Abstract
Retinoid X receptors (RXR) play a role as master regulators due to their capacity to form heterodimers with other nuclear receptors. Accordingly, retinoid signaling is involved in multiple biological processes, including development, cell differentiation, metabolism and cell death. However, the role and functions of RXR in different heterodimer complexes remain unsolved, mainly because most RXR drugs (called rexinoids) are not selective to specific heterodimer complexes. This also strongly limits the use of rexinoids for specific therapeutic approaches. In order to better characterize rexinoids at specific nuclear receptor complexes, we have developed and optimized luciferase protein complementation-based Bioluminescence Resonance Energy Transfer (BRET) assays, which can directly measure recruitment of a co-activator motif fused to yellow fluorescent protein (YFP) by specific nuclear receptor dimers. To validate the assays, we compared rexinoid modulation of co-activator recruitment by RXR homodimer, and heterodimers Nur77/RXR and Nurr1/RXR. Results reveal that some rexinoids display selective co-activator recruitment activities with homo- or hetero-dimer complexes. In particular, SR11237 (BMS649) has increased potency for recruitment of co-activator motif and transcriptional activity with the Nur77/RXR heterodimer compared to other complexes. This technology should prove useful to identify new compounds with specificity for individual dimeric species formed by nuclear receptors.
Keywords: nuclear receptors, pharmacological parameters, co-factor recruitment, receptor dimerization, protein conformation
Introduction
Nuclear receptors (NRs) constitute a large superfamily of transcription factors regulating the expression of genes and controlling a wide range of physiological processes through ligand signaling (1, 2). In addition to forming homo- and hetero-dimers with other NR partners, NRs rely on co-regulator proteins to modulate transcription of target genes (2, 3). NR co-regulators can be broadly subdivided into co-activators (CoA, e.g. SRC or p300/CBP), which potentiate transcription and co-repressors (CoR, e.g. NCoR or SMRT), which silence gene transcription. Co-repressors recruit multiprotein complexes implicated in transcriptional repression and histone deacetylation (HDAC). On the other hand, several co-activators possess histone acetyltransferase activity (HAT) required for chromatin remodeling and subsequent access of the transcriptional machinery to promoters or recruit other proteins essential for transactivation, such as the CREB binding protein (CBP). In the absence of agonist, many nuclear receptors interact with co-repressors. Upon agonist binding, conformational change in the receptor causes the shedding of co-repressor complexes and the binding of co-activators, often via a conserved short LXXLL motif (4). However, agonist-mediated co-repression has also been described (3).
NRs are defined by common structural motifs composed of four functional modules, the N-terminal transcription Activation Function domain (AF-1), DNA-binding domain (DBD), the ligand-binding domain (LBD) and the C-terminal Activation Function domain (AF-2) (5). The AF-2 transcription regulation domain supports ligand-dependent transactivation of the receptor, whereas the AF-1 domain activity can be modulated by phosphorylation (2). Co-regulator molecules (co-activators or co-repressors) that modulate the activity of NRs can interact with the AF-1 and/or AF-2 domains. NRs can bind DNA as monomers, homodimers, and heterodimers. DNA recognition sites, also referred to as NR response elements, contain one or two consensus core half-site sequences. For example, numerous NRs bind to elements containing the half-site consensus motif AGGTCA. For response elements involved in dimeric NR activity, the half-sites can be configured as inverted, everted or direct repeats (DR). In monomeric response elements, the 5′-flanking sequence, such as the A/T-rich sequence in the Nur77 responsive element (NBRE), can increase the affinity and specificity of interaction with NRs (6).
The retinoid X receptor (RXR) subgroup (NR2B) of NRs is composed of 3 members: RXRα (NR2B1), RXRβ (NR2B2), and RXRγ (NR2B3). RXRα is detected in multiple tissues including muscle, liver, lung, in skin, intestine, epidermis and kidney, whereas RXRβ (NR2B2) is ubiquitously expressed (1, 7). Unlike RXRα and β, RXRγ expression pattern is less widely spread. Indeed, RXRγ is expressed specifically in brain, pituitary, and cardiac and skeletal muscles (1, 7). RXRs mediate retinoid signaling through the action of their ligand 9-cis retinoic acid (9 cis-RA) (8). In addition, it has been shown that poly-unsaturated fatty acids, such as docosahexaenoic acid (DHA), represent endogenous ligands for RXR in the mature brain (9). The transcriptional activity of RXR mainly results from its capacity to act as a cognate partner for other NRs. RXR can be generally engaged in 3 types of partnerships, permissive, conditional and non-permissive heterodimers. Non-permissive heterodimers, such as RXR/VDR (vitamin D receptor) and RXR/TR (thyroid hormone receptor), are activated only by agonists of the partner. Conditional heterodimers, such as RXR/RAR (retinoic acid receptor), are not activated by RXR agonists, but the activity of agonists of the RXR partner receptor is enhanced by RXR agonists (synergistic effect). RXR agonists alone, partner receptor agonists alone or a combination of both can activate permissive heterodimers. Such complexes include heterodimers formed with PPAR (peroxisome proliferator-activated receptor), FXR (farnesoid X receptor), LXR (liver X receptor), and the orphan NRs Nur77 and Nurr1 (10, 11).
The NR4A subgroup of nuclear receptors includes Nur77 (NR4A1, also known as NGFI-B or TR3), Nurr1 (NR4A2) and Nor-1 (NR4A3). NR4A members are immediate early response genes and can be induced by a diverse range of signals, including growth factors, cytokines, hormones and neurotransmitters (12, 13). Accordingly, the NR4A family is implicated in multiple functions including cell cycle regulation, apoptosis, steroidogenesis, inflammation, carcinogenesis, atherogenesis, insulin resistance, neoglucogenesis, lipid metabolism, endocrine functions and neurotransmission (12, 13). Nur77 and the others NR4A members can bind to DNA in three different ways to regulate target gene expression. First, these receptors can bind to the Nerve-Growth-Factor Inducible gene B (NGFI-B)-responsive element (NBRE) as monomers (14). Second, they can bind to the Nur-response element (NurRE) as homodimers or heterodimers with another NR4A family member (15, 16). In addition, as mentioned before, Nurr1 and Nur77, but not Nor-1, can form heterodimers with RXR, to mediate the retinoid signaling on direct repeat (DR) responsive elements or through binding to a NBRE sequence (16–18). Thus, Nur77 and Nurr1 transcriptional activities can be indirectly manipulated through modulation of their heterodimeric partner RXR, using RXR selective drugs, called rexinoids (16–18).
Nur family receptors are members of the orphan nuclear receptor class, i.e. no known endogenous ligand has been described for these receptors. In fact, the NR4A LBD contains no apparent cavity due to bulky hydrophobic amino acid side chains, and lacks a classical co-activator-binding cleft (19, 20). However, co-activator recruitment is possible via the AF-1 region of Nur receptors and, within the context of Nur-RXR heterodimers, via the AF-2 activation function of RXR (21). Further, some reports have evidenced ligand-induced NR4A activity, although the mechanism of activation has not been clearly demonstrated (22–24). In the periphery, numerous genes involved in endocrine and metabolic processes have NR4A responsive elements (mainly NBRE) in their promoters that have been validated experimentally (15, 25–27). However, activities of Nur77 and Nurr1 in heterodimer complexes with RXR are still poorly characterized. In the central nervous system, Nurr1 plays a critical role in the development and maintenance of the dopaminergic phenotype of mesencephalic neurons (substantia nigra), whereas Nurr1/RXR heterodimer can drive a pro-survival activity of RXR compounds on dopamine cells in culture (28, 29). In addition, Nur77 is associated with dopamine-mediated locomotor activity (30). Using Nur77 knockout mice and RXR drug treatments, we observed that dopamine receptor antagonist-induced abnormal involuntary movements (dyskinesia) are sensitive to rexinoids but only in wild-type animals (31). Thus, activity of both receptors can be associated with RXR heterodimer activity in the brain, but it is not possible at present time to selectively target this activity. Indeed, numerous RXR compounds have been identified, but their activity at various RXR heterodimer species is not well characterized, due to the lack of assays to monitor cofactor recruitment by specific dimers in live cells. Interpretation of results from reporter assays are complicated by the expression of endogenous receptors with overlapping DNA binding patterns, and by the variety of co-activator proteins present in the host cell.
Here we describe new highly sensitive sensors that can detect recruitment of a co-activator by specific receptor dimers in live cells and in real time. These assays combine the Bioluminescence Resonance Energy Transfer (BRET) and luciferase Protein fragment Complementation Assay (PCA) technologies (32–35) to enable detection of simultaneous interactions between three partners (NR homo- or hetero-dimer and a co-regulator). We applied these assays to characterize presently available RXR ligands for their intrinsic activity for co-activator LXXLL motif recruitment to specific homo-(RXR/RXR) or hetero-dimer (Nur77/RXR and Nurr1/RXR) complexes.
Materials and methods
Drugs
Nine-cis retinoic acid (9-cis RA) (RAR and RXR agonist), bexarotene (RXR agonist), docosahexaenoic acid (RXR agonist), 6-mercaptopurine (Nur77 agonist), 1,1-bis(3′-Indolyl)-1-(p-methoxyphenyl)methane; 3,3′-[(4-Methoxyphenyl)methylene]bis-1H-indole (DIM-C-pPhOCH3) (Nur77 agonist) and cytosporone B (Nur77 agonist) were purchased from Sigma-Aldrich Canada Inc. (Oakville, ON, Canada), whereas fluorobexarotene (RXR agonist), SR11237 (BMS 649) (RXR agonist), LG1506 (PPAR/RXR agonist), LG268 (RXR agonist) and UVI3003 (RXR antagonist) were purchased from Tocris (Ellisville, MI, USA). XCT0135908 was custom synthetized by the Medicinal Chemistry Platform of the Institut de Recherche en Immunologie et Cancérologie (IRIC) of the University of Montreal. HX600, HX630 (RXR agonists) and HX531 (RXR antagonist) were kindly provided by Dr. Hiroyuki Kagechika and Koichi Shudo (School of Biomedical Science, Tokyo Medical and Dental University and Research Foundation Itsuu Laboratory, Tokyo, Japan).
Plasmids
Human Nur77, Nurr1 and RXRγ cDNAs were amplified by PCR. PCR products were then subcloned into a pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA) containing the Renilla luciferase (Luc) sequence. We generated constructs in which the Luc was fused at the N- or C-terminal portion of the NR. For protein fragment complementation assay (PCA), we used split luciferase Luc-F1 (a.a. 1–110) and Luc-F2 (a.a. 111–310) fragments, as previously described (32, 33). Again, we generated constructs in which the Luc-F1 or -F2 fragment was fused at N- and C-terminal portions for each NR. The construct encoding the co-activator motif (CoA-YFP) consisted in a LXXLL motif tandem repeat flanked at both extremities by a Yellow Fluorescent Protein (YFP). The construct also contains two Nuclear Localisation Signal sequences derived from the glucocorticoid receptor to ensure nuclear transport of the construct. This sequence was then subcloned into the pCMV-TOPAZ expression vector (Clonetech, Mountain View, CA, USA). To confirm the specificity of NR and CoA-YFP motif, we used similar CoA-YFP motif constructs in which leucine residues were replaced by alanine (AXXAA) or phenylalanine residues (FXXFF).
Cell culture and Transient transfection
Human embryonic kidney 293T (HEK-293T) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Wisent, St-Bruno, QC, Canada) supplemented with 10% foetal bovine serum (FBS) (Sigma-Aldrich, Oakville, ON, Canada), 2 mL/500 mL glutamine, and 100 U/ml penicillin-streptomycin (Wisent, St-Bruno, QC, Canada) at 37°C in 5% CO2. We transiently transfected HEK-293T cells in 96-well plates (White Optiplate; PerkinElmer, Waltham, MA) at a density of 9 × 104 cells per well (about 80% confluency) with a maximum of 120 ng of cDNA per well encoding the fusion proteins using the polyethylenimine (PEI; Polysciences Inc, Warrington, PA, USA) method (300 ng of linear PEI plus 100 ng of branched PEI for 100 ng of DNA). Preliminary experiments were performed in order to determine the amounts of constructs, in both simple and PCA-BRET configurations, which generate optimal net BRET signals. So, in a typical PCA-BRET dose-response curve assay, 35 ng of each NR partners and 30 ng of the CoA-YFP construct are transfected per well in 96-well plates. To maintain the same ratio of DNA in co-transfections, we used the empty vector pcDNA3.1, to equilibrate the amount of total DNA transfected.
BRET and PCA-BRET assays
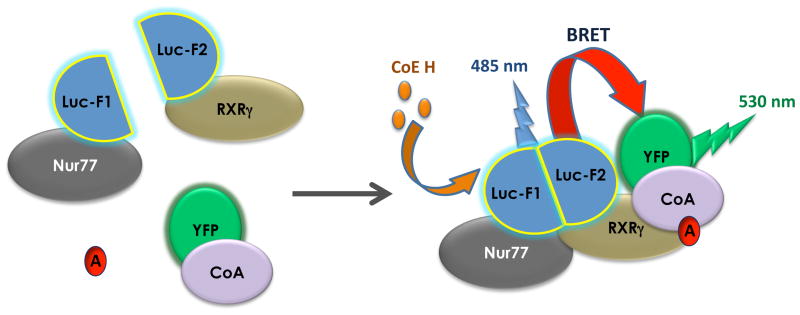
The BRET assays were performed essentially as described in previous reports (36–39). Adherent cells were stimulated by drugs for 20 min, 48 hours after transfection. All ligands used were solubilized in DMSO and then diluted in Hanks’ balanced salt solution (HBSS). BRET was read immediately after exposing the cells to 5 μM of the Luc substrate coelenterazine H (NanoLight Technology, Pinetop, AZ, USA) using the Mithras LB 940 microplate reader (Berthold Technologies, Oak Ridge, TN, USA). Signal detections were performed at a wavelength of 485 nm for Luc emission (energy donor) and at 530 nm for YFP emission (energy acceptor) (Fig. 1). Net BRET signals were derived from the emission detected with the energy acceptor filter divided by the emission detected with the energy donor filter (YFP/Luc), with subtraction of the background ratio obtained from cells expressing only the energy donor. In PCA-BRET experiments (see scheme in Fig. 1), BRET signals depend both on Luc fragment complementation due to receptor dimerization and on interaction of the Luc-fused proteins with at least one of the YFP-fused co-activator LXXLL motif (Fig. 1). This allows specific dimer activity measurements by fusion of receptor monomers with Luc fragments.
Figure 1. Schematic representation of the PCA-BRET assay.
In the protein fragment complementation assay (PCA-BRET), split Renilla Luciferase (Luc) fragments are fused to Nur77 and RXRγ, respectively, to generate Nur77-Luc-F1 (Nur77-F1) and RXRγ-Luc-F2 (RXRγ-F2) constructs. Upon hetero-dimerization, Luc fragments will complement and the reconstituted Luc will emit light in the presence of its substrate Coelenterazine H (CoE H) with an emission peak at 485 nm. In the presence of an agonist (A), the nuclear receptor complex will recruit the co-activator motif fused to the yellow fluorescent protein (CoA-YFP) and the energy will be transferred from the reconstituted Luc to the YFP, resulting in a second light emission peak at 530 nm. Since nuclear receptor dimerization is mandatory to trigger Luc reconstitution and initiate energy transfer, this assay detects ligand activation of a specific dimer.
Reporter assays
HEK-293T cells were transfected in 24-well plates by the calcium–phosphate co-precipitation method. Briefly, HEK-293T cells were plated at 150,000 cells/well prior to transfection. The next day, media were changed and cells were transfected with 0.6 μg of the DR-5 element from the RARβ promoter construct fused to the Firefly luciferase reporter gene (graciously provided by Dr. Jacques Drouin, IRCM), 100 ng of each transcription factor (Nur77, RXRγ, Nur77-F1 and/or F2-RXRγ), 100 ng pCMV-Topaz expression vector, used as an internal control, and pcDNA3.1 as carrier DNA to normalize transfected DNA quantity up to 1.5 μg/well, so that same amount of DNA was used in all experiments. One day later, HEK-293T cells were treated with either 1 μM 9-cis RA or with vehicle (DMSO, 0.001%), or increasing concentration of drugs for generation of dose-response curves for 24 h. Then, cells were harvested and luciferase activities were measured upon addition of luciferin (500 uM, NanoLight, Pinetop, AZ, USA) with the Veritas microplate luminometer (Turner Biosystems, Sunnyvale, CA, USA). Results were normalized using the Topaz signal with a Spectramax 190 plate reader (Molecular Devices, Sunnyvale, CA, USA). Data reported represent the average of at least three experiments, each performed in duplicate using different DNA preparations.
Results
Luciferase PCA assays for RXR dimer complex formation
In order to assess for co-activator (CoA) recruitment by specific dimers, we developed Renilla luciferase (Luc) protein complementation assays for receptor dimer formation. Reconstitution of Luc activity would then allow testing recruitment of a CoA LXXLL motif-containing peptide fused to two YFP acceptor proteins by BRET (Fig. 1). As both N-terminal and C-terminal fusions are possible for receptor monomers fused to either the Luc-F1 or Luc-F2 fragments, we first assessed the combinations that led to optimal Luc reconstitution.
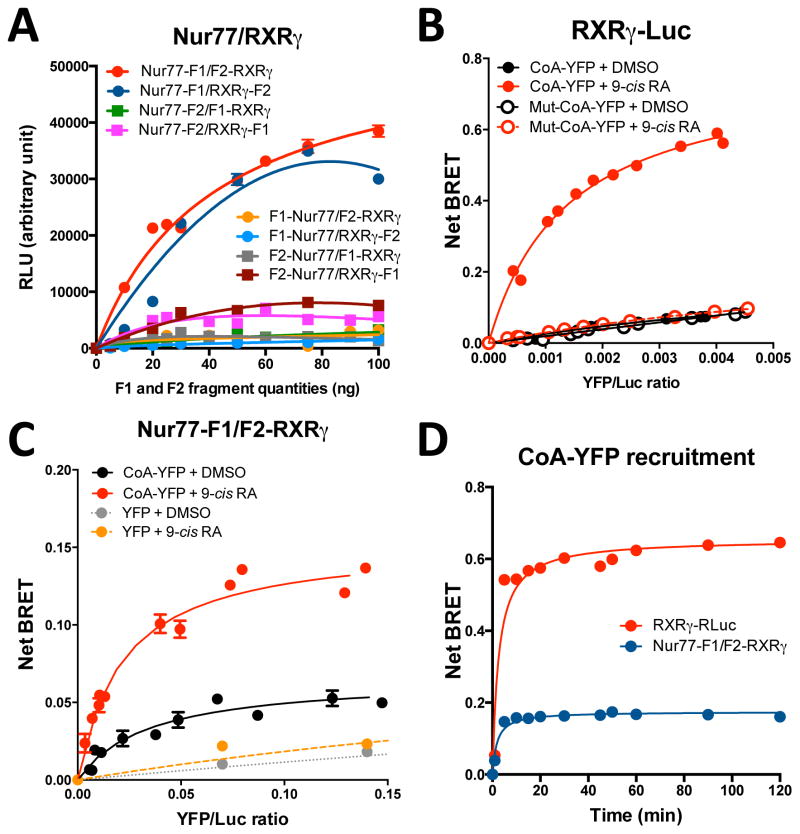
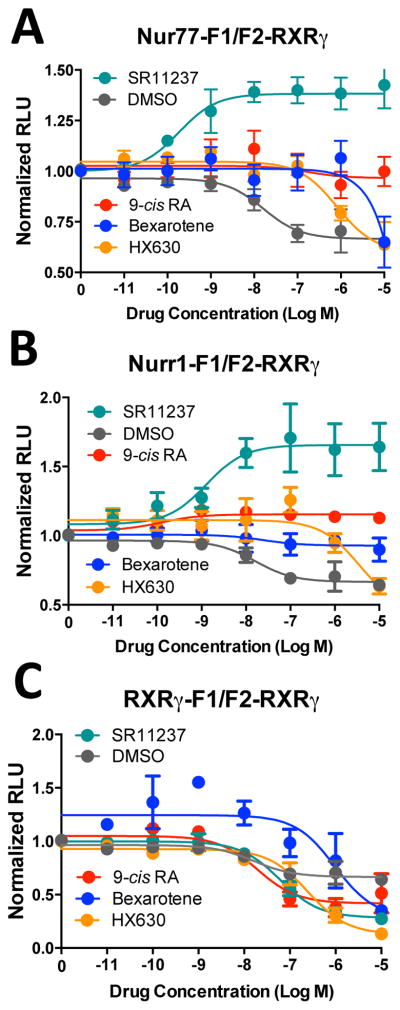
For the Nur77/RXRγ heterodimer, transient transfections of HEK-293T cells with the 8 possible combinations of F1 or F2 fragment fused at the N- or C-terminus of Nur77 or RXRγ were performed with increasing but equal amounts of both transfected plasmid (Fig. 2A). The results showed dose-dependent bioluminescent signals for multiple combinations, with optimal signals for the Nur77-F1/F2-RXRγ configuration (Fig. 2A), which was chosen for the following experiments. The Nur77-F1/RXRγ-F2 also yielded a good signal, but signals obtained with other pairs were 3–4 fold lower. Similarly, we also determined that the homodimeric RXRγ-F1/F2-RXRγ and heterodimeric Nurr1-F1/F2-RXRγ heterodimer configurations represent optimal fusion protein orientations for Luc reassembly (unpublished observations, X. Giner). Thus, Luc PCA assays enable live detection of RXRγ homodimers and of Nur77 or Nurr1 heterodimer formation with RXR.
Figure 2. A PCA assay for RXR dimer complexes.
(A) To identify the constructs that will give an optimal bioluminescent signal in the PCA-BRET assay, HEK-293T cells were transiently transfected with increasing amounts of the indicated nuclear receptors and split Renilla luciferase (Luc) constructs (combinations of Luc-F1 and -F2 fragments). Luc activity was measured 48 hours following transfection. Luc bioluminescent signals (relative light units, RLU) were measured upon coelenterazine H exposure and light detection at 485 nm. (B) In order to determine the specificity of the CoA recruitment of Nur77-F1/F2-RXRγ complex, HEK-293T cells were co-transfected with a constant amount of RXRγ-(RXRγ-Luc) (10 ng) and increasing amounts of the tagged co-activator motif (CoA-YFP) or a mutant version of the motif (Mut-CoA-YFP) (0 to 100 ng), in which leucine were replaced by alanine residues (AXXAA), in the presence of vehicle (DMSO) or RXR agonist 9-cis retinoic acid (9-cis RA, 1 μM). The YFP/Luc ratio represents the fluorescence emitted by the energy acceptor (Mut-CoA-YFP) over luminescence emitted by the energy donor (Nur77-F1/F2-RXRγ) (Fluorescence/Luminescence light emission). BRET signals were measured upon coelenterazine H exposure and light detection at 530 nm, 48 hours following transfection. (C) In the PCA-BRET assay, titrations of constant amounts of Nur77-F1 (35 ng) and F2-RXRγ (35 ng) constructs with increasing amounts of CoA-YFP or only the YFP protein (0 to 100 ng) in the presence of DMSO or 9-cis RA (1 μM) were performed. The YFP/Luc ratio represents the fluorescence emitted by the energy acceptor (CoA-YFP) over luminescence emitted by the energy donor (Nur77-F1/F2-RXRγ) (Fluorescence/Luminescence light emission). The curves were fitted using nonlinear regression analysis. (D) Comparison of RXRγ-Luc and Nur77-F1/F2-RXRγ co-activator recruitment kinetics. HEK-293T cells were treated 48h after transfection with 9-cis RA (1 μM). BRET (RXRγ-Luc) and PCA-BRET (Nur77-F1/F2-RXRγ) signals were measured from 0 to 120 min after addition of the Luc substrate coelenterazine H. Data represent means ± SEM of three experiments conducted in quadruplicata. BRET signals were normalized with the signal of each group treated with vehicle (DMSO). BRET signal generation is very fast and remains stable for up to 2 hours.
9-cis RA induces rapid co-activator recruitment to Nur-RXR heterodimers
BRET assays have been successfully used to monitor co-activator (CoA) peptide recruitment by activated NRs (39, 40). Here, we optimized BRET assays for CoA peptide recruitment by RXR. We tested constructs fusing RXRγ with Luc at its N- or C-terminus, and the LXXLL co-activator (CoA) motif with nuclear localization signals and YFP at both extremities in order to maximize the probability of energy transfer from NR-Luc complexes and to ensure nuclear localization (see supplementary Fig. S1 for sub-cellular localization). In BRET assays, we identified the RXRγ-Luc construct as being optimal for recruitment of CoA-YFP (unpublished observation, X. Giner).
In order to determine the optimal donor/acceptor ratio for signal detection, we performed titration curves in which a fixed amount of the energy donor (RXRγ-Luc or Nur77-F1/F2-RXRγ) was co-transfected with increasing amounts of the acceptor CoA-YFP motif. In a standard BRET assay (RXRγ-Luc and CoA-YFP), increasing amounts of CoA-YFP results in a linear curve, reflecting nonspecific energy transfer in the absence of ligand (Fig. 2B, treatment with DMSO vehicle). Similar results were obtained with a mutated, non-interacting CoA-YFP motif in which leucine residues were replaced by alanines, confirming the non-specific nature of these interactions. On the other hand, a strong and saturable increase in BRET signal was observed upon treatment with the RXR agonist 9-cis RA when using the wild type (LXXLL), but not the mutated (AXXAA) CoA-YFP motif (Fig. 2B). By plotting the net BRET signal in function of the amount of CoA-YFP plasmid transfected (see Fig. S2A), we estimate that the amount of CoA-YFP necessary to generate a BRET50 signal is 17 ng (with a RXRγ-Luc plasmid content fixed at 10 ng). We obtained a similar BRET50 value in the Nur77-F1/RXRγ-F2 PCA-BRET configuration (16 ng, Fig. S2B). For comparison, net BRET50 signals for CoA-YFP recruitment in the presence of DMSO are 51 and 55 ng for RXRγ-Luc and Nur77-F1/F2-RXRγ constructs, respectively (Fig. S2). Net BRET signal comparison of RXRγ-Luc and RXRγ-F1/F2-RXRγ assays also produced similar BRET50 values (Fig. S3A). However, Luc light emission at 485 nm is reduced by about 6-fold with the reconstituted split Luc (Fig. S3B). These data indicate that despite reduced Luc emission NR constructs containing the reconstituted split Luc are recruiting the co-activator with a similar affinity as the NR constructs containing the unsplit luciferase.
When using the Nur77-F1/F2-RXRγ complex as an energy donor, we observed basal CoA-YFP recruitment in the absence of ligand compared to unfused YFP (Fig. 2C). This suggests constitutive recruitment of the LXXLL motif-containing co-activators by the unliganted heterodimer. However, CoA-YFP motif recruitment was strongly upregulated upon addition of 9-cis RA (Fig. 2C). Co-transfection of unfused YFP generated identical linear, nonspecific signals in the presence as well as absence of 9-cis RA (Fig. 2C). Unexpectedly, basal recruitment (in the absence of 9-cis RA) of the mutated leucine/alanine CoA-YFP motif generated a BRET signal that is slightly superior to the CoA-YFP motif in basal condition, but this interaction was not affected by ligand addition (see supplementary Fig. S4A). These observations indicate that this mutated CoA motif is still able to interact with the NR complex, but that this interaction is not ligand-sensitive. Since we replaced leucine by alanine residues, which are structurally very close, it is possible that some constitutive interactions can still occur. In order to confirm the specificity of this CoA-YFP motif recruitment in the PCA-BRET configuration, we generated another mutant in which leucine were replaced by phenylalanine residues (which contain a larger side chain) (Fig. S4B). This mutant generated nonspecific signals both in basal and agonist-stimulated conditions, as compared to the native CoA-YFP motif, confirming the specificity of the agonist-induced interaction between the 3 partners.
We next compared kinetics of co-activator recruitment by RXRγ-Luc and Nur77-F1/F2-RXRγ (Fig. 2D). Both the BRET and PCA-BRET signals reached a plateau very fast (less than 5 min) after addition of 9-cis RA (10 μM) (Fig. 2D) and the net BRET signal remained stable for up to 2 hours. This indicates that Luc fragment complementation in the PCA-BRET assay did not reduce the velocity of CoA recruitment by the NR complex.
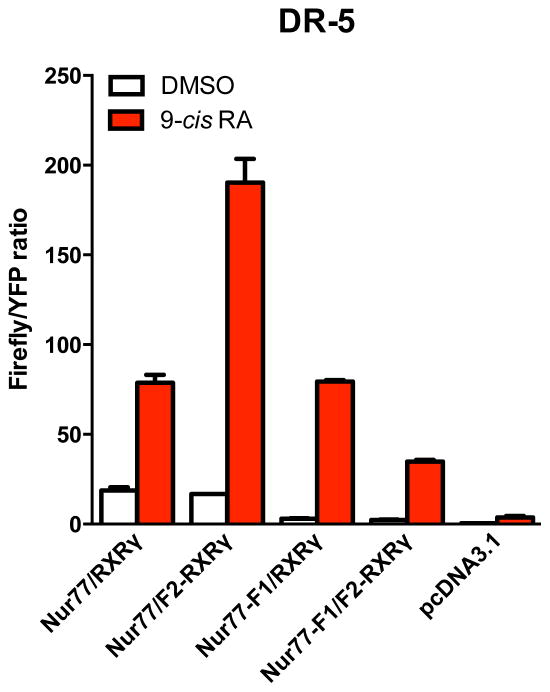
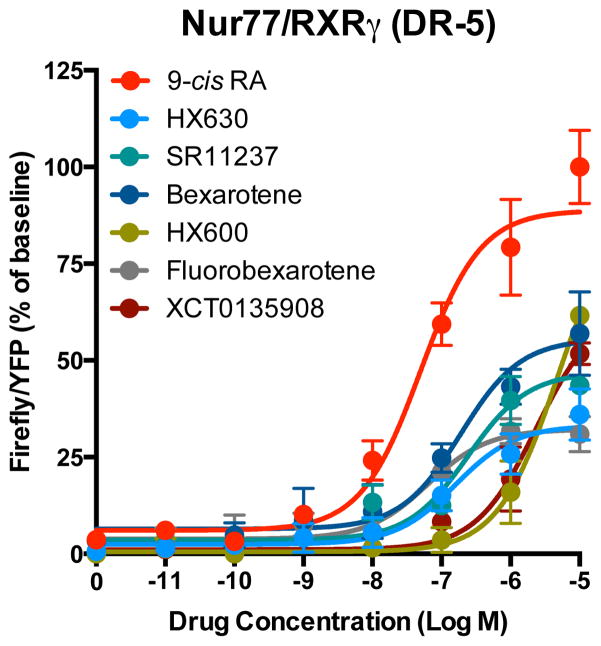
We also determined if the NR fused to Luc constructs are still able to transactivate a reporter gene using a classic direct repeat element that is sensitive to Nur77/RXR complex (DR-5) (Fig. 3). Although we noted a reduction of total firefly/YFP ratio signal with the Nur77-F1/F2-RXRγ constructs and basal transactivation activity with introduction of fused Luc fragments to Nur77 in the gene reporter assay, fusion proteins used for Luc PCA assays were able to transactivate a reporter gene through a classic DR-5 sequence in 9-cis RA-dependent manner (Nur77/RXRγ = 3.7 ± 0.4; Nur77/F2-RXRγ = 11.9 ± 0.5; Nur77-F1/RXRγ = 24.0 ± 2.0 and Nur77-F1/F2-RXRγ = 11.8 ± 2.0 fold induction) (Fig. 3).
Figure 3. Nuclear receptor-Luc fusion constructs remain transcriptionally active.
Histogram bars represent relative transcriptional activity of nuclear receptor constructs used to generate PCA-BRET sensor assays and native Nur77 and RXRγ upon exposure with DMSO or 9-cis RA (1 μM) at a previously characterized DR-5 response element in HEK-293T cells. As a control, cells were transfected with the same amount of pcDNA3.1 plasmid. Values represent mean Firefly/YFP ratios ± SEM (N = 3).
Differential activity of RXR ligands on specific RXR dimeric complexes
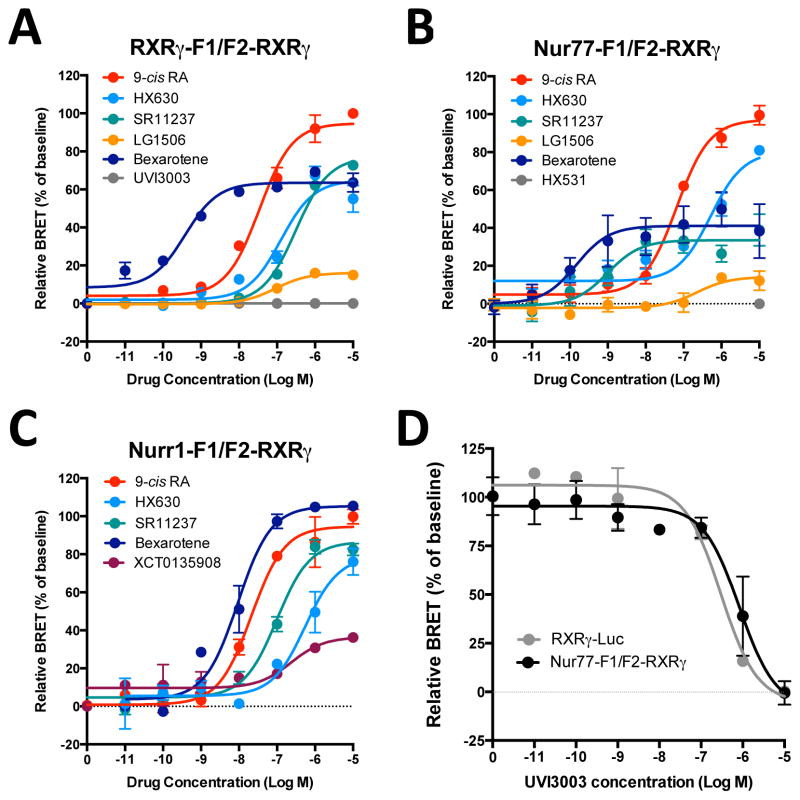
To compare the potency of known RXR ligands in the BRET sensor assays for specific RXR dimers, we performed dose-response curves (see examples in Fig. 4 and summarized data in Table 1). In the single RXRγ-Luc BRET assay, most drugs tested showed potencies (EC50) similar to values in the literature obtained using standard gene reporter assays for RXRγ, when available (Table 1). The natural agonist 9-cis RA displayed similar potencies in all BRET and PCA-BRET assays tested (Table 1). Interestingly however, although all rexinoids were active with each PCA-BRET assay, some displayed different potencies with various RXR dimeric complexes in the PCA-BRET assays (Table 1 and Fig. 4A–C). In particular, SR11237 had a markedly higher potency (5.6 nM) with the Nur77/RXRγ heterodimer assay than with the Nurr1/RXRγ heterodimer (144 nM) or the RXRγ/RXRγ homodimer (214 nM) in PCA-BRET assays (Table 1 and Fig. 4A–C). Of note, the EC50 observed in the RXR BRET assay is intermediate (30 nM). This suggests that SR11237 has selectivity for RXRγ bound to Nur77 vs Nurr1 and for Nur-RXR heterodimer vs RXRγ homodimer. This data also suggests potential contributions of other intracellular RXR partners present in the cell system when using the RXRγ-Luc BRET assay. The putative endogenous RXR ligand docosahexanoic acid (DHA) displayed a low micromolar activity (Table 1) in all the BRET and PCA-BRET assays tested, which is consistent with what is reported in the literature (9, 41). We also tested the activity of Nur ligands, including 6-mercaptopurine, DIM-C-pPhOCH3 and cytosporone B, in the present PCA-BRET assays. All the compounds remained inactive with RXRγ/RXRγ, Nur77/RXRγ and Nurr1/RXRγ complexes (unpublished observations, X. Giner).
Figure 4. Differential activity of RXR ligands on specific RXR dimeric complexes.
HEK-293T cells were co-transfected with RXRγ-F1 and F2-RXRγ (A), Nur77-F1 and F2-RXRγ (B) or Nurr1-F1 and F2-RXRγ (35 ng for all constructs) (C), and the CoA-YFP construct (30 ng). Cells were treated with increasing concentrations of the indicated RXR ligands for 20 min. BRET signals were measured at 530 nm immediately following addition of coelenterazine H. Data are expressed as percent (%) of basal net BRET signal (relative BRET). Each data point represents means ± SEM of quadruplicate. Each curve is representative of 3–5 independent experiments. Curves were fitted using nonlinear regression analysis to determine 50% effective concentration (EC50) and maximal capacity (Emax) of the compounds (pharmacological parameters can be found in Table 1). (D) For competition curves, HEK-293T cells were co-transfected with RXRγ-Luc or Nur77-F1 and F2-RXRγ, and the CoA-YFP construct. Cells were then treated with increasing concentrations of the RXR antagonist UVI3003 for 20 min in the presence of 9-cis RA at 50 nM. Data are expressed as percent (%) of baseline BRET signal and represent means ± SEM from 3 experiments conducted in quadruplicata. Curves were fitted using nonlinear regression analysis to determine 50% inhibitory concentration (IC50).
Table 1.
Pharmacological characterization of rexinoids for recruitment of a co-activator motif to RXRγ, RXRγ/RXRγ, Nur77/RXRγ Nurr1/RXRγ complexes in BRET and PCA-BRET assays.
| Drug | RXRγ-RLuc | RXRγ-F1/F2-RXRγ | Nur77-F1/F2-RXRγ | Nurr1-F1/F2-RXRγ | Literature a | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| EC50 (nM) | EMAX (%) | EC50 (nM) | EMAX (%) | EC50 (nM) | EMAX (%) | EC50 (nM) | EMAX (%) | EC50 (nM) | Ref. | |
| 9-cis RA | 22 ± 8 | 100 ± 1 | 40 ± 11 | 100 ± 2 | 27 ± 12 | 100 ± 2 | 26 ± 2 | 100 ± 2 | 10–150 | (41, 42, 51–53) |
| HX600 | 577 ± 164 | 56 ± 8 | 187 ± 13 | 33 ± 3 | 288 ± 151 | 57 ± 5 | ND | ND | 500–1000 | (42, 52) |
| HX630 | 727 ± 125 | 79 ± 7 | 966 ± 162 | 61 ± 2 | 584 ± 50 | 97 ± 12 | 356 ± 81 | 79 ± 1 | 620 | (42) |
| SR11237 | 30 ± 4 | 84 ± 4 | 214 ± 73 | 87 ± 7 | 5.6 ± 4.7 | 38 ± 8 | 144 ± 26 | 71 ± 5 | 232 | (54) |
| Bexarotene | 9 ± 7 | 67 ± 12 | 2 ± 1 | 70 ± 13 | 1.1 ± 0.5 | 50 ± 13 | 13 ± 6 | 94 ± 12 | 5–30 | (42, 54–56) |
| Fluorobexarotene | 0.7 ± 0.2 | 97 ± 13 | 5 ± 2 | 102 ± 14 | 1.5 ± 1.3 | 93 ± 7 | 8 ± 3 | 100 ± 10 | 43 | (57) |
| LG268 | 0.4 ± 0.2 | 95 ± 7 | 0.2 ± 0.1 | 75 ± 6 | 5.8 ± 5.7 | 69 ± 15 | 1.6 ± 1.2 | 38 ± 7 | 4–40 | (55, 58, 59) |
| XCT0135908 | ND | ND | 486 ± 19 | 18 ± 1 | 148 ± 90 | 31 ± 3 | 181 ± 54 | 32 ± 2 | NA | - |
| LG1506 | 196 ± 80 | 38 ± 4 | 76 ± 12 | 19 ± 3 | 93 ± 67 | 19 ± 5 | ND | ND | 4–11 | (58, 59) |
| DHA | > 5 000 | ND | > 5 000 | ND | > 5 000 | ND | ND | ND | >5000 | (9, 41) |
| HX531b | 2400 ± 200 | ND | > 5 000 | ND | > 5 000 | ND | ND | ND | 400–900 | (43, 44) |
| UVI3003b | 193 ± 37 | ND | 328 ± 160 | ND | 293 ± 63 | ND | ND | ND | ~300 | (45) |
EC50 or inhibitory constant (Ki) values were obtained by gene reporter assays for the RXRγ isoform from cited literature references;
values correspond to IC50 for 9-cis RA (50 nM) competition. Values represent means ± SEM from 3–5 independent experiments.
Abbreviations: EC50 = effective concentration at 50% of maximal activity; Emax = maximal response in percent (%) compared to 9-cis RA; NA, not available; ND = not determined; 9-cis RA = 9-cis retinoic acid; DHA = docosahexaenoic acid. Rexinoid pharmacological data can also be found in the in-depth review published by Dawson and Xia (60). EC50 > 5000 nM indicates that curve fitting did not converge at the concentration range used.
Of interest, some compounds had also different relative efficacies compared to 9-cis RA (Table 1 and Fig. 4A–C) with the dimer-specific assays versus the RXR BRET assay. LG1506, a selective PPAR/RXR ligand, displayed only low relative activities in the RXRγ/RXRγ homodimer or the Nurr77/RXRγ heterodimer PCA-BRET assay (19%), but higher partial activity in the RXRγ-Luc BRET assay (38% compared to 9-cis RA). HX630 displayed a partial relative maximal activity with the RXRγ/RXRγ homodimer (about 60%), which is consistent with the partial agonist activity reported in the literature (42), but it had near full relative maximal activity with the Nur77/RXRγ heterodimer selective PCA-BRET assay (see Fig. 4A,B and Table 1). On the other hand, SR11237, a pan RXR agonist, showed the opposite profile, with a partial maximal activity at the Nur77/RXRγ complex relative to 9-cis-RA (about 40%), while its maximal activity was comparable to that of 9-cis RA at RXRγ/RXRγ homodimer (see Fig. 4A,B and Table 1). Interestingly, bexarotene showed partial apparent efficacies except for Nurr1/RXRγ heterodimer to which it displayed full activity. Further, LG268 and HX600 have variable degrees of relative efficacy compared to 9-cis RA in the different PCA-BRET assays, whereas fluorobexarotene displayed full relative activity for all assays (Table 1 and Fig. 4A–C).
We also evaluated two RXR antagonists, HX531 and UVI3003, using 9-cis RA (50 nM) competition assays (Table 1 and Fig. 4D). Although the HX531 IC50 was somewhat lower, as compared to the literature (43, 44), IC50’s obtained with UVI3003 compound were similar in the BRET and PCA-BRET assays and are consistent with the literature (45) (Table 1 and Fig. 4D). Thus, these assays can also be used to monitor antagonist activities in competition assays.
For most rexinoids, relative transactivation efficacy data are not available in the literature. So, to directly compare the efficacy of co-activator recruitment as measured in the present Nur77/RXRγ PCA-BRET assay with transcriptional activation by the different rexinoids, we conducted a gene reporter assay using a DR-5 response element reporter construct previously described (16), combined with co-transfection of native Nur77 and RXRγ full-length proteins. All the compounds tested displayed an apparent partial maximal capacity compared to 9-cis RA (Fig. 5 and Table 2). Note that in a report by Lippert and colleagues (2009), SR11237 also displayed a partial maximal efficacy of 77% compared to 9-cis RA for a Gal4-RXRβ activation in Hela cells (46). Rexinoid potencies obtained in the reporter assay are generally somewhat lower compared to potencies obtained in Nur77/RXRγ PCA-BRET assays (compare Table 1 and Table 2 EC50 data), except for HX630 and XCT0123908 compounds, possibly due to the need for higher ligand concentrations to achieve transcriptional activation via full-length endogenous co-activators than for recruitment of overexpressed LXXLL motifs. Interestingly, SR11237 maintained its selectivity at Nur77/RXRγ over Nurr1/RXRγ using a DR-5 reporter gene assay combined with native Nurr1 and RXRγ (Nurr1/RXRγ EC50: 632 ± 207 nM and EMAX: 14 ± 4 %, as compared to EC50: 28 ± 9 nM and EMAX: 40 ± 10 % for Nur77/RXRγ, see Table II).
Figure 5. Rexinoid transactivation dose-response curves on a DR-5 element.
Dose-response curves were obtained by exposure to increasing concentrations of rexinoids in a gene reporter assay after co-transfection of Nur77, RXRγ and a reporter gene containing a DR-5 responsive element in HEK-293T cells. Data points represent means ± SEM expressed in percent of baseline Firefly/YFP ratios. These curves are representative of 3–5 independent experiments (pharmacological parameters can be found in Table 2).
Table 2.
Pharmacological characterization of rexinoids in a Nur77 + RXRγ gene reporter assay1.
| Drug | EC50 (nM) | Emax (%) |
|---|---|---|
| 9-cis RA | 60 ± 16 | 100 ± 4 |
| HX600 | 1180 ± 230 | 25 ± 9 |
| HX630 | 104 ± 33 | 30 ± 5 |
| SR11237 | 28 ± 9 | 40 ± 10 |
| Bexarotene | 45 ± 26 | 53 ± 10 |
| Fluorobexarotene | 28 ± 16 | 32 ± 4 |
| LG268 | 24 ± 23 | 44 ± 4 |
| XCT0135908 | 2980 ± 670 | 48 ± 15 |
for a DR-5 reporter element in HEK-293T cells. Values represent means ± SEM from 3–4 independent experiments. EC50 = effective concentration at 50% of maximal activity; Emax = maximal response in percent (%) compared to 9-cis RA.
Effect of rexinoids on dimer conformation
Differential relative maximal activity may reflect different capacity for co-activator recruitment (see above). Alternatively, specific conformational changes at different dimer constructs may alter the efficiency of energy transfer. The Luc PCA assay directly allows investigating the effect of drugs on protein conformation. The very fast recruitment of co-activator obtained in the PCA-BRET assays (Fig. 2D) is consistent with ligands inducing a conformational change in constitutively preformed homo- or hetero-dimers of NRs. To estimate the effect of agonists on NR complex conformation, we compared their impact on the Luc bioluminescent signals (Fig. 6). We observed that high concentrations of the vehicle (DMSO), as well as for some compounds like HX630 or bexarotene, reduced Luc bioluminescent signal (see examples in Fig. 6). This effect can be explained, at least in part, by nonspecific effects of DMSO at high concentrations on protein-protein interactions and by the fact that some of these compounds are colored, so that they can absorb a part of the emitted light. Note that while this reduction of Luc emission will also reduce fluorescence emission by the YFP acceptor, this effect has no impact on BRET results, which are expressed as YFP/Luc ratios.
Figure 6. Effect of rexinoids on dimer conformation.
HEK-293T cells were co-transfected with Nur77-F1 (A), Nurr1-F1 (B) or RXRγ-F1 (C), and F2-RXRγ. Cells were then treated with increasing concentrations of RXR ligands or vehicle (DMSO) for 20 min. Data represent means ± SEM from 3 independent experiments conducted in quadruplicata. Values are expressed as normalized relative light units (RLU) obtained at 485 nm.
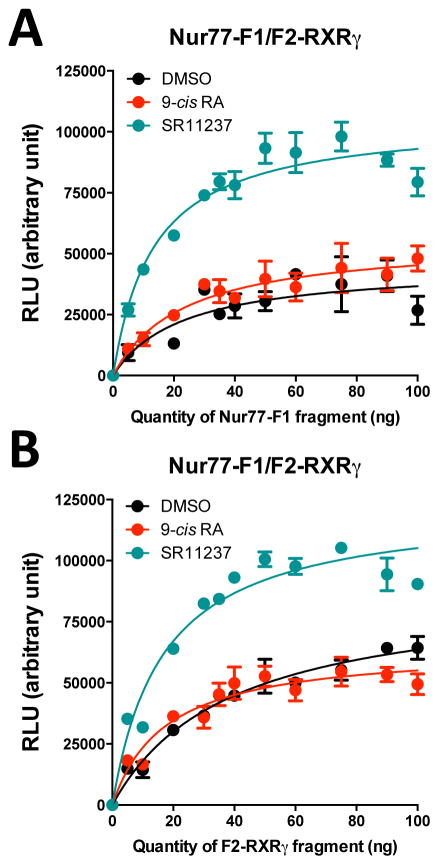
Interestingly, despite the fact that high concentration of DMSO reduce YFP emission, SR11237 was able to increase light emission of the reconstituted Luc compared to 9-cis RA in Nur77-F1/F2-RXRγ (9-cis RA = 1.03 ± 0.08 RLUmax; SR11237 = 1.41 ± 0.07 RLUmax, p < 0.01) and Nurr1-F1/F2-RXRγ (9-cis RA = 1.21 ± 0.06 RLUmax; SR11237 = 1.54 ± 0.15 RLUmax, p < 0.05), but not with the RXRγ-F1/F2-RXRγ (9-cis RA = 0.32 ± 0.08 RLUmax; SR11237 = 0.31 ± 0.07 RLUmax, NS) complex (Fig. 6A–C). This effect was confirmed in additional experiments, where we performed titration curves produced by increasing amounts of one Luc fragment of the Nur77-F1/F2-RXRγ complex in the presence of vehicle, 9-cis RA or SR11237 (Fig. 7). Both experimental conditions (increasing Nur77-F1 or F2-RXRγ fragment concentration) generated a higher light emission capacity in the presence of SR11237, as compared to 9-cis RA or vehicle (Fig. 7). However, plasmid concentrations at half-maximum light emission were similar (9-cis RA = 18 ± 4 ng; SR11237 = 16 ± 2 ng, Fig. 7B). This result suggests that SR11237 did not alter the affinity of NR complex formation, but instead induced a conformational change in the protein complex that facilitated Luc reconstitution. Interestingly, the relative potencies of these changes with the Nur77/RXR and Nurr1/RXR PCA assays mirrored those of co-activator recruitment in the PCA-BRET assays, suggesting that the variations in Luc PCA intensity detected are part of conformational rearrangements induced by SR11237 that lead to co-activator recruitment in the PCA-BRET assay.
Figure 7. Titration curves for Nur77-F1 and F2-RXRγ fragments.
Cells were co-transfected with a constant amount of F2-RXRγ (35 ng) and an increasing amount of Nur77-F1 (0 to 100 ng) (A) or a constant amount of Nur77-F1 (35 ng) and an increasing amount of F2-RXRγ (0 to 100 ng) (B) in the presence of vehicle (DMSO), SR11237 (1 μM) or 9-cis RA (1 μM) for 20 min. Data represent means ± SEM from 3 independent experiments conducted in triplicata. Values are expressed as relative light units (RLU) obtained at 485 nm. Curves were fitted using nonlinear regression analysis.
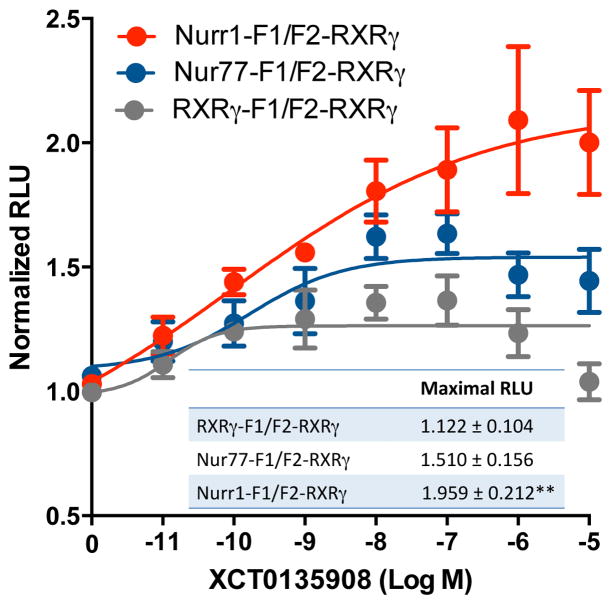
XCT0135908 has been shown to display a high selectivity for Nurr1/RXR heterodimer, as compared to other RXR complexes in Gal4-NR constructs co-transfected with full-length RXR in CV-1 cells (28). In the present Luc PCA BRET assays, XCT0135908 displayed similar potencies with Nur77-F1/F2-RXRγ and Nurr1-F1/F2-RXRγ, whereas potency with RXRγ-F1/F2-RXRγ was about 3-fold lower (Table 1). The compound also displayed higher apparent partial efficacies in Nur-F1/F2-RXRγ heterodimers as compared to the RXRγ-F1/F2-RXRγ complex (Table 1 and Fig. 4C). In addition, we observed a low micromolar transactivation activity and partial efficacy in our reporter assay for Nur77/RXRγ (Table 2 and Fig. 5). Interestingly, XCT0135908 increased light emission of the reconstituted Luc in Nurr1-F1/F2-RXRγ heterodimer (F2,20 = 6.074, p = 0.0087, N = 8; post hoc Tukey’s multiple comparisons test, p<0.01), but not RXRγ-F1/F2-RXRγ complexes (Fig. 8). The effect of XCT0135908 on the reconstitution of Luc activity with the Nur77-F1/F2-RXRγ heterodimer was of lower amplitude and did not reach statistical significance. Together, these data indicate that XCT0135908 induces selective conformational changes at the Nurr1/RXRγ complex and displays partial activation of Nur/RXR dimers compared to RXR/RXR homodimer.
Figure 8. XCT0135908 induces a distinct Nurr1/RXRγ complex conformation.
HEK-293T cells were co-transfected with Nur77-F1, Nurr1-F1 or RXRγ-F1, and F2-RXRγ. Cells were then treated with increasing concentrations of XCT0125908 for 20 min. Data represent means ± SEM from 5–8 independent experiments conducted in quadruplicata. Values are expressed as normalized relative light units (RLU) obtained at 485 nm.
Discussion
A central question in rexinoid biology and pharmacology research is the role and functions of RXR in specific heterodimers. In the present report, we have developed and optimized sensitive sensor assays to measure intrinsic ligand activity at specific NR dimer (homo- or hetero-dimer) complexes including RXRγ/RXRγ, Nur77/RXRγ and Nurr1/RXRγ. These novel assays monitor ligand-induced co-activator recruitment by a specific NR dimer complex activity in live cells. Although recent reports described BRET assays that are selective for nuclear receptor complexes (RXR/TRH, PPAR/RXR, RXR/RXR and Nurr1/RXR), BRET signals generated in these assays reflect receptor dimerization alone (47, 48). NR dimerization does not necessarily correlate with NR activity, as observed for instance with estrogen receptor agonists and antagonists (37). In the present assays, the BRET signal is generated by co-activator motif recruitment, while nuclear receptor selectivity is achieved by luciferase fragment complementation. Fusion proteins used for Luc PCA retain transcriptional activity and titration experiments can reveal ligand-induced effects on the conformation of specific RXR-containing complexes. For instance, SR11237 and XCT0135908 were unique among tested compounds in their ability to induce specific conformations of Nur/RXRγ heterodimers. Interestingly, the selectivity of SR11237 (both potency and efficacy) for Nur77/RXR complex, as evidence in the present PCA-BRET assay, was confirmed in a classic DR-5 reporter assay for Nur77/RXR and Nurr1/RXR complexes.
Pharmacological characterization of rexinoids in these assays confirmed activity of most rexinoids on the different RXR dimers, but revealed previously uncharacterized preferential activity of SR11237 (BMS649) on the Nur77/RXR heterodimer, which was activated with higher potency compared to Nurr1/RXR heterodimer and RXR/RXR homodimer. This suggests that the conformational changes observed in Luc PCA assays with SR11237, also with higher potency for Nur77/RXR vs Nurr1/RXR, reflect the activation of the dimers for recruitment of LXXLL motif peptides. High-resolution crystallography comparing RXR-9-cis RA and RXR-SR11237 complexes structural data indicated that the RXR-SR11237 complex can undergo conformational fluctuation attributed to the reorientation of one single side-chain residue (N306) that is sufficient to reduce the volume of the cavity by up to 10% (49). This side chain motion that generates additional stabilizing contacts is most probably the result of an attraction by the partial negative charge of the SR11237 oxygen atoms. As a result of the change of volume, the level of occupancy of the ligand binding pocket by the SR11237 ligand is higher than that with 9-cis RA (49). Thus, it is tempting to speculate that this additional interaction of SR11237 with RXR might confer a greater stability to selective heterodimer complexes and might be responsible for the higher apparent potency of SR11237 to recruit co-activator motif at Nur77/RXRγ (and to a lesser extent Nurr1/RXRγ), compared to the RXRγ/RXRγ complex.
Using our dimer-specific PCA-BRET assays, it is also possible to perform competition of agonist-induced co-activator recruitment by antagonists for specific NR homo- or hetero-dimer complex. Here we show that two previously characterized RXR antagonists, HX531 and UVI3003, can directly compete for 9-cis RA co-activator motif recruitments with IC50 values similar to those obtained for their antagonist activities in gene reporter transactivation assays (43–45). Both antagonists displayed similar activities at RXR/RXR and Nur77/RXR complexes (Table 1).
Nur77 compounds were found to be inactive in the Nur77/RXRγ PCA-BRET assay, suggesting that Nur77 remains silent for co-activator motif recruitment by Nur77/RXR complex. A similar suggestion has been made previously for Nurr1/RXR complex (28). We are aware however that co-activator recruitment via other determinants than the LXXLL binding motifs assessed in this study remains possible in the presence of these compounds.
Although most pharmacological parameters were compatible with published data obtained in reporter assays, discrepancies were noted between pharmacological parameters obtained in single RXR-Luc BRET assay and in dimer specific PCA-BRET assays. These might result from endogenously expressed components that can dimerize with RXR-Luc, as suggested by the use of PPAR selective ligands (Table 1). Similar limitations apply for reporter assays, although specificity is enhanced by the use of a specific binding motif that can recruit only certain types of dimers. On the other hand, a current limitation of the BRET assays is the assessment of recruitment of isolated LXXLL co-activator motif peptides rather than native co-activators, and may also explain differences in potency or efficacy between the two systems. In future developments of this technology, receptor interacting domains for different co-activators could also be tested alternatively to isolated LXXLL motifs.
In summary, direct measurements of co-activator recruitment potency and efficacy using PCA-BRET sensor assays provides rapid and sensitive measures of intrinsic pharmacological properties of ligands at specific NR complexes in live cells that can be complemented by observation of conformational changes in the PCA assays. These assays possess many advantages compared to presently available assays that monitor NR activity. Short exposure to ligands (20–30 min) minimizes interference caused by the toxicity of some compounds in assays requiring longer incubations. In addition, BRET-based assays do not require light excitation as opposed to fluorescence resonance energy transfer (FRET)-based assays, and so are not subjected to quenching or autofluorescence. They generate low background signals and high signal-to-noise ratios. The low variability and high reproducibility of PCA-BRET assays using spectrometric detection are important assets for future amenability of these assays to high throughput screening.
A new generation of selective modulators of heterodimers, such as LG1506, has been developed that separate the physiological activities of the RXR and its partner receptor and might be considered part of the so-called specific NR modulators group (50). However, no precise structural rules have yet emerged to guide chemical engineering of heterodimer-specific ligands for drug design. The present technology should prove useful to identify new compounds with specificity for individual dimeric species formed by RXRs.
Supplementary Material
Acknowledgments
We thank Drs Hyroyuki Kagechika and Koichi Shudo (School of Biomedical Science, Tokyo Medical and Dental University and Research Foundation Itsuu Laboratory, Tokyo, Japan) for generously providing HX600, HX630 and HX531 compounds and Dr Jacques Drouin (Institut de Recherche Clinique de Montréal, IRCM) for βRARE DR-5 element plasmid construct. We also thank Dr Michel Bouvier’s Lab members at the Institut de Recherche en Immunologie et Cancérologie (IRIC) of the University of Montreal for helpful discussions for BRET-based assay optimizations. This work was supported by a grant from the “Groupe de Recherche Universitaire sur le Médicament (GRUM)” at Université de Montréal to DL and by a grant from the Canadian Institute for Health Research to SM.
List of abbreviations
- 9-cis RA
9-cis retinoic acid
- AF-1
N-terminal transcription activation function domain
- AF-2
C-terminal transcription activation function domain
- BRET
bioluminescence resonance energy transfer
- CoA
co-activator
- DBD
DNA-binding domain
- DHA
docosahexaenoic acid
- DR
direct repeat
- LBD
ligand-binding domain
- Luc
luciferase
- NBRE
NGFI-B responsive element
- NGFI-B
nerve-growth factor inhibitor gene B
- NR
nuclear receptor
- PCA
protein fragment complementation assay
- PPAR
peroxisome proliferator-activated receptor
- RAR
retinoic acid receptor
- RXR
retinoid X receptor
- YFP
yellow fluorescent protein
Footnotes
Disclosure
The authors declare no conflict of interest.
References
- 1.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 3.Gurevich I, Flores AM, Aneskievich BJ. Corepressors of agonist-bound nuclear receptors. Toxicol Appl Pharmacol. 2007;223:288–298. doi: 10.1016/j.taap.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 5.Burris TP, Solt LA, Wang Y, Crumbley C, Banerjee S, Griffett K, Lundasen T, Hughes T, Kojetin DJ. Nuclear receptors and their selective pharmacologic modulators. Pharmacol Rev. 2013;65:710–778. doi: 10.1124/pr.112.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Bruemmer D. NR4A Orphan Nuclear Receptors in Cardiovascular Biology. Drug Discov Today Dis Mech. 2009;6:e43–e48. doi: 10.1016/j.ddmec.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol Rev. 2006;58:685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- 8.Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 9.Mata de Urquiza A, Liu S, Sjöberg M, Zetterström RH, Griffiths W, Sjövall J, Perlmann T. Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science. 2000;290:2140–2144. doi: 10.1126/science.290.5499.2140. [DOI] [PubMed] [Google Scholar]
- 10.Forman BM, Umesono K, Chen J, Evans RM. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 11.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 12.Maxwell MA, Muscat GEO. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2005;4:e002. doi: 10.1621/nrs.04002. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Safe S, Jin UH, Hedrick E, Reeder A, Lee SO. Minireview: role of orphan nuclear receptors in cancer and potential as drug targets. Mol Endocrinol. 2014;28:157–172. doi: 10.1210/me.2013-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson TE, Fahrner TJ, Johnston M, Milbrandt J. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science. 1991;252:1296–1300. doi: 10.1126/science.1925541. [DOI] [PubMed] [Google Scholar]
- 15.Philips A, Lesage S, Gingras R, Maira MH, Gauthier Y, Hugo P, Drouin J. Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Mol Cell Biol. 1997;17:5946–5951. doi: 10.1128/mcb.17.10.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maira M, Martens C, Philips A, Drouin J. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol Cell Biol. 1999;19:7549–7557. doi: 10.1128/mcb.19.11.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perlmann T, Jansson L. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev. 1995;9:769–782. doi: 10.1101/gad.9.7.769. [DOI] [PubMed] [Google Scholar]
- 18.Zetterström RH, Solomin L, Mitsiadis T, Olson L, Perlmann T. Retinoid X receptor heterodimerization and developmental expression distinguish the orphan nuclear receptors NGFI-B, Nurr1, and Nor1. Mol Endocrinol. 1996;10:1656–1666. doi: 10.1210/mend.10.12.8961274. [DOI] [PubMed] [Google Scholar]
- 19.Flaig R, Greschik H, Peluso-Iltis C, Moras D. Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain. J Biol Chem. 2005;280:19250–19258. doi: 10.1074/jbc.M413175200. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, Walker NP, Perlmann T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- 21.Wansa KDSA, Harris JM, Muscat GE. The activation function-1 domain of Nur77/NR4A1 mediates trans-activation, cell specificity, and coactivator recruitment. J Biol Chem. 2002;277:33001–33011. doi: 10.1074/jbc.M203572200. [DOI] [PubMed] [Google Scholar]
- 22.Zhan Y, Du X, Chen H, Liu J, Zhao B, Huang D, Li G, Xu Q, Zhang M, Weimer BC, Chen D, Cheng Z, Zhang L, Li Q, Li S, Zheng Z, Song S, Huang Y, Ye Z, Su W, Lin SC, Shen Y, Wu Q. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat Chem Biol. 2008;4:548–556. doi: 10.1038/nchembio.106. [DOI] [PubMed] [Google Scholar]
- 23.Dubois C, Hengerer B, Mattes H. Identification of a potent agonist of the orphan nuclear receptor Nurr1. ChemMedChem. 2006;1:955–958. doi: 10.1002/cmdc.200600078. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Lee SO, Safe S. Structure-dependent activation of NR4A2 (Nurr1) by 1,1-bis(3′-indolyl)-1-(aromatic)methane analogs in pancreatic cancer cells. Biochem Pharmacol. 2012;83:1445–1155. doi: 10.1016/j.bcp.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chao LC, Zhang Z, Pei L, Saito T, Tontonoz P, Pilch PF. Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol Endocrinol. 2007;21:2152–2163. doi: 10.1210/me.2007-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei LM, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med. 2006;12:1048–1055. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- 27.Martin LJ, Tremblay JJ. Nuclear receptors in Leydig cell gene expression and function. Biol Reprod. 2010;83:3–14. doi: 10.1095/biolreprod.110.083824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallen-Mackenzie A, de Urquiza AM, Petersson S, Rodriguez FJ, Friling S, Wagner J, Ordentlich P, Lengqvist J, Heyman RA, Arenas E, Perlmann T. Nurr1-RXR heterodimers mediate RXR ligand-induced signaling in neuronal cells. Genes & Development. 2003;17:3036–3047. doi: 10.1101/gad.276003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadkhodaei B, Ito T, Joodmardi E, Mattsson B, Rouillard C, Carta M, Muramatsu S, Sumi-Ichinose C, Nomura T, Metzger D, Chambon P, Lindqvist E, Larsson NG, Olson L, Bjorklund A, Ichinose H, Perlmann T. Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J Neurosci. 2009;29:15923–15932. doi: 10.1523/JNEUROSCI.3910-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lévesque D, Rouillard C. Nur77 and retinoid X receptors: crucial factors in dopamine-related neuroadaptation. Trends Neurosci. 2007;30:22–30. doi: 10.1016/j.tins.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ethier I, Kagechika H, Shudo K, Rouillard C, Lévesque D. Docosahexaenoic acid reduces haloperidol-induced dyskinesias in mice: Involvement of Nur77 and retinoid receptors. Biol Psychiatry. 2004;56:522–526. doi: 10.1016/j.biopsych.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 32.Stefan E, Aquin S, Berger N, Landry CR, Nyfeler B, Bouvier M, Michnick SW. Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo. Proc Natl Acad Sci USA. 2007;104:16916–16921. doi: 10.1073/pnas.0704257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paulmurugan R, Gambhir SS. Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal Chem. 2003;75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 35.Martel C, Dugre-Brisson S, Boulay K, Breton B, Lapointe G, Armando S, Trepanier V, Duchaine T, Bouvier M, Desgroseillers L. Multimerization of Staufen1 in live cells. RNA. 2010;16:585–597. doi: 10.1261/rna.1664210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llopis J, Westin S, Ricote M, Wang Z, Cho CY, Kurokawa R, Mullen TM, Rose DW, Rosenfeld MG, Tsien RY, Glass CK. Ligand-dependent interactions of coactivators steroid receptor coactivator-1 and peroxisome proliferator-activated receptor binding protein with nuclear hormone receptors can be imaged in live cells and are required for transcription. Proc Natl Acad Sci USA. 2000;97:4363–4368. doi: 10.1073/pnas.97.8.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lupien M, Jeyakumar M, Hebert E, Hilmi K, Cotnoir-White D, Loch C, Auger A, Dayan G, Pinard GA, Wurtz JM, Moras D, Katzenellenbogen J, Mader S. Raloxifene and ICI182,780 increase estrogen receptor-α association with a nuclear compartment via overlapping sets of hydrophobic amino acids in activation function 2 helix 12. Mol Endocrinol. 2007;21:797–816. doi: 10.1210/me.2006-0074. [DOI] [PubMed] [Google Scholar]
- 38.Pfleger KD, Eidne KA. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET) Nat Methods. 2006;3:165–174. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- 39.Duplessis TT, Koterba KL, Rowan BG. Detection of ERα-SRC-1 interactions using bioluminescent resonance energy transfer. Methods Mol Biol. 2009;590:253–263. doi: 10.1007/978-1-60327-378-7_16. [DOI] [PubMed] [Google Scholar]
- 40.Koterba KL, Rowan BG. Measuring ligand-dependent and ligand-independent interactions between nuclear receptors and associated proteins using Bioluminescence Resonance Energy Transfer (BRET) Nucl Recept Signal. 2006;4:e021. doi: 10.1621/nrs.04021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldstein JT, Dobrzyn A, Clagett-Dame M, Pike JW, DeLuca HF. Isolation and characterization of unsaturated fatty acids as natural ligands for the retinoid-X receptor. Arch Biochem Biophys. 2003;420:185–193. doi: 10.1016/j.abb.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 42.Umemiya H, Fukasawa H, Ebisawa M, Eyrolles L, Kawachi E, Eisenmann G, Gronemeyer H, Hashimoto Y, Shudo K, Kagechika H. Regulation of retinoidal actions by diazepinylbenzoic acids. Retinoid synergists which activate the RXR-RAR heterodimers. J Med Chem. 1997;40:4222–4234. doi: 10.1021/jm9704309. [DOI] [PubMed] [Google Scholar]
- 43.Morishita K, Yakushiji N, Ohsawa F, Takamatsu K, Matsuura N, Makishima M, Kawahata M, Yamaguchi K, Tai A, Sasaki K, Kakuta H. Replacing alkyl sulfonamide with aromatic sulfonamide in sulfonamide-type RXR agonists favors switch towards antagonist activity. Bioorg Med Chem Lett. 2009;19:1001–1003. doi: 10.1016/j.bmcl.2008.11.086. [DOI] [PubMed] [Google Scholar]
- 44.Sakaki J, Konishi K, Kishida M, Gunji H, Kanazawa T, Uchiyama H, Fukaya H, Mitani H, Kimura M. Synthesis and structure-activity relationship of RXR antagonists based on the diazepinylbenzoic acid structure. Bioorg Med Chem Lett. 2007;17:4808–4811. doi: 10.1016/j.bmcl.2007.06.079. [DOI] [PubMed] [Google Scholar]
- 45.Nahoum V, Perez E, Germain P, Rodriguez-Barrios F, Manzo F, Kammerer S, Lemaire G, Hirsch O, Royer CA, Gronemeyer H, de Lera AR, Bourguet W. Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function. Proc Natl Acad Sci USA. 2007;104:17323–17328. doi: 10.1073/pnas.0705356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lippert WP, Burschka C, Gotz K, Kaupp M, Ivanova D, Gaudon C, Sato Y, Antony P, Rochel N, Moras D, Gronemeyer H, Tacke R. Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology. ChemMedChem. 2009;4:1143–1152. doi: 10.1002/cmdc.200900090. [DOI] [PubMed] [Google Scholar]
- 47.Mulero M, Perroy J, Federici C, Cabello G, Ollendorff V. Analysis of RXR/THR and RXR/PPARG2 heterodimerization by bioluminescence resonance energy transfer (BRET) PLoS One. 2013;8:e84569. doi: 10.1371/journal.pone.0084569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McFarland K, Spalding TA, Hubbard D, Ma JN, Olsson R, Burstein ES. Low dose bexarotene treatment rescues dopamine neurons and restores behavioral function in models of Parkinson’s disease. ACS Chem Neurosci. 2013;4:1430–1438. doi: 10.1021/cn400100f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egea PF, Mitschler A, Moras D. Molecular Recognition of Agonist Ligands by RXRs. Mol Endocrinol. 2002;16:987–997. doi: 10.1210/mend.16.5.0823. [DOI] [PubMed] [Google Scholar]
- 50.de Lera AR, Bourguet W, Altucci L, Gronemeyer H. Design of selective nuclear receptor modulators: RAR and RXR as a case study. Nat Rev Drug Discov. 2007;6:811–820. doi: 10.1038/nrd2398. [DOI] [PubMed] [Google Scholar]
- 51.Farmer LJ, Marron KS, Canan Koch SS, Hwang CK, Kallel EA, Zhi L, Nadzan AM, Robertson DW, Bennani YL. Aza-retinoids as novel retinoid X receptor-specific agonists. Bioorg Med Chem Lett. 2006;16:2352–2356. doi: 10.1016/j.bmcl.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 52.Morita K, Kawana K, Sodeyama M, Shimomura L, Kagechika H, Makishima M. Selective allosteric ligand activation of the retinoid X receptor heterodimers of NGFI-B and Nurr1. Biochem Pharmacol. 2005;71:98–107. doi: 10.1016/j.bcp.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 53.Boehm MF, McClurg MR, Pathirana C, Mangelsdorf D, White SK, Hebert J, Winn D, Goldman ME, Heyman RA. Synthesis of high specific activity [3H]-9-cis-retinoic acid and its application for identifying retinoids with unusual binding properties. J Med Chem. 1994;37:408–414. doi: 10.1021/jm00029a013. [DOI] [PubMed] [Google Scholar]
- 54.Takamatsu K, Takano A, Yakushiji N, Morishita K, Matsuura N, Makishima M, Ali HI, Akaho E, Tai A, Sasaki K, Kakuta H. Reduction of lipophilicity at the lipophilic domain of RXR agonists enables production of subtype preference: RXRalpha-preferential agonist possessing a sulfonamide moiety. ChemMedChem. 2008;3:454–460. doi: 10.1002/cmdc.200700265. [DOI] [PubMed] [Google Scholar]
- 55.Boehm MF, Zhang L, Zhi L, McClurg MR, Berger E, Wagoner M, Mais DE, Suto CM, Davies JA, Heyman RA, Nadzan AM. Design and synthesis of potent retinoid X receptor selective ligands that induce apoptosis in leukemia cells. J Med Chem. 1995;38:3146–3155. doi: 10.1021/jm00016a018. [DOI] [PubMed] [Google Scholar]
- 56.Boehm MF, Zhang L, Badea BA, White SK, Mais DE, Berger E, Suto CM, Goldman ME, Heyman RA. Synthesis and structure-activity relationships of novel retinoid X receptor-selective retinoids. J Med Chem. 1994;37:2930–2941. doi: 10.1021/jm00044a014. [DOI] [PubMed] [Google Scholar]
- 57.Wagner CE, Jurutka PW, Marshall PA, Groy TL, van der Vaart A, Ziller JW, Furmick JK, Graeber ME, Matro E, Miguel BV, Tran IT, Kwon J, Tedeschi JN, Moosavi S, Danishyar A, Philp JS, Khamees RO, Jackson JN, Grupe DK, Badshah SL, Hart JW. Modeling, synthesis and biological evaluation of potential retinoid X receptor (RXR) selective agonists: novel analogues of 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)ethynyl]benzoic acid (bexarotene) J Med Chem. 2009;52:5950–5966. doi: 10.1021/jm900496b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leibowitz MD, Ardecky RJ, Boehm MF, Broderick CL, Carfagna MA, Crombie DL, D’Arrigo J, Etgen GJ, Faul MM, Grese TA, Havel H, Hein NI, Heyman RA, Jolley D, Klausing K, Liu S, Mais DE, Mapes CM, Marschke KB, Michellys PY, Montrose-Rafizadeh C, Ogilvie KM, Pascual B, Rungta D, Tyhonas JS, Urcan MS, Wardlow M, Yumibe N, Reifel-Miller A. Biological characterization of a heterodimer-selective retinoid X receptor modulator: potential benefits for the treatment of type 2 diabetes. Endocrinology. 2006;147:1044–1053. doi: 10.1210/en.2005-0690. [DOI] [PubMed] [Google Scholar]
- 59.Michellys PY, D’Arrigo J, Grese TA, Karanewsky DS, Leibowitz MD, Mais DA, Mapes CM, Reifel-Miller A, Rungta D, Boehm MF. Design, synthesis and structure-activity relationship of novel RXR-selective modulators. Bioorg Med Chem Lett. 2004;14:1593–1598. doi: 10.1016/j.bmcl.2003.12.089. [DOI] [PubMed] [Google Scholar]
- 60.Dawson MI, Xia Z. The retinoid X receptors and their ligands. Biochim Biophys Acta. 2012;1821:21–56. doi: 10.1016/j.bbalip.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.