyl-β-D-Galactopyranoside (ONPG) as substrate61. For conditions yielding low basal levels of LacZ, such as sublethal concentrations of chloramphenicol, a sensitive fluorescence substrate 4-methylumbelliferyl-D-galactopyranoside (MUG) was used62,63. Briefly, 100 μL cell sample harvested as described above, was added to 400 μL Z-buffer and warmed at 37°C water bath for several minutes. 50 μL of 2mg/mL MUG stock in DMSO was added and the reaction mixtures were incubated for 30 mins. The reaction was stopped with 250 μL of 1 M Na2CO3. The fluorescence intensity was measured with a micro-plate reader (365 nm excitation filter, 450 nm emission filters). LacZ induction curve was made by plotting the LacZ activity against the induction time and further analyzed using a square-root plot (Schleif plot) to obtain the lag time for the synthesis of the first LacZ molecule16 (Tfirst, Supplementary Fig. 2).
To correct for the time taken for the initiation steps (including IPTG penetration, LacI depression, Transcription initiation and translation initiation) of LacZ induction, LacZα induction kinetics was also performed following the above-described measurement of LacZ induction kinetics with modifications. Strain NQ1468 was grown to exponential phase at OD600 ~ 0.4. At 5–10 s interval, nine cell culture samples (200 μL) were immediately pipetted into 2 mL pre-cooled Eppendorf tubes which contain 5 μL chloramphenicol (34 mg/mL) for stopping translational elongation. LacZα-LacZω complement activity was always measured using the more sensitive MUG assay due to the much lower activity level of the complement compared to the full-length LacZ. Before performing the MUG assay, NQ1468 sample was first incubated at 37°C for 1 hour to let all the LacZα fragment complement with the LacZω fragment54. Samples were then subject to MUG assay in the same way as that of full-length LacZ activity measurement described above. LacZα induction kinetics was also made by plotting the activity versus time. The synthesis time of LacZα (Tα) was estimated using straight-line fit of the LacZα induction curve. The initiation time (Tinit) is calculated as Tinit = Tα − 90/k, where the 2nd term is the elongation time needed to complete the synthesis of the 90-residue long alpha fragment. As a first estimate of the elongation rate k, we used k = 934aa/(Tfirst − Tα), where Tfirst is the first appearance for the full length LacZ. The initiation time was found to be always around 10s (Supplementary Fig. 3). We therefore calculated all the LacZ translation elongation rate as 1024aa/( Tfirst − 10s).
To determine the translational elongation rate of several other proteins (AccA-LacZα, CarB-LacZα, CysK-LacZα and FbaA-LacZα) using the WE2015 series of strains, the same procedure and analysis procedure was followed as the LacZ induction assay except MUG substrate instead of ONPG was used for the assay. The synthesis time of the first LacZα-fused protein (Tfirst) was also obtained through Schleif plot (Supplementary Fig. 7B–E) as in LacZ induction. Therefore, the translation elongation rate of these four LacZα-fused protein equals to L/( Tfirst − 10s), where L is the length of each fused proteins (Supplementary Table 2), and 10s is the initiation time.
Translational elongation rate measured by pulse chase radioactive labeling
Translational elongation rates were also characterized for a number of proteins using the pulse-chase method as described by Pedersen11, albeit without dual labeling. Cells were grown to an OD600 of 0.5 at 37°C in 20 ml MOPS minimal media. A pulse of 0.03 mCi 35S methionine (1175 Ci/mmol) was added, and chased with 1 μmol unlabeled methionine after 10 seconds. At a 7-second interval, around 20 samples (600 μl) were drawn from the culture into tubes that contain 2 mg chloramphenicol, and were immediately frozen in liquid nitrogen. The samples were then thawed on ice, cells pelleted, and resuspended in 30 μl 2X Sodium Dodecyl Sulfate (SDS) Polyacrylamide gel electrophoresis (PAGE) loading buffer (100mM Tris-HCl, pH 6.8; 200mM Dithiotrietol; 4% SDS; 0.2% bromophenol blue and 20% glycerol). Samples were boiled for 1 minute, spun down and the proteins were resolved using 8% SDS-PAGE. The gels were dried and exposed to phosphorimager screens overnight. The screens were scanned using Typhoon FLA9000 and the bands were quantified using imagequant. In order to correct for loading errors, the band intensities were normalized with respect to the intensity of the free methionine at the front for the respective lanes. The band intensity of specific protein was plotted with time after adding 35S methionine and the turning time point at which the band intensity reaches plateau corresponds the time (T) needed for the translation elongation of specific protein. In this case, the translation elongation rate was obtained by using the length of specific protein to divide the elongation time. Note that according to the basic principle of pulse chase radioactive labeling. (Caption of Supplementary Fig. 10), there is no time cost of initiation step that is the case in LacZ induction assay.
Northern blotting under acidic conditions
Total aminoacyl-tRNA was extracted from cells under acidic conditions and subjected to acid gel electrophoresis as described in Janssen et al64 using tRNA-specific hybridization probes as listed in Dong et al33.
Polysome profiling by sucrose gradient
Polysome profiling was performed according to Balakrishnan et al, and Qin et al with modifications65,66. Briefly, cells in the mid-log phase (OD600 around 0.5) were rapidly chilled by pouring on crushed ice and harvested by centrifugation. The pellets were resuspended in lysis buffer (10 mM Tris-HCl pH 7.5, 10 mM MgCl2, 65 mM NH4Cl, 1 mg/ml lysozyme, 100 U/ml RNase-free DNase I (Roche), 400 U/ml Superase inhibitor (Ambion) and lysed by three freeze-thaw cycles. Clarified lysates were loaded on to 10–38% (w/v) sucrose gradients made in a buffer containing 10 mM Tris-HCl (pH 7.5), 10 mM MgCl2 and 65 mM NH4Cl. Gradients were spun in an SW41 rotor at 35000 rpm for 3.5 hours at 4°C and fractionated using a syringe-pump (Brandel) in conjunction with a ultraviolet absorbance detector (ISCO UA-6 type 11). ImageJ Software was used to quantify the relative fraction of the three ribosomes populations (polysomes, monosome, and subunit) from the raw figure of polysome profiling. For each peak of the three ribosomes populations in the profile plot, draw a line across the base of the peak to enclose the peak. Then the size of each peak was analyzed by the “Analyze → Measure” function of the ImageJ software.
Measurement of relative protein abundances using quantitative Mass Spectroscopy
The protein abundances of NCM3722 cells grown in five slow growth conditions including (1) 60 mM acetate+10 mM NH4Cl (λ: 0.46/h); (2) 0.075% mannose + 10 mM NH4Cl (λ: 0.29/h); (3) 0.2% glycerol + 10 mM arginine (λ: 0.21/h); (4) 20 mM glutamate + 10 mM NH4Cl (λ: 0.12/h); (5) 0.2% glycerol + 20 mM Threonine ((λ: 0.03/h) were obtained relative to those from the reference condition (0.2% glucose + 10 mM NH4Cl (λ: 0.98/h) using quantitative mass spectroscopy (qMS). The procedure is described below. The same procedure is used to determine the relative abundance between MG1655 cells obtained from Li et al3 and grown in MOPS glucose condition described in Li et al3 and NCM3722 cells grown in our reference condition.
Protein sample collection
14N/15N protein sample collection and TCA precipitation is similar as described in Hui et al4. The same amount of protein samples of NCM3722 strain growing in 0.2% glucose + 10 mM 15NH4Cl medium and 0.075% Mannose + 10 mM 15NH4Cl medium were mixed together and taken as the 15N reference sample. However, for the comparison between MG1655 strain and NCM3722 strain, only the protein sample of NCM3722 strain growing in 0.2% glucose + 10 mM 15NH4Cl medium was taken as 15N reference sample.
Peptide digestion
TCA-precipitated bacterial pellets containing ~200 μg protein were dissolved in 40 μL of 8M urea, 100 mM Tris-HCl, pH 7.5. Proteins were reduced with 10 mM DTT for 15 mins at 37°C. Cysteine alkylation was achieved with 40 mM iodoacetamide for 15 mins at 25°C in the dark. The protein solution was diluted 5-fold with 50 mM Tris-HCl, 10 mM CaCl2, pH 7.5 to a final urea concentration of 1.6M and 80 μL of this solution was digested with 2 μg trypsin (Pierce). After an overnight incubation at 37C, an additional 1 μg trypsin was added for 4h. Try tic peptides were desalted using PepClean reverse-phase columns (Pierce) per manufacturer instructions.
MS data acquisition and conversion
MS data were acquired using a AB Sciex 5600 TripleTOF after injecting 2 μg tryptic peptides, essentially as described by Hui et al, but with increased instrument scan times: MS1 accumulation time of 250 ms, MS2 accumulations of 150 ms. Vendor instrument files were converted to profile and centroided MZML formats using vendor software. Centroided mzML files were converted to mzXML using tools included in the Trans-Protemic Pipeline (TPP)67 67and searched using X!Tandem68 against the UniProt E. coli database (organism ID 83333) supplemented with common protein contaminants, enzymes, and reversed peptide decoy sequences. The peptide-spectrum match tolerances were set at 50 ppm and 100 ppm for the precursor and product ions, respectively. The TPP tools PeptideProphet and iProphet were used to score the peptide–spectrum matches and the search results were combined into a consensus library using SpectraST69.
MS data quantitation
As described in Hui et al, the 14N (light) experimental and 15N (heavy) reference peptide ion count intensities were quantified with least-squares Fourier transform convolution70 using Massacre71 modified to accept SpectraST libraries as input (Patsalo and Williamson, unpublished). In order to correct the imbalance in total experimental and reference protein amounts (approximately 20% deviation), the light/heavy quantitative ratios were corrected by the quotient of the cumulative 14N and 15N X!Tandem E. coli spectral counts for each sample.
The absolute proteome abundance of each individual r-proteins in each growth conditions was obtained by calibrating against the abundance of NCM3722 strain growing in the reference condition (0.2% glucose + 10 mM NH4Cl) as described in the captions of Supplementary Table 8–10.
Supplementary Material
Acknowledgments
We gratefully acknowledge discussion with numerous colleagues including Zoya Ignatova, Stefan Klumpp, Steen Pedersen, Severin Schink, Josh Silverman, Matthew Scott, R. Young, and members of the Hwa lab at various stages of this work. This research is supported by NIH grant R01GM109069 through TH; supported by grant 31530081 of the National Natural Science Fund of the People’s Republic of China (NSFC) through YPW. KF acknowledges NIH grant R01GM072528. JRW acknowledges the NIH grant GM118850. Manlu Zhu acknowledges the financial support from the China Scholarship Council (CSC, 201306010039).
Footnotes
AUTHOR CONTRIBUTIONS
XD, MZ, MW, and TH designed the study. XD, MZ, RB and VP performed experiments. XD, MZ, MW, HO, VP, JW, KF, Y-PW, and TH analyzed the data. XD, MZ, MW, Y-PW and TH wrote the paper and the supplement.
DATA AVAILABILITY
Supplementary information is available online. Reprints and permissions information is available online at http://www.nature.com/reprints/.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
References
- 1.Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 2.Gerosa L, Kochanowski K, Heinemann M, Sauer U. Dissecting specific and global transcriptional regulation of bacterial gene expression. Mol Syst Biol. 2013;9:658. doi: 10.1038/msb.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hui S, et al. Quantitative proteomic analysis reveals a simple strategy of global resource allocation in bacteria. Mol Syst Biol. 2015;11:784. doi: 10.15252/msb.20145697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.You C, et al. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature. 2013;500:301–306. doi: 10.1038/nature12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basan M, et al. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature. 2015;528:99–104. doi: 10.1038/nature15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maaløe O. Regulation of the protein-synthesizing machinery - ribosomes, tRNA, factors, and so on. In: Goldberger RF, editor. Biological Regulation and Development. Plenum; New York: 1979. [Google Scholar]
- 8.Neidhardt FC, Magasanik B. Studies on the role of ribonucleic acid in the growth of bacteria. Biochim Biophys Acta. 1960;42:99–116. doi: 10.1016/0006-3002(60)90757-5. [DOI] [PubMed] [Google Scholar]
- 9.Dalbow DG, Young R. Synthesis time of beta-galactosidase in Escherichia coli B/r as a function of growth rate. Biochem J. 1975;150:13–20. doi: 10.1042/bj1500013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young R, Bremer H. Polypeptide-chain-elongation rate in Escherichia coli B/r as a function of growth rate. Biochem J. 1976;160:185–194. doi: 10.1042/bj1600185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984;3:2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klumpp S, Scott M, Pedersen S, Hwa T. Molecular crowding limits translation and cell growth. Proc Natl Acad Sci U S A. 2013;110:16754–16759. doi: 10.1073/pnas.1310377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryals J, Little R, Bremer H. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J Bacteriol. 1982;151:1261–1268. doi: 10.1128/jb.151.3.1261-1268.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolter R, Siegele DA, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 15.Koch AL, Deppe CS. In vivo assay of protein synthesizing capacity of Escherichia coli from slowly growing chemostat cultures. J Mol Biol. 1971;55:549–562. doi: 10.1016/0022-2836(71)90336-6. [DOI] [PubMed] [Google Scholar]
- 16.Schleif R, Hess W, Finkelstein S, Ellis D. Induction kinetics of the L-arabinose operon of Escherichia coli. J Bacteriol. 1973;115:9–14. doi: 10.1128/jb.115.1.9-14.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Link H, Fuhrer T, Gerosa L, Zamboni N, Sauer U. Real-time metabolome profiling of the metabolic switch between starvation and growth. Nat Methods. 2015;12:1091–1097. doi: 10.1038/nmeth.3584. [DOI] [PubMed] [Google Scholar]
- 18.Zhu M, Dai X, Wang YP. Real time determination of bacterial in vivo ribosome translation elongation speed based on LacZα complementation system. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basan M, et al. Inflating bacterial cells by increased protein synthesis. Mol Syst Biol. 2015;11:836. doi: 10.15252/msb.20156178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. In: Neidhardt FC, editor. Escherichia coli and Salmonella. 2. Am Soc Microbiol; Washington, DC: 1996. pp. 1553–1569. [DOI] [PubMed] [Google Scholar]
- 21.Neidhardt FC, Ingraham JL, Schaechter M. Physiology of the bacterial cell — A molecular approach. Sinauer Associates; 1990. [Google Scholar]
- 22.Harvey RJ, Koch AL. How partially inhibitory concentrations of chloramphenicol affect the growth of Escherichia coli. Antimicrob Agents Chemother. 1980;18:323–337. doi: 10.1128/aac.18.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chopra I, Howe TG. Bacterial resistance to the tetracyclines. Microbiol Rev. 1978;42:707–724. doi: 10.1128/mr.42.4.707-724.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Day LE. Tetracycline inhibition of cell-free protein synthesis. II. Effect of the binding of tetracycline to the components of the system. J Bacteriol. 1966;92:197–203. doi: 10.1128/jb.92.1.197-203.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestka S. Binding of [14C]erythromycin to Escherichia coli ribosomes. Antimicrob Agents Chemother. 1974;6:474–478. doi: 10.1128/aac.6.4.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seo HS, et al. EF-G-dependent GTPase on the ribosome. conformational change and fusidic acid inhibition. Biochemistry. 2006;45:2504–2514. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- 27.Okura A, Kinoshita T, Tanaka N. Formation of fusidic acid-G factor-GDP-ribosome complex and the relationship to the inhibition of GTP hydrolysis. J Antibiot (Tokyo) 1971;24:655–661. doi: 10.7164/antibiotics.24.655. [DOI] [PubMed] [Google Scholar]
- 28.Uemura S, et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes J, Mellows G. Inhibition of isoleucyl-transfer ribonucleic acid synthetase in Escherichia coli by pseudomonic acid. Biochem J. 1978;176:305–318. doi: 10.1042/bj1760305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennis PP. Effects of chloramphenicol on the transcriptional activities of ribosomal RNA and ribosomal protein genes in Escherichia coli. J Mol Biol. 1976;108:535–546. doi: 10.1016/s0022-2836(76)80135-0. [DOI] [PubMed] [Google Scholar]
- 31.Ehrenberg M, Kurland CG. Costs of accuracy determined by a maximal growth rate constraint. Q Rev Biophys. 1984;17:45–82. doi: 10.1017/s0033583500005254. [DOI] [PubMed] [Google Scholar]
- 32.Zhang G, et al. Global and local depletion of ternary complex limits translational elongation. Nucleic Acids Res. 2010;38:4778–4787. doi: 10.1093/nar/gkq196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 34.Johansson M, Bouakaz E, Lovmar M, Ehrenberg M. The kinetics of ribosomal peptidyl transfer revisited. Molecular Cell. 2008;30:589–598. doi: 10.1016/j.molcel.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Siibak T, et al. Erythromycin- and chloramphenicol-induced ribosomal assembly defects are secondary effects of protein synthesis inhibition. Antimicrob Agents Chemother. 2009;53:563–571. doi: 10.1128/AAC.00870-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polikanov YS, Blaha GM, Steitz TA. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sin C, Chiarugi D, Valleriani A. Quantitative assessment of ribosome drop-off in E. coli. Nucleic Acids Res. 2016;44:2528–2537. doi: 10.1093/nar/gkw137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forchhammer J, Lindahl L. Growth rate of polypeptide chains as a function of the cell growth rate in a mutant of Escherichia coli 15. J Mol Biol. 1971;55:563–568. doi: 10.1016/0022-2836(71)90337-8. [DOI] [PubMed] [Google Scholar]
- 39.Harvey RJ. Fraction of ribosomes synthesizing protein as a function of specific growth rate. Journal of bacteriology. 1973;114:287–293. doi: 10.1128/jb.114.1.287-293.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Godson GN, Sinsheimer RL. Use of Brij lysis as a general method to prepare polyribosomes from Escherichia coli. Biochim Biophys Acta. 1967;149:489–495. doi: 10.1016/0005-2787(67)90176-1. [DOI] [PubMed] [Google Scholar]
- 41.Nath K, Koch AL. Protein degradation in Escherichia coli. I. Measurement of rapidly and slowly decaying components. J Biol Chem. 1970;245:2889–2900. [PubMed] [Google Scholar]
- 42.Mandelstam J. Turnover of protein in starved bacteria and its relationship to the induced synthesis of enzyme. Nature. 1957;179:1179–1181. doi: 10.1038/1791179a0. [DOI] [PubMed] [Google Scholar]
- 43.Milon P, et al. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc Natl Acad Sci U S A. 2006;103:13962–13967. doi: 10.1073/pnas.0606384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hauser R, et al. RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 2012;8:e1002815. doi: 10.1371/journal.pgen.1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Izutsu K, Wada A, Wada C. Expression of ribosome modulation factor (RMF) in Escherichia coli requires ppGpp. Genes Cells. 2001;6:665–676. doi: 10.1046/j.1365-2443.2001.00457.x. [DOI] [PubMed] [Google Scholar]
- 46.Shimizu Y. Biochemical aspects of bacterial strategies for handling the incomplete translation processes. Front Microbiol. 2014;5:170. doi: 10.3389/fmicb.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramaniam AR, Zid BM, O’Shea EK. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell. 2014;159:1200–1211. doi: 10.1016/j.cell.2014.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dresden MH, Hoagland MB. Polyribosomes of Escherichia coli. Re-formation during recovery from glucose starvation. J Biol Chem. 1967;242:1069–1073. [PubMed] [Google Scholar]
- 49.Koch AL. The adaptive responses of Escherichia coli to a feast and famine existence. Adv Microb Physiol. 1971;6:147–217. doi: 10.1016/s0065-2911(08)60069-7. [DOI] [PubMed] [Google Scholar]
- 50.Moore SD, Sauer RT. Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol Microbiol. 2005;58:456–466. doi: 10.1111/j.1365-2958.2005.04832.x. [DOI] [PubMed] [Google Scholar]
- 51.Soupene E, et al. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J Bacteriol. 2003;185:5611–5626. doi: 10.1128/JB.185.18.5611-5626.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyons E, Freeling M, Kustu S, Inwood W. Using genomic sequencing for classical genetics in E. coli K12. PLoS One. 2011;6:e16717. doi: 10.1371/journal.pone.0016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinsiek S, Bettenbrock K. Glucose transport in Escherichia coli mutant strains with defects in sugar transport systems. J Bacteriol. 2012;194:5897–5908. doi: 10.1128/JB.01502-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langley KE, Villarejo MR, Fowler AV, Zamenhof PJ, Zabin I. Molecular basis of beta-galactosidase alpha-complementation. Proc Natl Acad Sci U S A. 1975;72:1254–1257. doi: 10.1073/pnas.72.4.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1–I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morris DR, Hansen MT. Influence of polyamine limitation on the chain growth rate of beta-galactosidase and of its messenger ribonucleic acid. J Bacteriol. 1973;116:588–592. doi: 10.1128/jb.116.2.588-592.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cayley S, Lewis BA, Guttman HJ, Record MT., Jr Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J Mol Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- 59.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andersson DI, Bohman K, Isaksson LA, Kurland CG. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol Gen Genet. 1982;187:467–472. doi: 10.1007/BF00332630. [DOI] [PubMed] [Google Scholar]
- 61.Miller JH. Experiments in molecular genetics. 1972. [Google Scholar]
- 62.Vidal-Aroca F, et al. One-step high-throughput assay for quantitative detection of beta-galactosidase activity in intact gram-negative bacteria, yeast, and mammalian cells. Biotechniques. 2006;40:433–434. 436, 438. doi: 10.2144/000112145. passim. [DOI] [PubMed] [Google Scholar]
- 63.Martin L, Che A, Endy D. Gemini, a bifunctional enzymatic and fluorescent reporter of gene expression. PLoS One. 2009;4:e7569. doi: 10.1371/journal.pone.0007569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Janssen BD, Diner EJ, Hayes CS. Analysis of aminoacyl- and peptidyl-tRNAs by gel electrophoresis. Methods Mol Biol. 2012;905:291–309. doi: 10.1007/978-1-61779-949-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balakrishnan R, Oman K, Shoji S, Bundschuh R, Fredrick K. The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res. 2014;42:13370–13383. doi: 10.1093/nar/gku1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qin D, Fredrick K. Analysis of polysomes from bacteria. Methods Enzymol. 2013;530:159–172. doi: 10.1016/B978-0-12-420037-1.00008-7. [DOI] [PubMed] [Google Scholar]
- 67.Deutsh EW, Mendoza L, Shteynberg D, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 69.Lam H, et al. Building consensus spectral libraries for peptide identification in proteomics. Nature Methods. 2008;5:873–875. doi: 10.1038/nmeth.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sperling E, Bunner AE, Sykes MT, Williamson JR. Quantitative analysis of isotope distributions in proteomic mass spectrometry using least-square Fourier transform convolution. Analytical Chemistry. 2008;80:4906–4917. doi: 10.1021/ac800080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sykes MT, Sperling E, Chen SS, Williamson JR. Quantitation of the ribosomal protein autoregulatory network using mass spectrometry. Analytical Chemistry. 2010;82:5038–5045. doi: 10.1021/ac9028664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.