Abstract
Accumulating evidence indicates many brain functions are mediated by epigenetic regulation of neural genes, and their dysregulations result in neuronal disorders. Experiences such as learning and recall, as well as physical exercise, induce neuronal activation through epigenetic modifications and by changing the noncoding RNA profiles. Animal models, brain samples from patients, and the development of diverse analytical methods have broadened our understanding of epigenetic regulation in the brain. Diverse and specific epigenetic changes are suggested to correlate with neuronal development, learning and memory, aging and age-related neuronal diseases. Although the results show some discrepancies, a careful comparison of the data (including methods, regions and conditions examined) would clarify the problems confronted in understanding epigenetic regulation in the brain.
Keywords: Epigenetics, Learning and memory, Neurodegenerative diseases
INTRODUCTION
The term ‘epigenetics’ was initially used in the context of ‘beyond genetics’, to describe a phenomenon in development that could not be explained by genetics. However, it is currently used to describe gene expression changes that occur without changes in the DNA sequence (1). We now understand many components underpinning the epigenetic regulatory system, and this knowledge is widely applied in cancer studies to understand its mechanism of regulation (2). The field of epigenetic research has expanded, and for the last two decades there has been a corresponding progress in the field of neuroscience (3). It is not surprising that epigenetic regulation constitutes a large portion of the regulatory mechanism for brain function, as de novo transcription is required for the consolidation and reconsolidation of memory, which is one of the main functions of the brain (4, 5). When training stimuli is received, mRNA transcription and protein synthesis are required for structural changes in neurons, to establish synaptic plasticity and long-term memory storage (6). Chromatin, DNA, and RNA modifications constitute the majority of epigenetic regulations, and play important roles in regulating transcription and translation. Indeed, activity-dependent neuronal histone modifications have been found to regulate the learning and memory process (7). Mutations in lysine acetyltransferases (KATs) result in memory impairments in both mice and humans, whereas reduction of histone deacetylases (HDACs) by HDAC inhibitors enhances memory (8). The increasing relevance of epigenetics to neuroscience encompasses brain development, neuronal differentiation, cognition, neurodevelopmental disorders, psychiatric disorders, and neurodegenerative diseases (9, 10). However, due to space limitations, our main focus in this review will be the aging-dependent epigenetic mechanisms in the brain and in neurodegenerative diseases. Because epigenetic regulations are most extensive at the level of histones, DNA modifications, and non-coding RNAs, we will focus on these modifications during age-related physiological changes in the brain.
EPIGENETIC REGULATION FOR NORMAL BRAIN FUNCTION
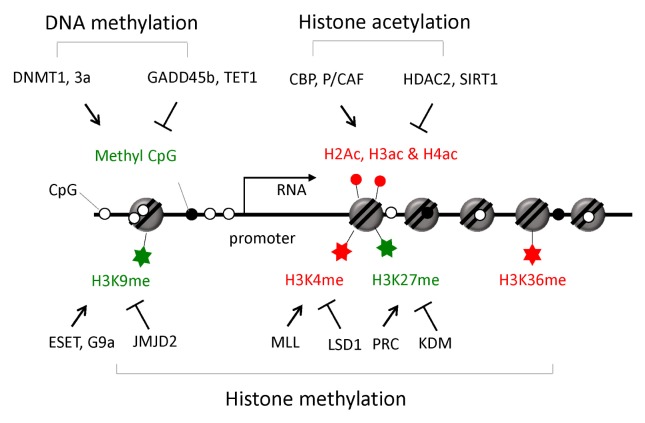
For proper homeostasis and functioning of the adult brain, a balanced control of neural stem/progenitor cell self-renewal, differentiation, production of neurons and glia (known as “adult neurogenesis”), repair, learning, and memory, are important (11). These neuronal processes accompany dynamic patterns of gene expression, and epigenetic control has emerged to have critical roles in generating specific gene expression patterns (Fig. 1). Generally, DNA methylation silences gene expression by preventing the formation of transcriptional machinery at the target sites. DNA methylation is catalyzed by DNA methyltransferases (DNMTs), and deletion of two major DNMTs (Dnmt1 and Dnmt3a) in the mouse forebrain excitatory neurons, caused deficits in synaptic plasticity, learning, and memory (12). There is additional direct evidence for neural activity-dependent DNA methylation changes. Transient activation of neuronal circuits trigger adult neurogenesis induced significant hypomethylation at CpG sites in the regulatory regions of brain-derived neurotrophic factor (Bdnf) and fibroblast growth factor-1 (Fgf-1). The immediately induced growth arrest and DNA damage inducible beta (Gadd45b), which is required for adult hippocampal neurogenesis, may promote the demethylation of DNAs at these sites (13). In addition, extensive analysis of the neuronal DNA methylome in adult mouse confirmed that neuronal activity can modify the DNA methylation landscape: upon electro-convulsive treatment, about 1.4% of 219,991 CpGs measured showed rapid active demethylation or de novo methylation in the hippocampus (14). These modified CpGs were significantly enriched in brain-specific genes related to neuronal plasticity. These results indicate that activity-induced DNA methylation changes are involved in neuronal function.
Fig. 1.
Epigenetic modifications and their role in gene expression. Major epigenetic modifications involved in neuronal gene expression around promoter regions are shown. Epigenetic markers stimulating transcription are shown in red, while inhibitory markers are shown in green. Enzymes involved in the generation (↑) or removal (⊤) of the epigenetic marks are indicated.
Chromatin exists in either a highly condensed or loosely packed state, which are associated with gene silencing or with active gene expression, respectively. More than 100 histone post-translational modifications have been identified so far, and the number continues to increase (15). Among them, methylation, acetylation, and phosphorylation are the most abundantly studied. Several types of neuronal activities modulate histones: facilitation of motor neuron synapses or memory training can enhance histone H3 and H4 acetylation and phosphorylation (16), and potassium chloride-mediated neuronal depolarization leads to H2B acetylation (17). BDNF is a well-known neurotrophin which triggers histone modification by regulating the dissociation of HDACs from chromatin; BDNF induces S-nitrosylation of HDAC2, which results in the release of HDAC2 from the chromatin, thus inducing the hyperacetylation of histones at neurotrophin-dependent gene promoters (18). A number of amino acids in the histone tail are targets of post-translational modification. Acetylation at histone H3 lysine 9 (H3K9), H3K14, H4K5, H4K8, and H4K12, methylation at H3K4, H3K9 and H3K36, and phosphorylation at H3S10 are examples of epigenetic modifications mediating synaptic plasticity, learning, and memory. Additional evidence of epigenetic regulation in learning-dependent synaptic plasticity is increasing, and epigenetic modifications can also be detected in a highly interwoven network in the brain: co-occurrence of H3K14 acetylation with H3S10 phosphorylation and H3K36 trimethylation, and H3 acetylation with DNA methylation have been observed (19, 20). The co-occurrence patterns of epigenetic modifications often result from crosstalk among epigenetic modifying enzymes that have differential preferences toward preexisting histone modification patterns at their binding sites.
Noncoding RNAs such as microRNAs, snoRNA, siRNAs, piRNAs, and lncRNAs, also function as epigenetic regulators by regulating mRNA transcription and translation. Among the microRNAs, the most extensively studied noncoding RNAs, miR-7, miR-9, miR-23, miR-124, miR-125a-b, miR-128, miR-132, miR-137, miR-139, miR-184, and miR-195 have been reported as being highly enriched in brain than in other tissues (21, 22). Neuronal function can be regulated by microRNAs, and miR-124 is a well-known example. Increasing levels of miR-124 promotes neuronal differentiation by downregulating a repressor of the neuron-specific splicing regulator polypyrimidine track binding protein 1 (PTBP1), a component of the RE-1 silencing transcription factor (REST)/neuron-restrictive silencing factor (NRSF) transcription repressor complex synaptonemal complex protein 1 (SCP1), and the glial cell specification factor SRY box 9 (Sox9) (22, 23). During adult neurogenesis, the epigenetically regulated targets of microRNAs include a number of transcription factors, indicating the importance of microRNA-mediated regulation in gene expression (24).
EPIGENETIC CHANGES IN BRAIN DURING NORMAL AGING PROCESS
Aging is characterized by a progressive decrease in the physiological capacity, reduced ability to respond adaptively to environmental stimuli, and increased susceptibility to diseases. It is a complex process influenced by multiple factors, including environmental conditions and genetics. Thus, a large degree of fluctuation can be detected among individuals, which makes it difficult to understand the exact mechanism underlying the aging process. Model organisms from the same genetic background and culture conditions are beneficial in defining this process, and an investigation of specific regions of the body, such as the brain, could identify specific genes vulnerable to aging (25, 26). In particular, with the recent development of next-generation sequencing technology, studies have investigated gene expression changes in the human brain. These studies all indicate that in addition to changes in the transcriptome profile, an increase in epigenetic variation has a strong correlation with the aging process (27). However, more recent studies revealed an interesting phenomenon: despite genetic and lifestyle heterogeneity in humans, the gene expression profile and epigenetic modifications of brain regions become similar among aged individuals in the later stages of life (older than 75 years); this phenomenon appears to be conserved across species (28).
Although aging induces diverse epigenetic changes in the brain, DNA methylation appears to play a prominent regulatory role. Global and gradual depletion of DNA methylation was initially suggested to correlate with aging (29), but age-related increases in the levels of 5-methylcytosine (5-mC), 5-hydroxymehtylcytosine (5-hmC), and DNA methyltransferase Dnmt3a have been reported in brain tissue (30). Indeed, the methylation status seems to change bidirectionally with aging: whereas DNA regions with repetitive elements tend to become hypomethylated, many developmental genes are hypermethylated as a function of age (31). Recently, a large-scale age-related gene expression study was performed using human whole blood cells from 14,983 individuals of European ancestry (32). Of the 1,497 genes that were found to be differentially expressed with chronological age, 19 to 26% were also identified to change concordantly in the cerebellum and frontal cortex of brain tissue. Analysis of the CpG sites in or near these age-associated genes revealed that changes in gene expression with chronological age correlated with changes in the DNA methylation levels at the regulatory regions of these genes.
Age-associated memory impairment is also found with altered hippocampal chromatin plasticity. Deregulation of histone H4K12 acetylation and failure in the expression of learning-induced genes is linked to age-related memory impairment (33). Age-related increase of HDAC2, which decreases the acetylation, has been reported in the mouse hippocampus. HDAC2 knockdown or HDAC inhibitors exhibit protective effects against age-related cognitive impairment (33). There seems to be an interplay between DNA methylation and histone modification, because co-localization and positive correlation of 5-mC with HDAC2 has been detected in the CA3 and CA1-2 regions of the hippocampus (34). Methylation on memory consolidation genes and its association with methyl-CpG binding proteins appear to recruit HDAC, which ultimately leads to chromatin remodeling and silencing of gene expression.
Along with changes in histone modifications and DNA methylation, noncoding RNAs appear to play a role in neuronal development. Although the role of noncoding RNAs in senescence is relatively poorly understood, microRNAs have been reported to contribute to loss of brain function during normal aging, based on studies in mice, rats, chimpanzees, and humans (35, 36). Although a general decline in microRNA abundance has been observed in peripheral blood mononuclear cells of aging individuals (37), recent studies by Yin et al. (38) did not observe a decrease in the overall microRNA expression with advancing age, when the rat whole brain was used. However, profiling of microRNAs according to specific regions of the brain identified differentially regulated microRNAs in aged brains. In general, microRNA levels remained relatively stable in the cortex, but showed an overall decline in the cerebellum during aging. However, specific microRNAs such as miR-144 were consistently upregulated in the cortex and cerebellum of aging chimpanzees and humans (35), whereas the level of miR-186 gradually decreased in the mouse brain cortex during aging (39). Response elements for miR-144 and miR-186 are found in a proapoptotic gene programmed cell death 4 (PDCD4), and a β-secretase encoding gene beta site APP-cleaving enzyme 1 (BACE1), respectively. The upregulation of miR-144 promotes tumorigenesis, while the downregulation of miR-186 contributes to the development of Alzheimer’s disease (AD) by the overproduction of amyloid beta (Aβ) protein. Therefore, diverse types of age-related pathogenesis in the brain may result from dysregulation of the epigenetic regulations.
EPIGENETIC CHANGES IN AGING-RELATED NEURODEGENERATIVE DISEASES
Alzheimer’s disease (AD)
AD is characterized by the progressive loss of memory and cognitive ability, aggregation (including extracellular deposition) of the Aβ peptide, and intracellular aggregation of phosphorylated tau protein. Early onset of AD is associated with genetic mutations in amyloid beta precursor protein (APP), presenilin 1 (PSEN1) and PSEN2, while late onset AD is known to be associated with various factors such as lifestyle, diet, stroke, and environmental factors, etc. As most AD cases are late-onset, sporadic, and highly related to aging, it has been suggested that epigenetic regulations may play an important role in increasing the risk of AD development. Based on the observation of decreased levels of 5-mC and DNA methyltransferase in AD patients, it has been suggested that global DNA hypomethylation is associated with AD (40). Also, the levels of essential methylation substrates such as the universal methyl donor S-adenosylmethionine (SAM), S-adenosylhomocystein (SAH), and folate, were also decreased in the brain of AD patients (41). However, other studies report no change, or even a global gain, in DNA methylation in AD patients (42). Similar contradicting results were also observed in the analysis of hydroxymethylation of DNA, which represents the demethylation process, in AD patients. These variations in observed AD-associated epigenetic changes could result from differential changes in DNA methylation in different regions of the genome in AD patients, involving global as well as local dysregulation of DNA methylation (43). Although the occurrence of familial AD is more infrequent than sporadic AD, methylation changes have been investigated in genes that are known to be associated with familial AD. APP, PSEN1, and microtubule associated protein tau (MAPT), which encode for precursors of Aβ, γ-secretase, and tau, respectively, have been predominantly used in the investigation of epigenetic alterations and in generating AD animal models. Some studies reported hypomethylation of the APP promoter in AD patients and in normal aging (44); however, Barrachina and Ferrer (45) reported no alteration in DNA methylation in the APP, PSEN1, and MAPT promoters when the frontal cortex and hippocampal area of various stage AD patients were examined. Wang et al. (46) also reported no alteration in DNA methylation in the promoters of APP, PSEN1, BACE1, and DNMT1. However, studies showed consistent hypermethylation of sorbin and SH3 domain containing 3 (SORBS3), ankyrin 1 (ANK1), and neutral endopeptidase (NEP), which encode the Aβ clearing enzyme neprilysin, in both AD animal models and AD patients (47, 48). Inter-individual variation are known to occur in AD patients, rendering it challenging to judge the role of DNA methylation in the pathogenesis of AD. Therefore, the importance of DNA methylation in AD pathology requires further investigation of specific loci or their regulatory relationship with the development of AD.
In addition to the involvement of DNA methylation, histone deacetylation could partially explain the etiology in the development of AD. The impairment in learning and memory observed in the AD mouse model was reversible and could be improved by treatment with HDAC inhibitors (9, 49, 50). An actual increase in HDAC2 level, and a decrease in histone acetylation, specifically at promoters of genes found to be involved in learning, memory, and synaptic plasticity, were detected in late-stage AD mouse models and AD patients (9, 49, 50). Knockdown of HDAC2 abolished neurodegeneration-associated memory impairment (50). Enrichment of hypoacetylation and HDAC2 negatively correlated with RNA polymerase binding and mRNA expression at the genes required for memory and learning. However, AD-related histone hypoacetylation appears to be enriched at specific histones: H4 acetylation was generally decreased in transgenic APP/PS1 mice, whereas global H3 acetylation remained unchanged. Increase in H3 acetylation was also observed at specific promoters such as BACE1, when Aβ was administered to both AD mice and AD patients (51).
Contrary to the general decrease in histone acetylation, histone methylation is reported to be upregulated in AD patients (52). Upregulation of phosphorylation in H3 was also observed in the hippocampus of AD patients in some studies, but could not be confirmed by others (52, 53). Brain region-, gene-, or disease stage-specificity might affect epigenetic modification patterns. Therefore, further investigation with comparable samples and genetic loci would reveal histone modification changes associated with AD.
MicroRNAs also regulate gene expression epigenetically, and the potential role of microRNA as a biomarker has been studied in AD. Upregulation of miR-34a and miR-181b, and downregulation of miR-9, miR-29a/b, miR-137, and miR-181c, were observed in blood mononuclear cells from AD patients (54). Serum can also serve as a source of cell-free microRNA; miR-23a, miR-26b and miR-125b were found to be down-regulated in serum (55). Changes in the microRNA profile were also observed in the cerebrospinal fluid and plasma exosome. In a review by Kumar and Reddy (56), which summarized 12 studies covering diverse biofluids from 1268 participants, 100 microRNAs were identified as differentially expressed in AD patients; they reported upregulation of 54 microRNAs and downregulation of 46 microRNAs. Although inconsistencies remain in microRNA profiling data between different studies, some microRNAs, such as miR-34a, which was overexpressed in blood cells of AD but down regulated in neurotoxic Aβ overexpressing mouse model, provide and insight into the triggering mechanism of cortical neuronal apoptosis in AD patients (57).
Parkinson’s Disease (PD)
PD is the second most common neurodegenerative disorder that lacks an effective treatment modality. It is characterized by the loss of dopaminergic neurons in the midbrain substantia nigra, a critical region for the initiation of motor events. The occurrence of cytoplasmic Lewy bodies in the midbrain and cortex, where α-synuclein is localized, and the malfunctioning of neurotransmitter systems, are linked to cognitive impairments and other PD symptoms. A genome-wide association study of familial forms of PD, although rarer than sporadic forms, identified synuclein α (SNCA) and leucine-rich repeat kinase 2 (LRRK2) as the main risk factors for PD (58). Most PD is sporadic, and emerging evidence identifies aberrant epigenetic modification as a possible mediator of environmental inputs (toxin as a major factor) that drive PD development. However, compared to AD cases, relatively little is known regarding epigenetic alterations in PD, with only a limited number of studies based on in vitro or animal PD models. So far, α-Synuclein is the best known target of abnormal epigenetic regulation in PD.
α-Synuclein is a natively soluble, but mutant protein, produced as a result of missense (A53T, A30P, E46, H50Q, and G51D) mutations, or its overexpression due to SNCA gene amplification, often resulting in abnormal aggregation of α-Synuclein in Lewy bodies in familial PD patients (59). Hundreds of SNPs of SNCA are associated with sporadic PD. In addition, hypomethylation at the promoter and introns of SNCA appear to increase the SNCA expression in PD patients (60, 61). Overexpressed α-Synuclein can also sequester DNMT1 to the cytoplasm; this protein is normally located in the nuclei of neurons in the healthy brain. Reduction in nuclear DNMT1 can explain the global DNA hypomethylation observed in PD model mice and patients (62).
MAPT, a major risk factor for AD, was also found to be a risk factor for sporadic PD. Tau protein is associated with microtubules and helps stabilize the axonal cytoskeleton. The methylation level of MAPT varies along different regions of the normal brain, but these levels are consistently in a negative correlation with MAPT expression levels, indicating their regulatory role in MAPT expression. Indeed, when the methylation level of MATP was examined in the leukocytes of PD patients, the age at PD onset positively correlated with the MAPT methylation levels (63). Therefore, MAPT methylation appears to have a protective function against PD. Consistently, MAPT methylation was significantly higher in females compared to males, thereby correlating with the lower incidence of PD in women. Intriguingly, MAPT methylation levels are different in different regions of the brain, and these regional differences correlate with the pathological severity of PD: the cerebellum was more effective in inducing compensatory MAPT hypermethylation against PD progress than the putamen, and hance more severe pathological PD development was observed in the putamen area (63).
Histone modifications are also involved in PD pathogenesis. Nuclear accumulation of α-Synuclein appears to play an important role in PD development: a significant amount of α-Synuclein is found in PD patients and animal models, and injection of herbicide induces the translocation of α-Synuclein into the nucleus of nigral neurons (64). The nuclear function of α-Synuclein appears to be the regulation of histone modification. α-Synuclein was shown to bind directly to histone proteins, which results in the reduction of H3 acetylation. In addition, the toxicity of α-Synuclein in PD models was rescued by the administration of HDAC inhibitors (65). In contrast to the pathological correlation of histone deacetylation in PD, the detrimental effect of histone acetylation in dopaminergic neuronal cells has also been reported (66). Pesticide-induced neuronal loss comprises a large fraction of PD development; thus, the effect of Dieldrin, a widely used insecticide originally developed as an alternative to DDT, on histone modification in dopaminergic neuronal degeneration was examined. Intriguingly, Dieldrin induced the H3 and H4 acetylation in a time-dependent manner, along with reduced HDAC activity (66). In addition, these Dieldrin-induced effects were attenuated effectively by KAT inhibitors. Further experiments are therefore required to clarify the functional association of histone acetylation status with PD.
In addition to epigenetic regulation via histone modification and DNA methylation, key risk factor genes for PD, such as SNCA, are also aberrantly targeted by microRNAs in PD. miR-7 and miR-153 are highly expressed in the brain, and can synergistically repress the SNCA translation (67). Overexpression of SNCA impairs the dopaminergic neuron function in PD, and a decrease in miR-7 expression was observed in the PD mouse model (68). Many microRNAs involved in neuronal function and survival were also shown to be dysregulated in PD. miR-124 and miR-133b play important roles in adult neurogenesis and dopaminergic neuronal functions, and they are significantly deficient in the brains of PD patients or mouse models (69, 70). On the other hand, miR-132 which targets a key regulator of neurite outgrowth, nuclear receptor related 1 (Nurr1), is significantly increased in affected PD model rats, and suppresses the NURR1 expression (71). MicroRNA profiles from blood of PD patients revealed downregulation of miR-1, miR-22p, and miR-29a, along with upregulation of miR-125a-3p, miR-137, miRmiR-181c, miR-193a-5p, miR-196b, miR-331-5p, and miR-454 (72, 73). In addition, extensive investigation of microRNA alterations in the prefrontal cortex region of postmortem patients identified 125 microRNAs that are differentially expressed in PD. Although only a few were matched to microRNAs known to be dysregulated in PD, the abnormal expression of 36 microRNAs appeared to correlate with dementia in PD, and miR-10b-5p expression level negatively correlated with PD onset (74), indicating their functional relevance in PD development. Therefore, a more thorough investigation into these microRNAs would help reveal epigenetic regulatory mechanisms mediated by microRNAs to influence PD pathogenesis.
Huntington’s Disease (HD)
HD is primarily a genetic disease initiated by the expansion of polyglutamine repeats in the huntingtin (HTT) coding region. Chorea, cognitive deterioration, and psychiatric disturbances are symptoms of HD, due to impairment of the basal ganglia and cerebral cortex. Although HTT has been identified as the main gene causing HD, variability in disease onset and severity has been reported, indicating that environmental factors and the epigenetic alterations caused by them are additional factors that influence the disease symptoms.
Several lines of evidence suggest the involvement of epigenetic modulation by HTT in HD patients. The polyglutamine-containing domain of HTT was shown to bind histone acetyltransferase, CREB binding protein (CBP) and P300/CBP-Associated Factor (P/CAF). The accumulation of CBP in the intracellular inclusion body was often found in the brains of HD patients and animal models (75, 76). In addition, overexpression of mutant HTT induced the global hypoacetylation of histone H3 and H4. Epigenetic alterations induced by HTT appear to play an important role in HD pathogenesis, as evidenced by the fact that HDAC inhibitor treatment arrested the progress of neuronal degeneration, in addition to reversing the aberrant histone acetylation pattern (76). These lines of evidence suggest that HTT with expanded polyglutamine downregulates the gene expression through epigenetic modifications in the affected region. However, other studies did not observe the global hypoacetylation of histone, although HDAC inhibitor treatment had a similar beneficial effect on disease symptoms (77). Instead, hypoacetylated H3 was found at the Drd2 gene locus encoding the dopamine D2 receptor, specifically in the striatum, but not in other regions of brain, such as the cortex, hippocampus and cerebellum. These results indicate that gene-specific epigenetic regulation is more important than global alterations in histone modifications, but further experiments are needed to clarify this discrepancy. H2A histone family member Y (H2AFY) is a histone variant that modulates transcription factor binding and transcription repression, and overexpression of H2AFY was found in the blood and frontal cortex from a number of HD patients. Because H2AFY levels correlated with the therapeutic effect of HDAC inhibitors against HD in clinical trials, chromatin regulation appears to be an important pathogenic mechanism in HD development (78). Histone methylation is also involved in the pathogenesis of HD. ERG-associated protein with SET domain (ESET), a histone H3K9 methyltransferase, mediates gene silencing; its expression is markedly increased in HD animals and patients (79). The downregulation of ESET by a pharmacological method not only suppressed the hypertrimethylation of H3K9, but also significantly ameliorated HD symptoms. Therefore, modifications in histone acetylation and methylation appear to play a significant role in the etiology of HD.
Although DNA methylation is often associated with disease, alterations in DNA methylation appear to be minimal in HD. Only recently, Ng et al. (80) raised the possibility of DNA methylation as one of the epigenetic modifications involved in HD, by showing that regions with low CpG content, where methylation changes occur in response to neuronal activity, undergo methylation changes with the overexpression of polyglutamine expanded HTT. When DNA methylation profiling in HD patients was examined by De Souza et al. (81), some evidence of HD-associated DNA methylation was found in the cortex region alone.
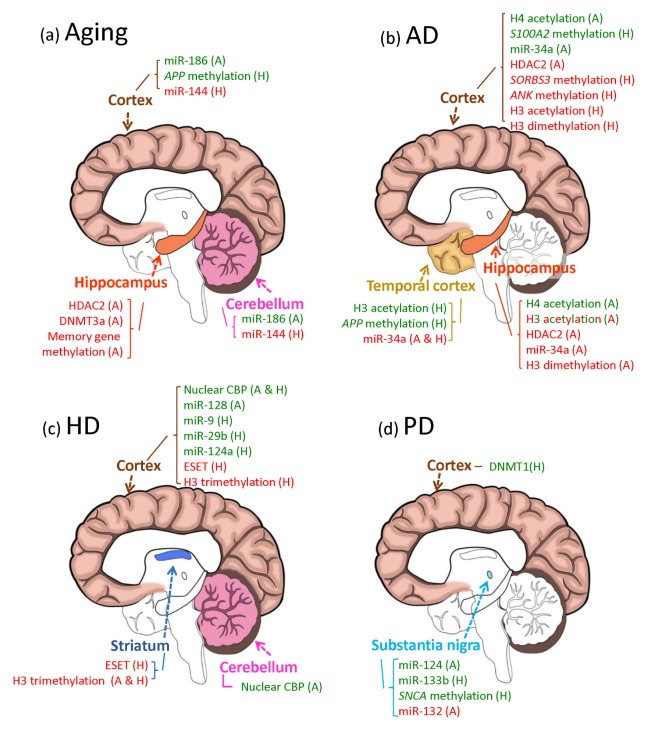
Changes in the expression profiles of microRNAs are also detected in HD. Of the 752 mature human microRNAs, 168 microRNAs were found to be altered in circulating plasma samples from symptomatic patients (82). Other research groups identified different altered microRNA profiles in HD models or patients. Some of these alterations are due to changes in the interaction between the mutant form of HTT and its partners to which wild-type HTT could bind under normal conditions. Under normal conditions, HTT interacts with REST and maintains it in the cytoplasm, but this association is disrupted in HD. A group of microRNAs are downregulated by REST, which has a repressor activity for many neuronal coding and non-coding genes. When translocated into the nucleus, REST represses the transcription of BDNF and several microRNAs, including miR-9, miR-29a, miR-29b, miR-124a, miR-132, and miR-330 (83, 84). Ago2, a component of the RISC complex, is another protein known to interact with HTT, and its dissociation from HTT leads to an increase in microRNA-mediated gene silencing in HD (85). Thus, the maintenance of selected microRNA expression could be a default protective function in the brain, but its deregulation may contribute to neurodegenerative disease. The three major epigenetic modifications mentioned above that control gene expression in the brain during aging, and the three neurodegenerative processes, are summarized in Fig. 2.
Fig. 2.
Epigenetic modifications observed in specific brain areas of aged or neurodegenerative disease-affected animal models or patients. Modifications in DNA and histones, and changes in microRNA abundance associated with aging (A), or the three neurodegenerative diseases AD (B), HD (C), and PD (D), are shown. Brain regions studied in each disease model are marked with specific colors: cerebral cortex (brown), temporal cortex (golden rod), hippocampus (orange), cerebellum (hot pink), striatum (royal blue), and substantia nigra (sky blue). Up (red) and down (green) regulated epigenetic markers in animal models (A) or in human patients (H) were mapped to the region where they were examined. H3 acetylation in the hippocampus of AD is written in both colors to indicate conflicting results reported. Results that did not show any change are not mentioned in this figure, but are mentioned in the main text.
EPIGENETIC MODULATION OF THE BLOOD MILIEU AND ITS REFLECTION OF NEURONAL FUNCTION
As neurodegenerative diseases affect neuronal cells in the brain, examination of gene expression changes is not easily achieved prior to mortality. Detection of microRNAs in circulatory biofluids such as blood, urine, and saliva, and their stability in extracellular fluids (86), opened up the possibility and provided accessibility for the detection of gene expression changes occurring inside tissues. MicroRNAs encapsulated with exosomes are capable of crossing the blood-brain barrier and are secreted in the cerebrospinal fluid or blood (87); they would therefore reflect the microRNA profile inside the brain. The identification of biomarkers in blood or blood components facilitates less invasive procedures, allowing an assessment of neurodegenerative diseases at early stages. In addition to microRNA profiles, many studies researching epigenetic modifications in the brain were performed using peripheral blood mononuclear cells, and disease-associated epigenetic alterations were successfully identified. Peters et al. (32) examined whole-blood gene expression in 14,983 individuals, and identified 1,497 genes that are differentially expressed with chronological age. The genes whose expression levels changed in accordance with age were enriched with CpG methylation sites at locations that are tied to gene expression regulation, such as promoters, enhancers, and insulators. They even found that transcriptomic age and epigenetic age are positively correlated. Indeed, many epigenetic alterations associated with AD, PD, and HD have been identified using blood samples. The use of blood samples renders the study of aging and neurodegenerative diseases applicable beyond animal models, and extended to humans.
PERSPECTIVES FOR DRUG DEVELOPMENT
Neurodegenerative diseases are among the most feared disorders especially, in aging or aged societies. When PD patients are diagnosed based on typical motor symptoms, about 50% of the dopaminergic neurons have already been irreversibly lost, and the same applies for other neurodegenerative diseases. Diagnosis before the loss of intellectual or cognitive impairment would be beneficial; therefore, the development of specific diagnostic tests or effective treatments is in demand. Although research is focused on identifying biomarkers that can detect diseases early in their pathogenesis, no biomarkers or drugs have been developed that can completely delay or prevent the progression of AD, PD and HD. Because histone acetylation affects the regulation of gene transcription, the acetylation or deacetylation at specific histone sites represents biomarkers for the disease state. HDAC inhibitors show great potential not only in treating cognitive impairment in neurodegenerative disorders, but also in enhancing cognitive function in healthy people (8). Therefore, the use of HDAC inhibitors or DNMT could be considered for the treatment of neurodegenerative diseases. MicroRNAs are secreted into the blood, and exosome-associated microRNAs are known to be stable. Several upregulated or downregulated microRNAs have been identified in all the three neurodegenerative diseases mentioned in this review. Therefore, manifestation of disease can be easily measured based on microRNA levels, and they could also be promising components in developing drugs for neurodegenerative diseases. Because epigenetic regulations are reversible, drugs targeted to epigenetic alterations would be easier to access, and they can also be used to measure the effectiveness of the treatment. Diseases would be manageable if the onset of diseases could be diagnosed at the very early stages. Therefore, more efforts have to be made to select biomarkers to detect disease onset as early as possible, to enable relevant preventive treatment.
FINANCIAL DISCLOSURE AND ACKNOWLEDGEMENTS
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (grant number: NRF-2012M3A9B4028272), and Collaborative Genome Program for Fostering New Post-Genome industry through the National Research Foundation of Korea (NRF) funded by the Ministry of Science ICT and Future Planning (grant number: 2015M3C9A4053251 and 2016M3C9A4921712) to Y-J Kim and by the Mid-Career Researcher Program (NRF-2012R1A2A2A01010176) and Basic Science Research Program (NRF-2011-0010593) of the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning to J. Kim-Ha. We thank JB Moon for his help with figures.
REFERENCES
- 1.Choudhuri S. From Waddington’s epigenetic landscape to small noncoding RNA: some important milestones in the history of epigenetics research. Toxicol Mech Methods. 2011;21:252–274. doi: 10.3109/15376516.2011.559695. [DOI] [PubMed] [Google Scholar]
- 2.Lee SM, Kim-Ha J, Choi WY, et al. Interplay of genetic and epigenetic alterations in hepatocellular carcinoma. Epigenomics. 2016;8:993–1005. doi: 10.2217/epi-2016-0027. [DOI] [PubMed] [Google Scholar]
- 3.Fischer A. Epigenetic memory: the Lamarckian brain. EMBO J. 2014;33:945–967. doi: 10.1002/embj.201387637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedreira ME, Dimant B, Maldonado H. Inhibitors of protein and RNA synthesis block context memory and long-term habituation in the crab Chasmagnathus. Pharmacol Biochem Behav. 1996;54:611–617. doi: 10.1016/0091-3057(95)02206-6. [DOI] [PubMed] [Google Scholar]
- 5.Arguello AA, Ye X, Bozdagi O, et al. CCAAT enhancer binding protein δ plays an essential role in memory consolidation and reconsolidation. J Neurosci. 2013;33:3646–3658. doi: 10.1523/JNEUROSCI.1635-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alberini CM, Kandel ER. The regulation of transcription in memory consolidation. Cold Spring Harb Perspect Biol. 2014;7:a021741. doi: 10.1101/cshperspect.a021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr Res. 2007;61:58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- 8.Graff J, Tsai LH. The potential of HDAC inhibitors as cognitive enhancers. Annu Rev Pharmacol Toxicol. 2013;53:311–330. doi: 10.1146/annurev-pharmtox-011112-140216. [DOI] [PubMed] [Google Scholar]
- 9.Fischer A, Sananbenesi F, Wang XY, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 10.Gräff J, Kim D, Dobbin MM, Tsai LH. Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiol Rev. 2011;91:603–649. doi: 10.1152/physrev.00012.2010. [DOI] [PubMed] [Google Scholar]
- 11.Kempermann G, Song H, Gage FH. Neurogenesis in the Adult Hippocampus. Cold Spring Harb Perspect Biol. 2015;7:a018812. doi: 10.1101/cshperspect.a018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng J, Zhou Y, Campbell SL, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma DK, Jang MH, Guo JU, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo JU, Ma DK, Mo H, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chwang WB, Arthur JS, Schumacher A, Sweatt JD. The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J Neurosci. 2007;27:12732–12742. doi: 10.1523/JNEUROSCI.2522-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maharana C, Sharma KP, Sharma SK. Depolarization induces acetylation of histone H2B in the hippocampus. Neuroscience. 2010;167:354–360. doi: 10.1016/j.neuroscience.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 18.Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 19.Tweedie-Cullen RY, Brunner AM, Grossmann J, et al. Identification of combinatorial patterns of posttranslational modifications on individual histones in the mouse brain. PLoS One. 2012;7:e36980. doi: 10.1371/journal.pone.0036980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14:97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 21.Adlakha YK, Saini N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol Cancer. 2014;13 doi: 10.1186/1476-4598-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh J, Zhao X. Genetics and Epigenetics in Adult Neurogenesis. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a018911. pii: a018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santos MC, Tegge AN, Correa BR, et al. miR-124, -128, and -137 Orchestrate Neural Differentiation by Acting on Overlapping Gene Sets Containing a Highly Connected Transcription Factor Network. Stem Cells. 2016;34:220–232. doi: 10.1002/stem.2204. [DOI] [PubMed] [Google Scholar]
- 25.Kim SN, Rhee JH, Song YH, et al. Age-dependent changes of gene expression in the Drosophila head. Neurobiol Aging. 2005;26:1083–1091. doi: 10.1016/j.neurobiolaging.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 26.Berchtold NC, Sabbagh MN, Beach TG, Kim RC, Cribbs DH, Cotman CW. Brain gene expression patterns differentiate mild cognitive impairment from normal aged and Alzheimer’s disease. Neurobiol Aging. 2014;35:1961–1972. doi: 10.1016/j.neurobiolaging.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talens RP, Christensen K, Putter H, et al. Epigenetic variation during the adult lifespan: cross-sectional and longitudinal data on monozygotic twin pairs. Aging Cell. 2012;11:694–703. doi: 10.1111/j.1474-9726.2012.00835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh G, Ebrahimi S, Wang SC, et al. Epigenetic assimilation in the aging human brain. Genome Biol. 2016;17:76. doi: 10.1186/s13059-016-0946-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bollati V, Schwartz J, Wright R, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chouliaras L, van den Hove DL, Kenis G, et al. Age-related increase in levels of 5-hydroxymethylcytosine in mouse hippocampus is prevented by caloric restriction. Curr Alzheimer Res. 2012;9:536–544. doi: 10.2174/156720512800618035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maegawa S, Hinkal G, Kim HS, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters MJ, Joehanes R, Pilling LC, et al. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. doi: 10.1038/ncomms9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peleg S, Sananbenesi F, Zovoilis A, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 34.Chouliaras L, van den Hove DL, Kenis G, et al. Histone deacetylase 2 in the mouse hippocampus: attenuation of age-related increase by caloric restriction. Curr Alzheimer Res. 2013;10:868–876. doi: 10.2174/1567205011310080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Persengiev S, Kondova I, Otting N, Koeppen AH, Bontrop RE. Genome-wide analysis of miRNA expression reveals a potential role for miR-144 in brain aging and spinocerebellar ataxia pathogenesis. Neurobiol Aging. 2011;32:2316.e17–27. doi: 10.1016/j.neurobiolaging.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 36.Inukai S, de Lencastre A, Turner M, Slack F. Novel microRNAs differentially expressed during aging in the mouse brain. PLoS One. 2012;7:e40028. doi: 10.1371/journal.pone.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. microRNA expression patterns reveal differential expression of target genes with age. PLoS One. 2010;5:e10724. doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin L, Sun Y, Wu J, et al. Discovering novel microRNAs and age-related nonlinear changes in rat brains using deep sequencing. Neurobiol Aging. 2015;36:1037–1044. doi: 10.1016/j.neurobiolaging.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Kim J, Yoon H, Chung DE, Brown JL, Belmonte KC, Kim J. miR-186 is decreased in aged brain and suppresses BACE1 expression. J Neurochem. 2016;137:436–445. doi: 10.1111/jnc.13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chouliaras L, Mastroeni D, Delvaux E, et al. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer’s disease patients. Neurobiol Aging. 2013;34:2091–2099. doi: 10.1016/j.neurobiolaging.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrison LD, Smith DD, Kish SJ. Brain S-adenosylmethionine levels are severely decreased in Alzheimer’s disease. J Neurochem. 1996;67:1328–1331. doi: 10.1046/j.1471-4159.1996.67031328.x. [DOI] [PubMed] [Google Scholar]
- 42.Coppieters N, Dieriks BV, Lill C, Faull RL, Curtis MA, Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer’s disease human brain. Neurobiol Aging. 2014;35:1334–1344. doi: 10.1016/j.neurobiolaging.2013.11.031. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez-Mut JV, Gräff J. Epigenetic Alterations in Alzheimer’s Disease. Front Behav Neurosci. 2015;9:347. doi: 10.3389/fnbeh.2015.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West RL, Lee JM, Maroun LE. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer’s disease patient. J Mol Neurosci. 1995;6:141–146. doi: 10.1007/BF02736773. [DOI] [PubMed] [Google Scholar]
- 45.Barrachina M, Ferrer I. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J Neuropathol Exp Neurol. 2009;68:880–891. doi: 10.1097/NEN.0b013e3181af2e46. [DOI] [PubMed] [Google Scholar]
- 46.Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS One. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Jager PL, Srivastava G, Lunnon K, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lunnon K, Smith R, Hannon E, et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat Neurosci. 2014;17:1164–1170. doi: 10.1038/nn.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ricobaraza A, Cuadrado-Tejedor M, Perez-Mediavilla A, Frechilla D, Del Rio J, Garcia-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer’s disease mouse model. Neuropsychopharmacology. 2009;34:1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- 50.Gräff J, Rei D, Guan JS, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marques SC, Lemos R, Ferreiro E, et al. Epigenetic regulation of BACE1 in Alzheimer’s disease patients and in transgenic mice. Neuroscience. 2012;220:256–266. doi: 10.1016/j.neuroscience.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 52.Lithner CU, Lacor PN, Zhao WQ, et al. Disruption of neocortical histone H3 homeostasis by soluble Aβ: implications for Alzheimer’s disease. Neurobiol Aging. 2013;34:2081–2090. doi: 10.1016/j.neurobiolaging.2012.12.028. [DOI] [PubMed] [Google Scholar]
- 53.Ogawa O, Zhu X, Lee HG, et al. Ectopic localization of phosphorylated histone H3 in Alzheimer’s disease: a mitotic catastrophe? Acta Neuropathol. 2003;105:524–528. doi: 10.1007/s00401-003-0684-3. [DOI] [PubMed] [Google Scholar]
- 54.Geekiyanage H, Jicha GA, Nelson PT, Chan C. Blood serum miRNA: non-invasive biomarkers for Alzheimer’s disease. Exp Neurol. 2012;235:491–496. doi: 10.1016/j.expneurol.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galimberti D, Villa C, Fenoglio C, et al. Circulating miRNAs as potential biomarkers in Alzheimer’s disease. J Alzheimers Dis. 2014;42:1261–1267. doi: 10.3233/JAD-140756. [DOI] [PubMed] [Google Scholar]
- 56.Kumar S, Reddy PH. Are circulating microRNAs peripheral biomarkers for Alzheimer’s disease? Biochim Biophys Acta. 20161862:1617–1627. doi: 10.1016/j.bbadis.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Modi PK, Jaiswal S, Sharma P. Regulation of Neuronal Cell Cycle and Apoptosis by MicroRNA 34a. Mol Cell Biol. 2015;36:84–94. doi: 10.1128/MCB.00589-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 59.Xu W, Tan L, Yu JT. Link between the SNCA gene and parkinsonism. Neurobiol Aging. 2015;36:1505–1518. doi: 10.1016/j.neurobiolaging.2014.10.042. [DOI] [PubMed] [Google Scholar]
- 60.Ai SX, Xu Q, Hu YC, et al. Hypomethylation of SNCA in blood of patients with sporadic Parkinson’s disease. J Neurol Sci. 2014;337:123–128. doi: 10.1016/j.jns.2013.11.033. [DOI] [PubMed] [Google Scholar]
- 61.Tan YY, Wu L, Zhao ZB, et al. Methylation of α-synuclein and leucine-rich repeat kinase 2 in leukocyte DNA of Parkinson’s disease patients. Parkinsonism Relat Disord. 2014;20:308–313. doi: 10.1016/j.parkreldis.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 62.Desplats P, Spencer B, Coffee E, et al. Alpha-synuclein sequesters Dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. J Biol Chem. 2011;286:9031–9037. doi: 10.1074/jbc.C110.212589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coupland KG, Mellick GD, Silburn PA, et al. DNA methylation of the MAPT gene in Parkinson’s disease cohorts and modulation by vitamin E in vitro. Mov Disord. 2014;29:1606–1614. doi: 10.1002/mds.25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goers J, Manning-Bog AB, McCormack AL, et al. Nuclear localization of alpha-synuclein and its interaction with histones. Biochemistry. 2003;42:8465–8471. doi: 10.1021/bi0341152. [DOI] [PubMed] [Google Scholar]
- 65.Kontopoulos E, Parvin JD, Feany MB. Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet. 2006;15:3012–3023. doi: 10.1093/hmg/ddl243. [DOI] [PubMed] [Google Scholar]
- 66.Song C, Kanthasamy A, Anantharam V, Sun F, Kanthasamy AG. Environmental neurotoxic pesticide increases histone acetylation to promote apoptosis in dopaminergic neuronal cells: relevance to epigenetic mechanisms of neurodegeneration. Mol Pharmacol. 2010;77:621–632. doi: 10.1124/mol.109.062174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285:12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009;106:13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanagaraj N, Beiping H, Dheen ST, Tay SS. Downregulation of miR-124 in MPTP-treated mouse model of Parkinson’s disease and MPP iodide-treated MN9D cells modulates the expression of the calpain/cdk5 pathway proteins. Neuroscience. 2014;272:167–179. doi: 10.1016/j.neuroscience.2014.04.039. [DOI] [PubMed] [Google Scholar]
- 70.Kim J, Inoue K, Ishii J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lungu G, Stoica G, Ambrus A. MicroRNA profiling and the role of microRNA-132 in neurodegeneration using a rat model. Neurosci Lett. 2013;553:153–158. doi: 10.1016/j.neulet.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 72.Margis R, Margis R, Rieder CRM. Identification of blood microRNAs associated to Parkinson’s disease. J Biotechnol. 2011;152:96–101. doi: 10.1016/j.jbiotec.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 73.Cardo LF, Coto E, de Mena L, et al. Profile of microRNAs in the plasma of Parkinson’s disease patients and healthy controls. J Neurol. 2013;260:1420–1422. doi: 10.1007/s00415-013-6900-8. [DOI] [PubMed] [Google Scholar]
- 74.Hoss AG, Labadorf A, Beach TG, Latourelle JC, Myers RH. microRNA Profiles in Parkinson’s Disease. Prefrontal Cortex Front Aging Neurosci. 2016;8:36. doi: 10.3389/fnagi.2016.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nucifora FC, Jr, Sasaki M, Peters MF, et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 76.Steffan JS, Bodai L, Pallos J, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 77.Sadri-Vakili G, Bouzou B, Benn CL, et al. Histones associated with downregulated genes are hypo-acetylated in Huntington’s disease models. Hum Mol Genet. 2007;16:1293–1306. doi: 10.1093/hmg/ddm078. [DOI] [PubMed] [Google Scholar]
- 78.Hu Y, Chopra V, Chopra R, et al. Transcriptional modulator H2A histone family, member Y (H2AFY) marks Huntington disease activity in man and mouse. Proc Natl Acad Sci U S A. 2011;108:17141–17146. doi: 10.1073/pnas.1104409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ryu H, Lee J, Hagerty SW, et al. ESET/SETDB1 gene expression and histone H3 (K9) trime-thylation in Huntington’s disease. Proc Natl Acad Sci U S A. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ng CW, Yildirim F, Yap YS, et al. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc Natl Acad Sci U S A. 2013;110:2354–2359. doi: 10.1073/pnas.1221292110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De Souza RA, Islam SA, McEwen LM, et al. DNA methylation profiling in human Huntington’s disease brain. Hum Mol Genet. 2016;25:2013–2030. doi: 10.1093/hmg/ddw076. [DOI] [PubMed] [Google Scholar]
- 82.Díez-Planelles C, Sánchez-Lozano P, Crespo MC, et al. Circulating microRNAs in Huntington’s disease: Emerging mediators in metabolic impairment. Pharmacol Res. 2016;108:102–110. doi: 10.1016/j.phrs.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 83.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soldati C, Bithell A, Johnston C, Wong KY, Stanton LW, Buckley NJ. Dysregulation of REST-regulated coding and non-coding RNAs in a cellular model of Huntington’s disease. J Neurochem. 2013;124:418–430. doi: 10.1111/jnc.12090. [DOI] [PubMed] [Google Scholar]
- 85.Savas JN, Makusky A, Ottosen S, et al. Huntington’s disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc Natl Acad Sci U S A. 2008;105:10820–10825. doi: 10.1073/pnas.0800658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Machida T, Tomofuji T, Ekuni D, et al. MicroRNAs in Salivary Exosome as Potential Biomarkers of Aging. Int J Mol Sci. 2015;16:21294–21309. doi: 10.3390/ijms160921294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jung HJ, Suh Y. Circulating miRNAs in ageing and ageing-related diseases. J Genet Genomics. 2014;41:465–472. doi: 10.1016/j.jgg.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]