Abstract
Fructose 1,6-bisphosphate aldolase (FBA) is important for both glycolysis and gluconeogenesis in life. Class II (zinc dependent) FBA is an attractive target for the development of antibiotics against protozoa, bacteria, and fungi, and is also widely used to produce various high-value stereoisomers in the chemical and pharmaceutical industry. In this study, the crystal structures of class II Escherichia coli FBA (EcFBA) were determined from four different crystals, with resolutions between 1.8 Å and 2.0 Å. Native EcFBA structures showed two separate sites of Zn1 (interior position) and Zn2 (active site surface position) for Zn2+ ion. Citrate and TRIS bound EcFBA structures showed Zn2+ position exclusively at Zn2. Crystallographic snapshots of EcFBA structures with and without ligand binding proposed the rationale of metal shift at the active site, which might be a hidden mechanism to keep the trace metal cofactor Zn2+ within EcFBA without losing it.
Keywords: Class II fructose 1,6-bisphosphate aldolase; Inorganic cofactor; Metal ion; X-ray crystallography
INTRODUCTION
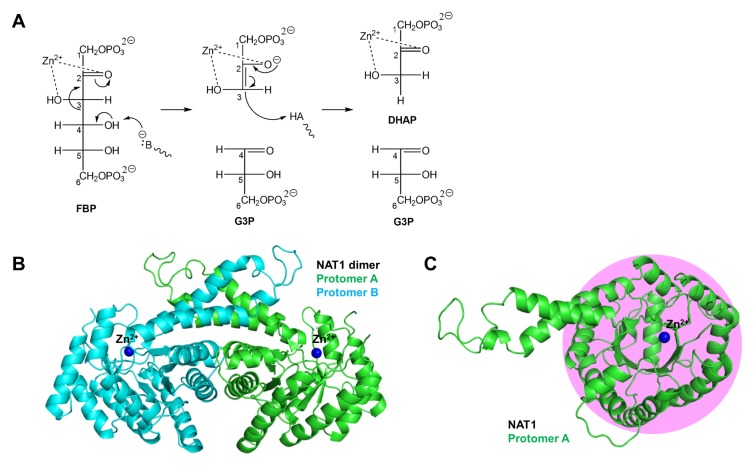
Fructose 1,6-bisphosphate aldolase (FBA; E.C. 4.1.2.13) catalyzes the second reversible step of the glycolytic pathway, which is the hydrolysis of the open-chain form of fructose 1,6-bisphosphate (FBP) to glyceraldehyde 3-phosphate (G3P) and dihydroxyacetone phosphate (DHAP). There are two known families of FBA - class I and class II. Although both classes adopt similar α/β folds, they have different catalytic components at the active site. Class I FBAs use a Schiff-base (imine, containing carbon-nitrogen double bond) linkage between the active site Lys residue and the substrate C(2) carbonyl group (1). Class II FBAs use inorganic cofactor of zinc ion at the active site to bind and catalyze substrates; the divalent zinc ion coordinates both C(2) carbonyl and C(3) hydroxyl oxygen atoms of substrate (2) (Fig. 1A).
Fig. 1.
Mechanism and structure of EcFBA. (A) Catalytic mechanism of EcFBA. (B) Overall native structure of EcFBA dimer (NAT1). (C) Protomer EcFBA structure showing the TIM barrel fold (pink shade) viewed from the top.
The class I FBAs are mostly distributed in mammals, whereas class II FBAs are only found in protozoa, bacteria, fungi, and blue-green algae. The necessity of class II FBAs in Mycobacterium tuberculosis, Escherichia coli (E. coli), and Streptomyces galbus (3–6) makes this class an attractive target for the development of antibiotics. In addition to the essential roles of FBAs in diverse catabolic and anabolic processes within cells, they have also been widely used in the chemical and pharmaceutical industry to produce various high value stereoisomers of carbohydrates, amino acids, and hydroxylated molecules (7–9).
Previously determined crystal structures of EcFBA alone, and in complex with transition-state analog (phosphoglycolohydroxamate, PGH), showed two mutually exclusive metal binding sites at the active site (2, 10). In Giardia lamblia, the substrate fructose 1,6-bisphosphate (FBP)-bound FBA structure was determined, and a substrate-binding induced metal shift was proposed (11). Studies of developing specific inhibitors against FBAs have been carried out (12, 13). In both mechanistic studies and inhibitor development for new antibiotics, the position and coordination of the catalytic metal ion is essential.
In this study, we determined four different crystal structures (1.8 Å to 2.0 Å) of class II EcFBA. The crystallographic snapshots of EcFBA structures show three different metal-binding states at two sites, having different metal-coordinating environments, which provide the detail mechanism of active site metal shift.
RESULTS
Overall structure of EcFBA
Apo-EcFBA is present as a dimer in the asymmetric unit; its structure is well conserved, when compared to the previously published structure (10). Each protomer has the conserved (α/β)8 barrel fold of other class II FBAs (Fig. 1B and 1C). Dimerization takes place on the surface of outer helices (α2 and α4) of the TIM barrel fold with the additional help of two protruded long helices (α10 and α11) (Fig. S1). Most of the 359 amino acids in EcFBA show good electron density, except for residues 178–195 and 224–235 (Table S1).
As expected, the active site of EcFBA was located at the C-terminal end of the TIM barrel. Till date, no substrate or product bound EcFBA structures are available, and only the reaction intermediate analog phosphoglycolohydroxamate (PGH)-bound EcFBA structure has been determined (2). Since past few years, our group has been studying to determine the substrate, product, or reaction intermediate-bound EcFBA structures. From the trials, we collected several tens of X-ray diffraction datasets from EcFBA crystals soaked with substrate or ligands such as fructose 1,6-bisphosphate, tagatose 1,6-phosphate, and dihydroxyacetone. However, most turned out to be just native EcFBA structures without ligand binding, and we were unable to obtain the reaction related ligand bound EcFBA structure. Nonetheless, EcFBA structures in complex with either citrate or TRIS were unexpectedly determined.
The FBA (GlFBA) structures of native, FBP-bound, and tagatose 1,6-bisphosphate-bound were determined in Giardia lamblia (11), and the citrate-bound FBA (SaFBA) structure was determined in Staphylococcus aureus (14); the GlFBA and SaFBA have sequence identities of 24.3% and 27.5%, respectively, with EcFBA (Fig. S1). When we superimposed EcFBA structure onto GlFBA (PDB ID: 3GB6) and SaFBA (PDB ID: 4TO8) structures, the overall fold and TIM barrel structure was well conserved, but the loop regions showed differences (Fig. S2). The RMSD value of the overall protomer structure between EcFBA and GlFBA, and between EcFBA and SaFBA, are 1.67 Å in 238 residues and 1.60 Å in 250 residues, respectively. The RMSD value of core TIM barrel structure between EcFBA and GlFBA, and EcFBA and SaFBA, are 1.76 Å in 139 residues and 1.50 Å in 139 residues, respectively.
In EcFBA, two metal sites were located at the C-terminal side of the β-barrel structure of each protomer, separated by a distance of 3.7 Å. One site (termed ‘Zn1’) was at the interior of protein and the other site (termed ‘Zn2’) existed on the active site surface.
Metal binding and coordination in FBA structures
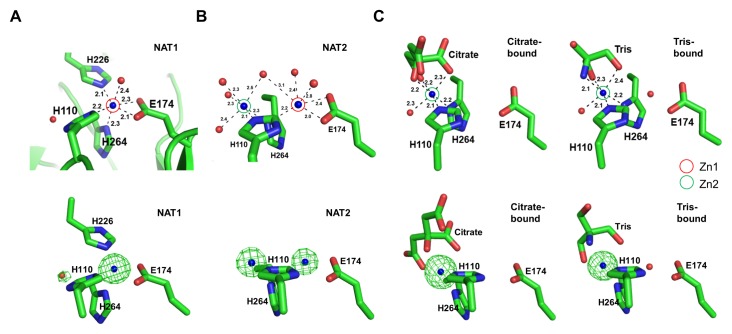
Previously, Blom et al. reported an EcFBA structure (PDB ID: 1DOS), which had two mutually exclusive Zn2+ binding sites (10). In this study, we determined almost the same two metal-binding sites in EcFBA structures from four different datasets. The crystallographic snapshots of EcFBA structures show the clear tendency that the metal site having more coordinations has the stronger electron density of metal. We classified the datasets into three different metal-binding states: 1) Zn2+ ion exclusively bound at the interior (Zn1) of EcFBA, 2) Zn2+ ion partially bound at both the interior (Zn1) and the active site surface (Zn2) of EcFBA, and 3) Zn2+ ion exclusively bound at the active site surface (Zn2) of EcFBA (Fig. 2).
Fig. 2.
Crystallographic snapshots of three different metal-binding states of EcFBA. (A) Only Zn1 was occupied in NAT1 structure. (B) Both Zn1 and Zn2 were partially occupied in NAT2 structure. (C) Only Zn2 was occupied in citrate (left) or TRIS-bound (right) structures. Upper figures show stick representations of Zn1 and Zn2 in active site, and lower figures show omit maps (contoured at 10.0 σ; green) of Zn2+ in Zn1 and Zn2.
Native EcFBA structures
Both metal-binding sites were observed in two native EcFBA structures (NAT1 and NAT2). In NAT1, Zn1 consisted of three His residues (His110, His226, and His264) and Glu174 (Fig. 2A). In NAT2, Zn2 consisted of two His residues (His110 and His264) (Fig. 2B). The three His and one Glu residues are strictly conserved in class II FBAs (Fig. S1). The imidazole rings of His110 and His264 could form a H-bond with Zn2+ ion at both sites. The electron density of the metal ion was stronger than that expected for sodium ion, and weaker than that for zinc ion at full occupancy. We calculated the occupancy of zinc ion at both sites, and placed zinc ions with partial occupancy at both sites (Table S2).
In the NAT1 structure, zinc ion was hexacoordinated; His110, His226, and Glu174 (bifurcated hydrogen bonds) coordinated the zinc ion in-plane, while a water molecule and His264 coordinated above and below (Fig. 2A). In the NAT2 structure, two zinc ions were positioned at Zn1 and Zn2, with only two coordinating His residues; the His226 was missing from disordered structure (Fig. 2B). The coordination at Zn2 involved His110 and a ‘rotated conformation’ of His264, together with four water molecules. In order to coordinate both Zn1 and Zn2, His110 sidechain could adopt a mixed ‘flip-flop’ state and His264 showed dual rotated conformations. Both of His110 and His264 residues were positioned as dual conformations with partial occupancy.
Citrate-bound and TRIS-bound EcFBA structures
Citrate was present at a high concentration of 0.1 M in the crystallization condition of citrate-bound EcFBA structure. Changing the buffer to TRIS also led to the TRIS bound structure. In the citrate-bound structure, zinc ion was bound at Zn2, where it was hexacoordinated with two His residues (His110 and His264), three oxygen atoms from the citrate molecule, and a water molecule (Fig. 2C). One conformation of His264 existed that faced the Zn2. His226 was not visible in the electron density maps. In the TRIS-bound structure, zinc ion at Zn2 was bound in a similar way as that in the citrate-bound structure, except that the two oxygen atoms and one nitrogen atom of TRIS replaced the three oxygen atoms of citrate (Fig. 2C).
Comparison of EcFBA structures
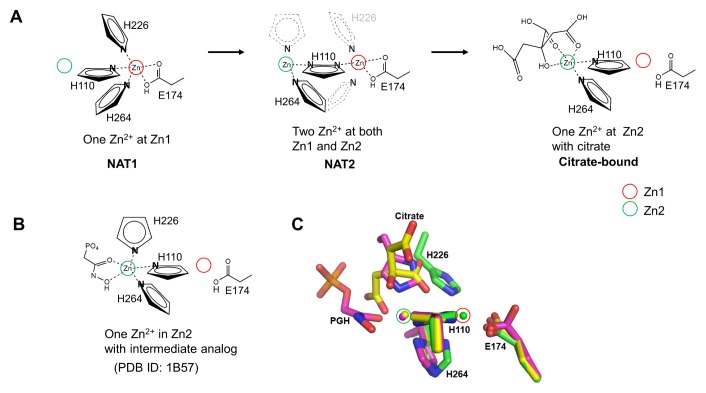
We superimposed the determined EcFBA structures to identify any conformational changes (Table S3). We observed that the overall structures and positions of both Zn2+ binding sites were well conserved in all structures. The imidazole ring plane of His110 was parallel to the two metal sites. Two other coordinating His residues of His264 (below the metal ion) and His226 (above the metal ion) changed their conformation depending on the corresponding coordinating metal ion; His264 changed the conformation towards the coordinating Zn2+ ion, and His226 was visible only when coordinating Zn1. The citrate or TRIS-binding site partially overlapped with His226 coordinating Zn2 (Fig. 3C). When we superimposed the PGH-bound EcFBA structure [PDB ID: 1B57 (2)], His226 coordinated Zn2 at the same position occupied by either citrate or TRIS. In the PGH-bound structure, Zn2 was penta-coordinated by three His residues (His110, His226, and His264) and two oxygen atoms from PGH (Fig. 3B).
Fig. 3.
Metal-binding site structures of EcFBA. (A) Schematic drawing of metal shift in the EcFBA active site with corresponding conformational changes of coordinating residues. Only coordinating nitrogen atom in histidine residues is shown. In NAT2 structure, flip-flop dual conformations of His110 are shown as two nitrogen atoms in a single imidazole ring. His226 residue is not shown due to disordered conformations, and the proposed positions are represented with grey dashed lines. His264 residue is shown as dual conformations to coordinate Zn2+ at Zn1 or Zn2. (B) Schematic drawing of phosphoglycolohydroxamate (PGH)-bound EcFBA structure (2). (C) Comparison between native and ligand-bound EcFBA structures. Superimposed active site structures between NAT1 (green), citrate-bound (yellow), and phosphoglycolohydroxamate (PGH)-bound (purple) EcFBA structures.
DISCUSSION
A divalent metal ion plays an essential role in the catalysis of class II FBAs. In this study, three different states of metal binding and coordination were observed in the EcFBA structures, accompanied by conformational changes in the corresponding coordinating residues (Fig. 2 and 3A). The crystallographic snapshots of EcFBA structures suggest the detail mechanism of metal shift between two sites.
Zn1 and Zn2 intrinsically have five and three coordinating electron donors, respectively. Three coordinators were the same His residues shared at both Zn1 and Zn2, showing conformational changes. The additional two coordinators at Zn1 were the bifurcated side chain of Glu174 residue. Therefore, the preferential coordination of the three His residues at one site will diminish the coordination at the other site. When we compared NAT1 structure with NAT2 structure, one coordinating residue (His226) in Zn1 was lost in the NAT2 structure, thus decreasing the metal-binding affinity at Zn1, and making it easier for Zn2+ from Zn1 to move to Zn2 with the rotation of His264 to Zn2. We determined two ligand (citrate or TRIS)-bound EcFBA structures, and both substrate-mimicking ligands coordinated Zn2. Zn2 existed on the active site surface of the protein and had only two possible coordinating His residues of His110 and His264 in apo-EcFBA, considering flexible His226. In the Zn2 of apo-EcFBA, additional metal coordinating positions should be left unoccupied for substrate binding. Binding of substrate at the active site provides two additional metal coordinating electron donors at Zn2. When Zn2+ moves from Zn1 to Zn2 with conformational changes of His110 and His264, the metal coordination at Zn2 gets stronger; at the same time, that of Zn1 gets weaker (Fig. 4). The enhanced Zn2+ binding at Zn2 diminishes the metal-binding affinity of Zn1. In the ligand-bound EcFBA structures, no Zn2+ was bound at Zn1. In both GlFBA and SaFBA structures, Zn1 and Zn2 were conserved; the zinc ion binds at Zn1 without ligand and at Zn2 with ligand (Fig. S3), which corresponds with the EcFBA structures in this study.
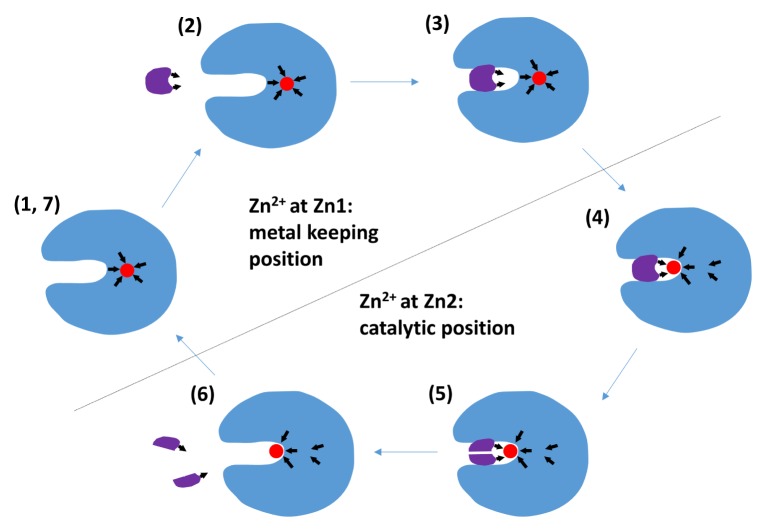
Fig. 4.
Proposed mechanism of active site Zn2+ movement in EcFBA. (1) Native EcFBA structure with a metal ion at the interior protein (Zn1). (2) The substrate contains a metal-coordinating motif. The active site surface also contains a metal-binding site (Zn2). (3) When substrate binds at active site, Zn2 is formed with more electronegative coordinators. (4) The completely formed Zn2 recruits a metal ion from Zn1 to Zn2. (5) The active form of EcFBA performs catalytic reaction. (6) Products are released from the active site and metal ion at Zn2 loses parts of coordination that had been provided by the substrate. (7) Metal ion at Zn2 returns to Zn1, which has more complete coordination at the interior of the protein. The arrows show the direction of coordination by His and Glu residues and substrate.
We speculate that Zn1 might be a reservoir for trace metal Zn2+. Zn2 is exposed to solution because Zn2+ should directly interact with substrate and perform catalysis, of which the minimal coordination geometry without substrate-binding could risk the loss of the valuable trace metal Zn2+ from EcFBA. The conditional shift of the metal cofactor to the inner position with more secure coordination of EcFBA, could be helpful to keep the essential metal cofactor within the enzyme. The detail metal shift mechanism will be useful for better understanding the catalytic mechanism of EcFBA, and possibly of general FBAs.
MATERIALS AND METHODS
Gene cloning
The d-fructose-1,6-bisphosphate aldolase (fbaA, GI: 16130826) gene encodes 359 amino acid residues. The cloning of fbaA gene was carried out with the normally used method (15). fbaA was amplified by PCR using genomic DNA isolated from E. coli K-12, using a genomic DNA extraction kit (GeneAll, Seoul, Korea). The forward and reverse primers for fbaA contained SalI and NotI restriction sites (underlined): 5′-GTCGACATGTCTAAGATTTT TGATTTCGTAAAACC-3′ and 5′-ATAATGCGGCCGCTTACAGAACGTCGATCGCGTT-3′. DNA fragments obtained by PCR amplification with Taq polymerase (Bioneer, Alameda, CA, USA) were purified using a PCR purification kit (Qiagen, Valencia, CA, USA) and ligated into the pRSF-duet-1 vector (Novagen, Madison, WI, USA), which was then transformed into E. coli ER2566 cells and plated on Luria-Bertani (LB) agar containing 0.1 mM kanamycin. A kanamycin-resistant colony was selected, and its plasmid was sequenced using a DNA analyzer (ABI Prism 3730xl; Perkin-Elmer, Waltham, MA, USA).
Overexpression and purification
The EcFBA was expressed and purified with the normally used recombinant protein expression and purification method (16). The recombinant E. coli cells were cultured at 37°C in a 2 L-flask containing 500 ml of LB medium and 0.1 mM kanamycin, while being shaken at 200 rpm. When the optical density of the bacterial culture reached 0.6 at 600 nm, isopropyl-β-d thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM, inducing EcFBA expression. Following this, the culture was incubated on a shaker for a further 16 h at 16°C, at 150 rpm. Cells were harvested from the culture broth by centrifugation at 6,000×g for 30 min (4°C) and re-suspended in 50 mM phosphate buffer (pH 8.0) containing 300 mM NaCl, 10 mM imidazole, and a protease inhibitor (0.1 mM phenylmethylsulfonyl fluoride). The re-suspended cells were disrupted by sonication, on an ice bath. The cell debris was removed by centrifugation at 1,3000×g for 20 min (4°C), and the supernatant was filtered through a 0.45-μm filter. The filtrate was applied to an immobilized metal ion affinity chromatography (IMAC) cartridge (Bio-Rad) equilibrated with 50 mM phosphate buffer (pH 8) containing 300 mM KCl. The column was washed extensively with 50 mM phosphate buffer (pH 8) containing 300 mM KCl and 30 mM imidazole, and the bound protein was eluted with 50 mM phosphate buffer (pH 8) containing 300 mM KCl and 300 mM imidazole, at a flow rate of 1 ml/min. The active fractions were collected and dialyzed in 50 mM Tris-HCl buffer (pH 8.5). After dialysis, the resulting solution contained the purified enzyme that was used for crystallization trials. The affinity purification tag from the expression vector was not cleaved off, and was retained in the purified enzyme. A standard Bradford assay was used to determine protein concentration. Protein expression was evaluated by SDS-PAGE.
Crystallization and X-ray data collection
Initial crystallization was carried out at 287 K in 96-well plates (Intelli-Plates, Art Robbins, USA), using the sitting-drop vapor diffusion method. An automated pipetting system (Hydra II E-drop, Matrix) was used with screening kits from Hampton Research and Emerald Biosystems. The drops consisted of 0.5 μl protein solution (10 mg/ml in 20 mM Tris, pH 7.0) and 0.5 μl reservoir solution; these were then equilibrated against 70 μl reservoir solution at 14°C. After 3 days, nascent thin planar EcFBA crystals were observed in ‘Hampton Crystal Screen Lite condition No. 09’ (15% PEG 4000, 0.2 M ammonium acetate, and 0.1 M sodium citrate tribasic dihydrate, pH 5.6). These crystals were reproduced using hanging drops consisting of 0.9 μl of protein solution mixed with 0.9 μl of reservoir solution. Each hanging drop was positioned over 1 ml of the reservoir solution. Crystals were optimized by varying the PEG concentration and by assessing four buffers (sodium citrate, HEPES, TRIS, Bis-TRIS, and MES). Thick planar EcFBA crystals were obtained in reservoir solutions consisting of 0.2 M ammonium acetate, 15% PEG 4000 and 0.1 M of any the four buffers listed (above), covering a range of pH values from 5.5 to 7.0. However, most fully grown crystals were seen at pH 7.0. Potassium chloride (10 mM) was added to the reservoir solutions for soaking ligands of substrate fructose 1,6-bisphosphate or inhibitor tagatose 1,6-bisphosphate into apo-EcFBA crystals. All crystals were cryoprotected with 20% (v/v) glycerol, and then flash-cooled at 100 K in liquid nitrogen. X ray diffraction data were collected from the crystals on beamline 5C, Pohang Accelerator Laboratory (PAL, Republic of Korea), using an ADSC Q315r CCD detector. The data were integrated and scaled using DENZO and SCALEPACK, respectively.
Structure determination and refinement
The apo structure of EcFBA was determined by molecular replacement using Phaser-MR from the CCP4 software package (17), using the 1.6 Å structure of EcFBA (PDB: 1DOS) having a 99.7% sequence identity, as a template model. Construction of both the model and electron density was performed in COOT (18). The resulting model was refined using Refmac5 in the CCP4 package (19). The apo-EcFBA structure was employed as a template for determining the other EcFBA structures by molecular replacement. The occupancy of metal ion was calculated in PHENIX (20). The resulting structures were validated in PROCHECK (21). The statistics of structure refinement are summarized in Table S1. Root-mean-square deviation values for the superimposed structures were calculated using the GESAMT tool (22) in ‘Superpose Molecules’ in the CCP4 software suite. Structures were displayed using PyMOL (23). Structure-based multiple sequence alignment was performed in T-Coffee (24), and then presented using the ESPript server (25). The determined EcFBA structures were deposited in the Protein Data Bank (PDB): NAT1 structure (crystal I) was deposited as 5GK3, NAT2 structure (crystal II) as 5GK4, Citrate-bound structure as 5GK6, and Tris-bound structure as 5GK7.
Supplementary Information
ACKNOWLEDGEMENTS
We are grateful to the staff members of Beamline 5C at the Pohang Accelerator Laboratory (PAL), Republic of Korea. This paper was supported by Konkuk University in 2014.
REFERENCES
- 1.Lorentzen E, Siebers B, Hensel R, Pohl E. Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins. Biochemistry. 2005;44:4222–4229. doi: 10.1021/bi048192o. [DOI] [PubMed] [Google Scholar]
- 2.Hall DR, Leonard GA, Reed CD, Watt CI, Berry A, Hunter WN. The crystal structure of Escherichia coli class II fructose-1, 6-bisphosphate aldolase in complex with phosphoglycolohydroxamate reveals details of mechanism and specificity. J Mol Biol. 1999;287:383–394. doi: 10.1006/jmbi.1999.2609. [DOI] [PubMed] [Google Scholar]
- 3.Marsh JJ, Lebherz HG. Fructose-bisphosphate aldolases: an evolutionary history. Trends Biochem Sci. 1992;17:110–113. doi: 10.1016/0968-0004(92)90247-7. [DOI] [PubMed] [Google Scholar]
- 4.Pegan SD, Rukseree K, Franzblau SG, Mesecar AD. Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis. J Mol Biol. 2009;386:1038–1053. doi: 10.1016/j.jmb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerdes SY, Scholle MD, Campbell JW, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wehmeier UF. Molecular cloning, nucleotide sequence and structural analysis of the Streptomyces galbus DSM40480 fda gene: the S. galbus fructose-1,6-bisphosphate aldolase is a member of the class II aldolases. FEMS Microbiol Lett. 2001;197:53–58. doi: 10.1111/j.1574-6968.2001.tb10582.x. [DOI] [PubMed] [Google Scholar]
- 7.Wagner J, Lerner RA, Barbas CF. Efficient aldolase catalytic antibodies that use the enamine mechanism of natural enzymes. Science. 1995;270:1797–1800. doi: 10.1126/science.270.5243.1797. [DOI] [PubMed] [Google Scholar]
- 8.Von der Osten CH, Sinskey AJ, Barbas CF, Pederson RL, Wang YF, Wong CH. Use of a recombinant bacterial fructose-1, 6-diphosphate aldolase in aldol reactions: preparative syntheses of 1-deoxynojirimycin, 1-deoxymannojirimycin, 1, 4-dideoxy-1, 4-imino-D-arabinitol, and fagomine. J Am Chem Soc. 1989;111:3924–3927. doi: 10.1021/ja00193a025. [DOI] [Google Scholar]
- 9.Liu J, Hsu C-C, Wong C-H. Sequential aldol condensation catalyzed by DERA mutant Ser238Asp and a formal total synthesis of atorvastatin. Tetrahedron Lett. 2004;45:2439–2441. doi: 10.1016/j.tetlet.2004.01.110. [DOI] [Google Scholar]
- 10.Blom NS, Tetreault S, Coulombe R, Sygusch J. Novel active site in Escherichia coli fructose 1,6-bisphosphate aldolase. Nat Struct Biol. 1996;3:856–862. doi: 10.1038/nsb1096-856. [DOI] [PubMed] [Google Scholar]
- 11.Galkin A, Li Z, Li L, et al. Structural insights into the substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase. Biochemistry. 2009;48:3186–3196. doi: 10.1021/bi9001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daher R, Coincon M, Fonvielle M, et al. Rational design, synthesis, and evaluation of new selective inhibitors of microbial class II (zinc dependent) fructose bis-phosphate aldolases. J Med Chem. 2010;53:7836–7842. doi: 10.1021/jm1009814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capodagli GC, Sedhom WG, Jackson M, Ahrendt KA, Pegan SD. A noncompetitive inhibitor for Mycobacterium tuberculosis’s class IIa fructose 1,6-bisphosphate aldolase. Biochemistry. 2014;53:202–213. doi: 10.1021/bi401022b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capodagli GC, Lee SA, Boehm KJ, Brady KM, Pegan SD. Structural and functional characterization of methicillin-resistant Staphylococcus aureus’s class IIb fructose 1,6-bisphosphate aldolase. Biochemistry. 2014;53:7604–7614. doi: 10.1021/bi501141t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SJ, Jang JH, Yoon GY, et al. Mycobacterium abscessus D-alanyl-D-alanine dipeptidase induces the maturation of dendritic cells and promotes Th1-biased immunity. BMB Rep. 2016;49:554–559. doi: 10.5483/BMBRep.2016.49.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JH, Choi JS, Kim S, et al. Synergistic effect of two E2 ubiquitin conjugating enzymes in SCF(hFBH1) catalyzed polyubiquitination. BMB Rep. 2015;48:25–29. doi: 10.5483/BMBRep.2015.48.1.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murshudov GN, Skubak P, Lebedev AA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Afonine PV, Grosse-Kunstleve RW, Echols N, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laskowski R, MacArthur M, Moss D, Thornton J. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 22.Krissinel E. Enhanced fold recognition using efficient short fragment clustering. J Mol Biochem. 2012;1:76–85. [PMC free article] [PubMed] [Google Scholar]
- 23.Schrodinger L. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- 24.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 25.Gouet P, CE, SD, Metoz F. ESPript: multiple sequence alignments in PostScript. Bioinformatic. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.