Abstract
ARF is an alternative reading frame product of the INK4a/ARF locus, inactivated in numerous human cancers. ARF is a key regulator of cellular senescence, an irreversible cell growth arrest that suppresses tumor cell growth. It functions by sequestering MDM2 (a p53 E3 ligase) in the nucleolus, thus activating p53. Besides MDM2, ARF has numerous other interacting partners that induce either cellular senescence or apoptosis in a p53-independent manner. This further complicates the dynamics of the ARF network. Expression of ARF is frequently disrupted in human cancers, mainly due to epigenetic and transcriptional regulation. Vigorous studies on various transcription factors that either positively or negatively regulate ARF transcription have been carried out. However, recent focus on posttranslational modifications, particularly ubiquitination, indicates wider dynamic controls of ARF than previously known. In this review, we discuss the role and dynamic regulation of ARF in senescence and cancer.
Keywords: ARF, ARF knockout mice, Cancer, Posttranslational regulation, Senescence, Tumorigenesis, Ubiquitination
INTRODUCTION
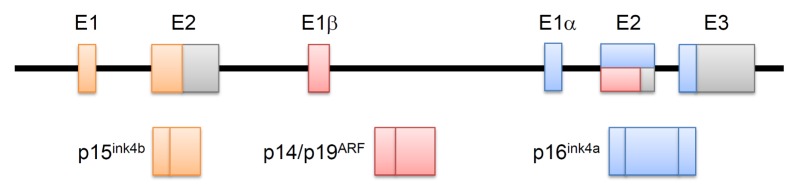
The tumor suppressors p15ink4b, p14ARF, and p16ink4a, are well-characterized products of the INK4b/ARF/INK4a gene locus. Corresponding to the region of mouse chromosome 4, this locus is situated on chromosome 9 of the human genome, within a 35 kb region. As expected for potent tumor suppressors, the INK4b/ARF/INK4a locus is frequently targeted for deletion or epigenetic suppression in numerous cancers. The coding regions of p14ARF and p16ink4a start from exons 1β and 1α, respectively. Moreover, owing to an alternative reading frame, they comprise of completely different amino acids despite sharing exons 2 and 3. Although p15ink4b has a similar role in cell cycle inhibition, it has a coding region physically separated from that of p14ARF and p16ink4a (Fig. 1) (1, 2).
Fig. 1.
The INK4b/ARF/INK4a locus. The INK4b/ARF/INK4a locus encodes three proteins, including p15INK4b, p14ARF, and p16INK4a. The coding regions of p14ARF and p16ink4a start in different exons—exon 1, 1β, and 1α, respectively. Exon 2 of the INK4b/ARF/INK4a locus is shared by p14ARF and p16ink4a. They comprise completely different amino acids, despite sharing the exons 2 and 3, through an alternative reading frame.
As a tumor suppressor, ARF is intimately related to p53 stabilization, which induces cellular senescence and prevents tumor cell growth. ARF releases p53 from MDM2, a well-known E3-ubiquitin ligase of p53, by trapping MDM2 in the nucleolus via direct interaction, which physically separates p53 from MDM2 (3–6). Interaction of ARF with protein partners other than MDM2, also results in suppression of tumor growth via induction of either cellular senescence or apoptosis, in a p53-independent manner. These have been evidenced using p53-deficient cell lines and knockout (KO) mice models. Moreover, recent identification of posttranslational regulatory mechanisms further complicates the dynamics of the ARF mechanism (7–9). In this review, we discuss the transcriptional as well as posttranslational regulatory mechanisms of ARF, in an attempt to provide comprehensive understanding of the same.
REGULATION OF ARF
Transcriptional regulation of ARF
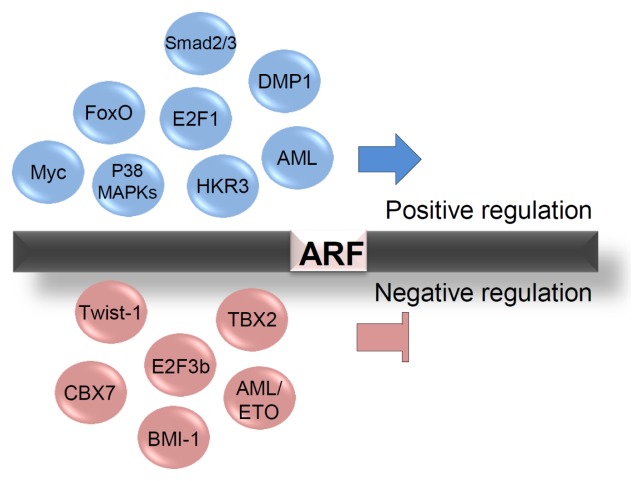
Since the emergence of ARF as a tumor suppressor, the regulation of its expression has been one of the most pursued areas of research. Numerous transcription factors (TFs) regulate ARF either positively or negatively (Fig. 2). In particular, the E2F1 transcription factor (which controls genes involved in cell cycle regulation) induces ARF transcription by directly interacting with its binding site upstream of the exon 1β, thus activating apoptosis and cell growth arrest (10). This is one of the most well characterized pathways of oncogene-induced senescence, activated upon oncogenic stress. However, this process is modulated differently by the various E2F isoforms. For instance, E2F3b, an isoform of E2F3, represses ARF transcription and stimulates cellular growth. The observation that the loss of ARF can rescue E2F3b depletion-mediated cell growth arrest clearly suggests anti-reciprocal correlation between these two proteins (11). Myc, another well-established and vigorously investigated oncogene, exhibits regulatory patterns similar to E2F1. To prevent hyper-proliferation of cells under oncogenic stress, it also activates fail-safe programs such as apoptosis and cellular senescence, by inducing ARF transcription (12). Frederique and colleagues reported that Myc overexpression induces accumulation of ARF both at the mRNA and protein levels in mouse embryonic fibroblasts (MEFs) (13). They showed that Myc-induced ARF activates p53 signaling, thus preventing immortalization of the MEFs. Further, Myc also activates FoxO-mediated ARF transcription. Caroline and colleagues reported that Myc signaling increases nuclear FoxO, which in turn binds to the ARF promoter and thus suppresses Myc-driven lymphomagenesis in mice (14). A cyclin D-binding Myb-like protein, DMP1, also induces ARF. Kazushi and colleagues demonstrated that DMP1 binds to the ARF promoter and activates its transcription in MEFs, which results in cell cycle arrest (15). Furthermore, the ARF promoter also provides several binding sites for acute myeloid leukemia-1 (AML1), which activates its transcription leading to cellular senescence in MEFs. However, the t(8;21) fusion protein AML1-ETO, which is frequently expressed in acute leukemia, represses ARF expression (16). Meanwhile, oncogenic signaling by RAS influences ARF expression in a DMP1-dependent manner. RAS signaling induces Jun-mediated DMP1 transcription, thus increasing ARF expression (17). Next, Yanbin and colleagues suggested that TGFβ signaling directly induces ARF expression. They found that TGFβ signaling elevates the ARF mRNA levels through Smad2/3 and p38 MAPKs in MEFs. Smad2/3 binds directly to the ARF promoter upon stimulation by TGFβ. However, it is yet to be addressed how Smad and p38 MAPK signaling cooperate to induce ARF expression (18). p38 MAPK has previously been reported to regulate ARF expression. Dmitry and colleagues found that decreased Wip1 phosphatase increases ARF expression in a p38 MAPK-dependent manner in Ppm1d−/− MEFs (19). Moreover, a recent study suggests that HKR3 (Human Krüppel-related 3) activates ARF transcription by binding to the ARF promoter with coactivator p300, which induces acetylation of the histones H3 and H4 (20).
Fig. 2.
Transcriptional regulation of ARF. Positive and negative regulators of ARF transcription. Transcriptional activators and repressors are shown in the upper part and lower parts, respectively.
Several repressors of ARF transcription have also been reported (Fig. 2). For example, the polycomb group gene BMI-1 suppresses cellular senescence through repression of ARF transcription. BMI-1-deficient MEFs show impaired cell cycle progression and enter premature senescence, which are rescued by ARF depletion (21). BMI-1 also requires the EZH2-containing Polycomb-Repressive Complex 2 (PRC2) to repress ARF transcription. PRC2 maintains the levels of H3K27Me3 as well as the BMI-1/PRC1 complex at the ARF locus (22). Another polycomb group gene, CBX7, increases the lifespan of normal human cells and MEFs through suppression of ARF expression, independent of BMI-1 (23). Further, TBX2 immortalizes MEFs and decreases senescence in normal human cells by repression of ARF transcription (24). Basic helix-loop-helix (bHLH) transcription factor Twist-1 activates the recruitment of EZH2 to the ARF transcription start site. Thus, it increases the levels of H3K27Me3 on the ARF locus, followed by repression of ARF transcription (25).
Although many factors that regulate the transcription of ARF are well described, its posttranslational regulation is largely unknown. Recently, some posttranslational regulators of ARF have been reported, thus magnifying its importance in senescence and tumorigenesis.
Posttranslational regulation of ARF
Although the functional importance of ARF in cellular senescence and tumor suppression is well characterized, knowledge about its posttranslational regulation is limited. A previous report suggested that ARF, which has no lysine sites, is polyubiquitinated at its N-terminus followed by proteasomal degradation by an unknown E3-ubiquitin ligase (26). Five years later, the first E3-ubiquitin ligase for ARF (TRIP12) was identified and named ULF (ubiquitin ligase for ARF) (Fig. 3) (7). ULF induces polyubiquitination and proteasomal degradation of ARF, thus activating cell proliferation. Interestingly, the ULF-mediated degradation of ARF is further regulated by NPM and c-Myc, suggesting that Myc regulates ARF both transcriptionally and translationally. Subsequently, Ko and colleagues reported a second E3-ubiquitin ligase, Makorin 1 (MKRN1), which targets ARF. MKRN1 KO MEFs presented retarded cell growth and senescence with concomitant increase in the ARF protein levels (8). Corroborating these results, MKRN1 was shown to induce ubiquitination and proteasomal degradation of ARF. Moreover, its ablation decreased tumor growth through induction of the ARF-dependent senescence in xenograft models using p53-positive and -negative gastric cancer cell lines. Further, tumor tissues from gastric cancer patients showed negative correlation between MKRN1 and ARF proteins. Siva1, an E3-ubiquitin ligase for ARF, induces the proteasomal degradation of ARF, thus inhibiting p53 function (9). The existence of several E3 ligases for ARF is intriguing. Systemic studies on the cellular localization of these E3 ligases might help to shed some light. It is also possible that each ligase is a mediator that links the different signaling pathways involving ARF. Extensive studies on each ligase are required for the complete elucidation of ARF regulation.
Fig. 3.
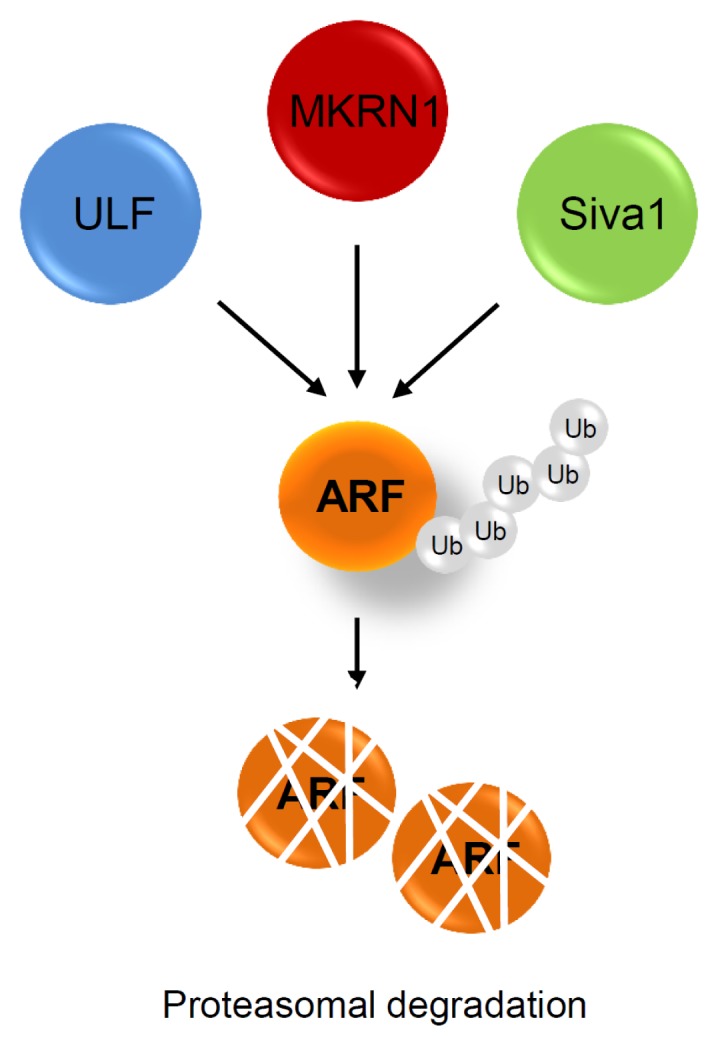
Posttranslational regulation of ARF. Three ubiquitin E3 ligases have been reported. ULF, MKRN1, and Siva1 bind directly to, and induce ubiquitination of ARF, resulting in the induction of its proteasomal degradation.
ARF IN SENESCENCE AND CANCER
Tumor suppressive function of ARF
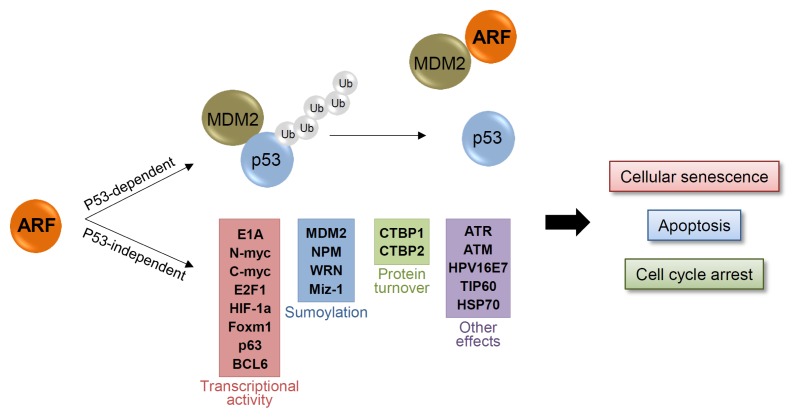
Senescence is an irreversible cell growth arrest that prevents the hyper-proliferation of cancer cells. As oncogenic signal stimulation always carries the risk of cancer cell eruption, normal cells require senescence as a fail-safe program to prevent this hyper-proliferation and tumorigenesis. ARF plays a key role in this fail-safe program through activation of the p53 pathway (Fig. 4). ARF binds and sequesters MDM2 in the nucleolus, leading to the activation of p53 in the nucleoplasm (3–6). The activated p53 then promotes transcription of numerous target genes such as BAX or p21, thus inducing either cellular senescence or apoptosis (27–29). Mice lacking p19ARF are susceptible to early tumors. In fact, ARF and p53 double KO mice have tumor incidences similar to those with p53 KO alone. This suggests that the tumor suppressive function of ARF is entirely p53-dependent (30).
Fig. 4.
p53 dependent and independent tumor suppressive functions of ARF. ARF plays a key role in cellular senescence and tumorigenesis through activation of the p53 pathway. ARF binds and sequesters MDM2 in the nucleolus, leading to the activation of p53 in the nucleoplasm. p53 transcribes numerous target genes and activates cellular senescence or apoptosis. ARF also functions in a p53-independent manner through interaction with numerous target proteins. ARF regulates sumoylation, transcription activities, or protein turnover of target proteins, which lead to tumor suppression and senescence.
While there is no argument on the dependency of ARF on p53, some evidence shows that ARF might have a p53-independent tumor suppressive function as well. Weber and colleagues reported that triple KO (TKO) mice for MDM2, p53, and p19ARF show greater frequency of multiple tumors than do double KO mice for MDM2 and p53. Moreover, reintroduction of ARF into TKO MEFs induces cell cycle arrest. These results suggest that ARF has an additional tumor suppressive role that is p53/MDM2-independent (31). Genetic deficiency in ARF, but not p53, accelerates the development of melanoma in a genetically engineered mouse model, while senescence is induced in p53- but not in ARF-deficient primary melanocytes (32). The induction of cell cycle arrest, cellular senescence, apoptosis, and regression in xenograft tumors by ARF in p53-deficient cell lines indicates the obvious p53-independent roles for ARF (33–38). Possibly, p53-independent functions of ARF are through its interactions with numerous regulatory proteins (Fig. 4). For example, Tip60 directly binds to and induces acetylation of retinoblastoma (RB), leading to its destabilization through proteasomal degradation. ARF prevents the Tip60-mediated RB acetylation and leads to accumulation of hyper-phosphorylated RB, which then triggers anti-proliferative functions (39). Further, through MALDI-TOF analyses, NPM is identified as a binding partner of ARF. ARF binds and sequesters NPM in the nucleolus, thus blocking its nucleocytoplasmic shuttling, and resulting in subsequent cell growth arrest (40, 41). ARF also inhibits the activities of various TFs such as E2F1, HIF-1α, Foxm1, c-Myc and n-Myc through direct interaction, thus preventing cell proliferation (33, 42–45). This demonstrates the negative feedback regulation of ARF induced by some of these TFs. ARF also induces the ATR/CHK1-dependent RelA (NF-kB) phosphorylation and reduces its transcriptional activity (46). Furthermore, CtBP1 and 2, transcription co-repressors that have anti-apoptotic functions, are also reported to interact with ARF. Binding of ARF to CtBP1 and 2 promotes their proteasomal degradation (47). In addition, ARF also stimulates the sumoylation of several target proteins including NPM, MDM2, WRN, and Miz-1 (48–52). While its mechanism remains unclear, the observation that ARF interacts with UBC9, a sumo E2 conjugating enzyme, indicates that ARF might facilitate sumo transfer from the E2 complex to its target proteins (52). In addition, there are many other proteins that interact with ARF (such as HPV16E7, HSP70 and others) and assist in its p53-dependent or -independent functions (53, 54).
In short, ARF has numerous interacting partners other than MDM2, which facilitate and maximize its tumor suppressive effect via induction of cellular senescence or apoptosis in p53-dependent or -independent manner (Fig. 4). Given its p53-dependent and -independent functions, elaborate studies on the context-dependent roles of ARF are required, since most cancer cells are either deprived of p53 or carry mutant p53.
ARF KO mice
The first KO mice for ARF were developed even before it was found that INK4a and ARF coexist on the same genomic locus. As a result, the first KO mice had both the INK4a and ARF sites eliminated. These mice spontaneously developed various types of cancers within the first year. KO MEFs also displayed significantly faster growth rate than wild type MEFs. Furthermore, they were transformed by oncogenic stimulus of RASV12 (55). Later, mice lacking p19ARF but not p16INK4a were generated by targeting exon 1β. These mice also developed numerous tumors, spontaneously as well as upon carcinogen treatment, leading to death within the first year. In several mouse strains (C57BL6, 129svj X C57BL6 and FVB), ARF-specific KO mice develop sarcoma, squamous cell carcinoma, lung carcinoma, and metastatic lymphoma. Moreover, exposure to carcinogens such as DMBA, X-rays, and irradiation make ARF KO mice more prone to tumors (Table 1) (56–58). The fact that mice and MEFs lacking ARF alone show similar features of cancer development and cell growth, elicited a curiosity about the characteristics of mice KO for Ink4a alone. In contrast to p19ARF-deficient MEFs, MEFs that lack p16Ink4a but retain p19ARF show normal cell growth features and are susceptible to RAS-induced senescence (59). Meanwhile, p16-null MEFs exhibit increased immortalization compared with wild type MEFs, though less compared with p19ARF KO and p19ARF/p16ink4a KO MEFs. KO mice lacking p16 alone also develop spontaneous and carcinogen-induced tumors, which indicate that p16Ink4a functions as a tumor suppressor in association with p19ARF.
Table 1.
Knockout mice of ARF and INK4a
| Strain | Targeting site | Treatment | Type of tumor formation | Latency | Ref | |
|---|---|---|---|---|---|---|
| ARF−/− | 129svj X C57BL6 | Exon1β | None | Sarcoma (43%) Lymphoid malignancies (29%) Carcinoma (17%) /Squamous cell carcinoma /Pulmonary adenocarcinoma /Poorly differentiated carcinoma in pancreas /Salivary gland carcinoma Tumors of the nervous system (11%) /Spinal cord neural sheath tumor/Glioma |
~38 weeks | 56 |
| DMBA | Squamous cell carcinoma /+Lymphoma /+Sarcoma and adnexal tumor /+Sarcoma and lymphomax /+sarcoma |
~24 weeks | ||||
| X-ray | Sarcomas Lymphoma Meningeal tumor |
|||||
| ARF−/− | FVB | Exon1β | None | Small lymphoma (33%) Malig. Sp. Cell neo (30%) Lung carcinoma (12%) Osteogenic sarcoma (9%) Tumors of the nervous system (9%) Carcinoma/HCC (6%) |
~62 weeks | 57 |
| DMBA | Small lymphoma (60%) Malig. Sp. Cell neo (40%) Lung carcinoma (15%) |
~24 weeks | ||||
| ARF−/− | C57Bl/6 | Exon1β | None | Fibrosarcoma (33.3%) Metastatic salivary gland carcinoma (16.7%) Thymoma (16.7%) Malignant fibrous histocytoma (16.7%) Lymphoma (brain) (16.7%) |
~21 weeks | 58 |
| DMBA | Epidermal papilloma (55.5%) Lymphoma, Epidermal papilloma Fibrosarcoma, malignant adenexal tumor Fibrosarcoma, epidermal papilloma Invasive epidermoid carcinoma |
~20 weeks | ||||
| Irradiation | Fibrosarcoma (50%) Lymphoma (brain) (50%) |
~19 weeks | ||||
| INK4a−/− | FVB | Exon1α | None | Soft-tissue sarcoma (12.8%) Splenic lymphoma (10.25%) Melanoma (2.56%) |
~44 weeks | 59 |
| DMBA | Thymic lymphoma (13%) Splenic lymphoma (6%) Soft-tissue sarcoma (10%) Malignant spindle-cell neoplasma (6%) Lung adenoma (6%) Squamous papilloma (29%) |
~23 weeks | ||||
| INK4a−/−, ARF−/− | C57BL/6 | Exon2, 3 | None | Fibrosarcoma (33.3%) Sarcoma Liposarcoma, Lyphoma Angiosarcoma B-cell lymphoma Lymphoma |
~36 weeks | 55 |
| UV | Fibrosarcoma (50%) Squamous cell carcinoma Lymphoma |
~36 weeks | ||||
| DMBA+UV | Fibrosarcoma (50%) B-cell lymphoma Lymphoma Low grade Fibrosarcoma Squamous cell carcinoma |
~14 weeks |
Immunoglobulin promoter enhancer (Eμ)-driven Myc expression promotes B-cell lymphoma in mice. The B-cell lymphoma latency is significantly shortened in mice lacking both Ink4a and ARF, or ARF alone (27, 60, 61). Moreover, the role of the INK4a/ARF locus in HRASV12-induced melanoma has been reported using melanocyte-specific HRASV12 transgenic mice. Mice lacking p19ARF and p16ink4a develop melanoma with short latency and high penetrance (62).
The tumor suppressive role of Ink4a/ARF was also studied in a super Ink4a/ARF mouse strain (carrying a transgenic copy of the entire INK4a/ARF locus), wherein increased activities of these tumor suppressors were observed (63). Furthermore, cells derived from these mice showed increased resistance to oncogene-induced transformation. Surprisingly, the super Ink4a/ARF mice showed higher resistance to carcinogen-induced tumor development without affecting normal viability and aging. This suggests that restricted increase of tumor suppressors at systemic levels might enhance an individual’s ability to resist cancer development.
Status of ARF in human cancers
Given that ARF plays an important role in tumor suppression, its deregulation in numerous cancers is also reported (Table 2). Studies on the deregulation of ARF have focused on gene loss or silencing by promoter hyper methylation and mutation. Its expression patterns are mainly studied at the transcript level because of its low expression, and availability constraints of its antibodies for immunohistochemistry. However, ARF protein expression in human cancers has been constantly pursued, since the importance of its posttranslational regulation was postulated. Low mRNA levels of ARF are frequently observed in human cancers, including those of the breast, colon, and liver. This has chiefly been attributed to gene silencing by promoter methylation and deletion of the gene locus. Both homologous deletion and loss of heterozygosity for this locus have been detected in human cancers (35, 44, 64–69). These alterations mostly result in deletion of the entire INK4a/ARF locus affecting the expressions of both ARF and Ink4a. The best-known genetic alteration that specifically affects ARF expression is promoter hyper methylation. In this context, the ARF promoter is a CpG island that can be silenced by DNA hyper methylation. Silencing of the gene by this mechanism is reported in numerous human cancers including those of the colon, liver, breast, and lung (64–66, 69–82). Hyper-methylation of the ARF promoter occurs independently of the INK4a promoter methylation, suggesting specific alteration of the ARF expression in these cancers. Conversely, elevated levels of ARF mRNA are reported in tumors (68). ARF might also be upregulated in cancers by various oncogenic stimuli such as c-Myc, RAS, and E2F1. Furthermore, mutations such as short deletions or insertions in the ARF-specific exon 1β, and missense mutations in exon 2 have been frequently reported in familial melanoma syndromes (83–86).
Table 2.
ARF status in human cancers
| Tissue type | ARF status | Frequency (%) | Ref |
|---|---|---|---|
| Breast | Low mRNA | 77 | 70 |
| Promoter methylation | 50 | ||
| HD | 11.5 | ||
| LOH | 27 | ||
| High mRNA | 17 | ||
| No alteration | 88 | ||
| Promoter methylation | 24 | 71 | |
| Bladder | Promoter methylation | 56 | 72 |
| LOH | 22 | 68 | |
| HD | 14 | ||
| Mutation | 2 | ||
| Colon | Promoter methylation | 33 | 73 |
| Promoter methylation | 50.8 | 74 | |
| Promoter methylation | 28.2 | 80 | |
| Promoter methylation | 32 | 75 | |
| Liver | Promoter methylation | 25 | 76 |
| HD | 4 | ||
| LOH | 16 | ||
| Promoter methylation | 12 | 77 | |
| HD | 2.3 | 67 | |
| Mutation | 4.6 | ||
| High mRNA | 93.2 | ||
| Gastric | Promoter methylation (diffuse type) | 45.5 | 78 |
| Promoter methylation (intestinal) | 25 | ||
| Lung | Promoter methylation | 30 | 69 |
| LOH | 26 | ||
| Promoter methylation | 8 | 79 | |
| Oral | Promoter methylation | 18 | 66 |
| HD | 12 | ||
| Promoter methylation | 43.8 | 64 | |
| HD | 26.5 | ||
| Prostate | Promoter methylation | 6.25 | 65 |
| HD | 6.25 | ||
| Kidney | HD | 23.5 | 81 |
| Brain | Promoter methylation | 22 | 79 |
HD: Homologus Deletion, LOH: Loss Of Heterozygosity.
Sylvie and colleagues reported that protein expression of ARF is low in lung cancer, and that there is frequent uncoupling between transcript and protein levels (87). Thus, they suggested that ARF also gets inactivated at the posttranslational level in cancers. In a recent study, the authors also reported that the ARF protein in gastric cancer is expressed at low levels, and is negatively correlated with MKRN1 expression (an E3 ubiquitin ligase that induces ARF degradation) (8). These results also support the importance of the posttranslational regulation of ARF in tumor suppression/progression.
CONCLUSION
The role of ARF in tumor suppression is relatively well established through various experiments using cancer cells, mice models, and human patients with cancer. As is well known, the key mechanism of tumor suppression by ARF is induction of cellular senescence via activation of the p53 pathway. ARF binds and sequesters MDM2 in the nucleolus, thus preventing the degradation of p53. However, the p53-independent role of ARF is still to be clearly delineated, and its targets verified. Toward this, KO mice models have proven to be useful, as shown by Weber and colleagues (discussed above). These results led to the discovery of novel ARF interacting partners that regulate tumor suppression in a p53-independent manner. However, the discovery of too many interacting partners has now caused considerable confusion. ARF interacts with numerous partners and regulates their sumoylation, transcriptional activities, or protein turnover, resulting in p53-independent inhibition of tumor cell growth. As p53 is frequently mutated in many cancers, a clear understanding of this p53-independent function of ARF might provide crucial clues to finding therapeutic targets in p53-mutated cancers. To this end, the downstream and upstream factors of ARF, along with their regulatory mechanisms, must be addressed extensively. In addition, the differential posttranslational regulation of ARF associated with tumor suppression and tumorigenesis must be elucidated using mouse models and human patient samples.
ACKNOWLEDGEMENTS
This work is supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (2014R1A1A1002589) and the Korea Healthcare Technology R&D Project, Ministry for Health & Welfare Affairs, Republic of Korea (A121387).
REFERENCES
- 1.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 3.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 4.Pomerantz J, Schreiber-Agus N, Liegeois NJ, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/S0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 5.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci U S A. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 7.Chen D, Shan J, Zhu WG, Qin J, Gu W. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature. 2010;464:624–627. doi: 10.1038/nature08820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ko A, Shin JY, Seo J, et al. Acceleration of gastric tumorigenesis through MKRN1-mediated posttranslational regulation of p14ARF. J Natl Cancer Inst. 2012;104:1660–1672. doi: 10.1093/jnci/djs424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Zha M, Zhao X, et al. Siva1 inhibits p53 function by acting as an ARF E3 ubiquitin ligase. Nat Commun. 2013;4:1551. doi: 10.1038/ncomms2533. [DOI] [PubMed] [Google Scholar]
- 10.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci U S A. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aslanian A, Iaquinta PJ, Verona R, Lees JA. Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev. 2004;18:1413–1422. doi: 10.1101/gad.1196704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitt CA. Senescence, apoptosis and therapy--cutting the lifelines of cancer. Nat Rev Cancer. 2003;3:286–295. doi: 10.1038/nrc1044. [DOI] [PubMed] [Google Scholar]
- 13.Zindy F, Eischen CM, Randle DH, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouchard C, Lee S, Paulus-Hock V, Loddenkemper C, Eilers M, Schmitt CA. FoxO transcription factors suppress Myc-driven lymphomagenesis via direct activation of Arf. Genes Dev. 2007;21:2775–2787. doi: 10.1101/gad.453107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue K, Roussel MF, Sherr CJ. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci U S A. 1999;96:3993–3998. doi: 10.1073/pnas.96.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linggi B, Muller-Tidow C, van de Locht L, et al. The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat Med. 2002;8:743–750. doi: 10.1038/nm726. [DOI] [PubMed] [Google Scholar]
- 17.Sreeramaneni R, Chaudhry A, McMahon M, Sherr CJ, Inoue K. Ras-Raf-Arf signaling critically depends on the Dmp1 transcription factor. Mol Cell Biol. 2005;25:220–232. doi: 10.1128/MCB.25.1.220-232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Y, Zhao YD, Gibbons M, et al. Tgfβ signaling directly induces Arf promoter remodeling by a mechanism involving Smads 2/3 and p38 MAPK. J Biol Chem. 2010;285:35654–35664. doi: 10.1074/jbc.M110.128959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulavin DV, Phillips C, Nannenga B, et al. Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16(Ink4a)-p19(Arf) pathway. Nat Genet. 2004;36:343–350. doi: 10.1038/ng1317. [DOI] [PubMed] [Google Scholar]
- 20.Yoon JH, Choi WI, Jeon BN, et al. Human Kruppel-related 3 (HKR3) is a novel transcription activator of alternate reading frame (ARF) gene. J Biol Chem. 2014;289:4018–4031. doi: 10.1074/jbc.M113.526855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 22.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs JJ, Keblusek P, Robanus-Maandag E, et al. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat Genet. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 25.Cakouros D, Isenmann S, Cooper L, et al. Twist-1 induces Ezh2 recruitment regulating histone methylation along the Ink4A/Arf locus in mesenchymal stem cells. Mol Cell Biol. 2012;32:1433–1441. doi: 10.1128/MCB.06315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo ML, den Besten W, Bertwistle D, Roussel MF, Sherr CJ. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 2004;18:1862–1874. doi: 10.1101/gad.1213904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Blandino G, Givol D. Induced p21waf expression in H1299 cell line promotes cell senescence and protects against cytotoxic effect of radiation and doxorubicin. Oncogene. 1999;18:2643–2649. doi: 10.1038/sj.onc.1202632. [DOI] [PubMed] [Google Scholar]
- 29.Chang BD, Xuan Y, Broude EV, et al. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene. 1999;18:4808–4818. doi: 10.1038/sj.onc.1203078. [DOI] [PubMed] [Google Scholar]
- 30.Moore L, Venkatachalam S, Vogel H, et al. Cooperativity of p19ARF, Mdm2, and p53 in murine tumorigenesis. Oncogene. 2003;22:7831–7837. doi: 10.1038/sj.onc.1206985. [DOI] [PubMed] [Google Scholar]
- 31.Weber JD, Jeffers JR, Rehg JE, et al. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 2000;14:2358–2365. doi: 10.1101/gad.827300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha L, Ichikawa T, Anver M, et al. ARF functions as a melanoma tumor suppressor by inducing p53-independent senescence. Proc Natl Acad Sci U S A. 2007;104:10968–10973. doi: 10.1073/pnas.0611638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eymin B, Karayan L, Seite P, et al. Human ARF binds E2F1 and inhibits its transcriptional activity. Oncogene. 2001;20:1033–1041. doi: 10.1038/sj.onc.1204220. [DOI] [PubMed] [Google Scholar]
- 34.Tsuji K, Mizumoto K, Sudo H, Kouyama K, Ogata E, Matsuoka M. p53-independent apoptosis is induced by the p19ARF tumor suppressor. Biochem Biophys Res Commun. 2002;295:621–629. doi: 10.1016/S0006-291X(02)00723-4. [DOI] [PubMed] [Google Scholar]
- 35.Yarbrough WG, Bessho M, Zanation A, Bisi JE, Xiong Y. Human tumor suppressor ARF impedes S-phase progression independent of p53. Cancer Res. 2002;62:1171–1177. [PubMed] [Google Scholar]
- 36.Hemmati PG, Gillissen B, von Haefen C, et al. Adenovirus-mediated overexpression of p14(ARF) induces p53 and Bax-independent apoptosis. Oncogene. 2002;21:3149–3161. doi: 10.1038/sj.onc.1205458. [DOI] [PubMed] [Google Scholar]
- 37.Eymin B, Leduc C, Coll JL, Brambilla E, Gazzeri S. p14ARF induces G2 arrest and apoptosis independently of p53 leading to regression of tumours established in nude mice. Oncogene. 2003;22:1822–1835. doi: 10.1038/sj.onc.1206303. [DOI] [PubMed] [Google Scholar]
- 38.Kelly-Spratt KS, Gurley KE, Yasui Y, Kemp CJ. p19Arf suppresses growth, progression, and metastasis of Hras-driven carcinomas through p53-dependent and -independent pathways. PLoS Biol. 2004;2:E242. doi: 10.1371/journal.pbio.0020242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leduc C, Claverie P, Eymin B, et al. p14ARF promotes RB accumulation through inhibition of its Tip60-dependent acetylation. Oncogene. 2006;25:4147–4154. doi: 10.1038/sj.onc.1209446. [DOI] [PubMed] [Google Scholar]
- 40.Itahana K, Bhat KP, Jin A, et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151–1164. doi: 10.1016/S1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 41.Brady SN, Yu Y, Maggi LB, Jr, Weber JD. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol Cell Biol. 2004;24:9327–9338. doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fatyol K, Szalay AA. The p14ARF tumor suppressor protein facilitates nucleolar sequestration of hypoxia-inducible factor-1alpha (HIF-1alpha) and inhibits HIF-1-mediated transcription. J Biol Chem. 2001;276:28421–28429. doi: 10.1074/jbc.M102847200. [DOI] [PubMed] [Google Scholar]
- 43.Kalinichenko VV, Major ML, Wang X, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;431:712–717. doi: 10.1038/nature02958. [DOI] [PubMed] [Google Scholar]
- 45.Amente S, Gargano B, Diolaiti D, Della Valle G, Lania L, Majello B. p14ARF interacts with N-Myc and inhibits its transcriptional activity. FEBS Lett. 2007;581:821–825. doi: 10.1016/j.febslet.2007.01.062. [DOI] [PubMed] [Google Scholar]
- 46.Rocha S, Garrett MD, Campbell KJ, Schumm K, Perkins ND. Regulation of NF-kappaB and p53 through activation of ATR and Chk1 by the ARF tumour suppressor. EMBO J. 2005;24:1157–1169. doi: 10.1038/sj.emboj.7600608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paliwal S, Pande S, Kovi RC, Sharpless NE, Bardeesy N, Grossman SR. Targeting of C-terminal binding protein (CtBP) by ARF results in p53-independent apoptosis. Mol Cell Biol. 2006;26:2360–2372. doi: 10.1128/MCB.26.6.2360-2372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xirodimas DP, Chisholm J, Desterro JM, Lane DP, Hay RT. P14ARF promotes accumulation of SUMO-1 conjugated (H)Mdm2. FEBS Lett. 2002;528:207–211. doi: 10.1016/S0014-5793(02)03310-0. [DOI] [PubMed] [Google Scholar]
- 49.Chen L, Chen J. MDM2-ARF complex regulates p53 sumoylation. Oncogene. 2003;22:5348–5357. doi: 10.1038/sj.onc.1206851. [DOI] [PubMed] [Google Scholar]
- 50.Woods YL, Xirodimas DP, Prescott AR, Sparks A, Lane DP, Saville MK. p14 Arf promotes small ubiquitin-like modifier conjugation of Werners helicase. J Biol Chem. 2004;279:50157–50166. doi: 10.1074/jbc.M405414200. [DOI] [PubMed] [Google Scholar]
- 51.Rizos H, Woodruff S, Kefford RF. p14ARF interacts with the SUMO-conjugating enzyme Ubc9 and promotes the sumoylation of its binding partners. Cell Cycle. 2005;4:597–603. doi: 10.4161/cc.4.4.1588. [DOI] [PubMed] [Google Scholar]
- 52.Tago K, Chiocca S, Sherr CJ. Sumoylation induced by the Arf tumor suppressor: a p53-independent function. Proc Natl Acad Sci U S A. 2005;102:7689–7694. doi: 10.1073/pnas.0502978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Damalas A, Velimezi G, Kalaitzakis A, et al. Loss of p14(ARF) confers resistance to heat shock- and oxidative stress-mediated cell death by upregulating beta-catenin. Int J Cancer. 2011;128:1989–1995. doi: 10.1002/ijc.25510. [DOI] [PubMed] [Google Scholar]
- 54.Pan W, Datta A, Adami GR, Raychaudhuri P, Bagchi S. P19ARF inhibits the functions of the HPV16 E7 oncoprotein. Oncogene. 2003;22:5496–5503. doi: 10.1038/sj.onc.1206857. [DOI] [PubMed] [Google Scholar]
- 55.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/S0092-8674(00)81079-X. [DOI] [PubMed] [Google Scholar]
- 56.Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–2222. [PubMed] [Google Scholar]
- 57.Sharpless NE, Ramsey MR, Balasubramanian P, Castrillon DH, DePinho RA. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis. Oncogene. 2004;23:379–385. doi: 10.1038/sj.onc.1207074. [DOI] [PubMed] [Google Scholar]
- 58.Kamijo T, Zindy F, Roussel MF, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/S0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 59.Sharpless NE, Bardeesy N, Lee KH, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 60.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chin L, Pomerantz J, Polsky D, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matheu A, Pantoja C, Efeyan A, et al. Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes Dev. 2004;18:2736–2746. doi: 10.1101/gad.310304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shintani S, Nakahara Y, Mihara M, Ueyama Y, Matsumura T. Inactivation of the p14(ARF), p15(INK4B) and p16(INK4A) genes is a frequent event in human oral squamous cell carcinomas. Oral Oncol. 2001;37:498–504. doi: 10.1016/S1368-8375(00)00142-1. [DOI] [PubMed] [Google Scholar]
- 65.Konishi N, Nakamura M, Kishi M, Nishimine M, Ishida E, Shimada K. Heterogeneous methylation and deletion patterns of the INK4a/ARF locus within prostate carcinomas. Am J Pathol. 2002;160:1207–1214. doi: 10.1016/S0002-9440(10)62547-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sailasree R, Abhilash A, Sathyan KM, Nalinakumari KR, Thomas S, Kannan S. Differential roles of p16INK4A and p14ARF genes in prognosis of oral carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:414–420. doi: 10.1158/1055-9965.EPI-07-0284. [DOI] [PubMed] [Google Scholar]
- 67.Ito T, Nishida N, Fukuda Y, Nishimura T, Komeda T, Nakao K. Alteration of the p14(ARF) gene and p53 status in human hepatocellular carcinomas. J Gastroenterol. 2004;39:355–361. doi: 10.1007/s00535-003-1302-9. [DOI] [PubMed] [Google Scholar]
- 68.Berggren P, Kumar R, Sakano S, et al. Detecting homozygous deletions in the CDKN2A(p16(INK4a))/ARF(p14(ARF)) gene in urinary bladder cancer using real-time quantitative PCR. Clin Cancer Res. 2003;9:235–242. [PubMed] [Google Scholar]
- 69.Hsu HS, Wang YC, Tseng RC, et al. 5′ cytosinephospho-guanine island methylation is responsible for p14ARF inactivation and inversely correlates with p53 overexpression in resected non-small cell lung cancer. Clin Cancer Res. 2004;10:4734–4741. doi: 10.1158/1078-0432.CCR-03-0704. [DOI] [PubMed] [Google Scholar]
- 70.Silva J, Silva JM, Dominguez G, et al. Concomitant expression of p16INK4a and p14ARF in primary breast cancer and analysis of inactivation mechanisms. J Pathol. 2003;199:289–297. doi: 10.1002/path.1297. [DOI] [PubMed] [Google Scholar]
- 71.Silva J, Dominguez G, Silva JM, et al. Analysis of genetic and epigenetic processes that influence p14ARF expression in breast cancer. Oncogene. 2001;20:4586–4590. doi: 10.1038/sj.onc.1204617. [DOI] [PubMed] [Google Scholar]
- 72.Dominguez G, Carballido J, Silva J, et al. p14ARF promoter hypermethylation in plasma DNA as an indicator of disease recurrence in bladder cancer patients. Clin Cancer Res. 2002;8:980–985. [PubMed] [Google Scholar]
- 73.Dominguez G, Silva J, Garcia JM, et al. Prevalence of aberrant methylation of p14ARF over p16INK4a in some human primary tumors. Mutat Res. 2003;530:9–17. doi: 10.1016/S0027-5107(03)00133-7. [DOI] [PubMed] [Google Scholar]
- 74.Lee M, Sup Han W, Kyoung Kim O, et al. Prognostic value of p16INK4a and p14ARF gene hypermethylation in human colon cancer. Pathol Res Pract. 2006;202:415–424. doi: 10.1016/j.prp.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 75.Esteller M, Tortola S, Toyota M, et al. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res. 2000;60:129–133. [PubMed] [Google Scholar]
- 76.Tannapfel A, Sommerer F, Benicke M, et al. Genetic and epigenetic alterations of the INK4a-ARF pathway in cholangiocarcinoma. J Pathol. 2002;197:624–631. doi: 10.1002/path.1139. [DOI] [PubMed] [Google Scholar]
- 77.Tannapfel A, Busse C, Geissler F, Witzigmann H, Hauss J, Wittekind C. INK4a-ARF alterations in liver cell adenoma. Gut. 2002;51:253–258. doi: 10.1136/gut.51.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iida S, Akiyama Y, Nakajima T, et al. Alterations and hypermethylation of the p14(ARF) gene in gastric cancer. Int J Cancer. 2000;87:654–658. doi: 10.1002/1097-0215(20000901)87:5<654::AID-IJC6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 79.Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]
- 80.Chaar I, Amara S, Elamine OE, et al. Biological significance of promoter hypermethylation of p14/ARF gene: relationships to p53 mutational status in Tunisian population with colorectal carcinoma. Tumour Biol. 2014;35:1439–1449. doi: 10.1007/s13277-013-1198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kasahara T, Bilim V, Hara N, Takahashi K, Tomita Y. Homozygous deletions of the INK4a/ARF locus in renal cell cancer. Anticancer Res. 2006;26:4299–4305. [PubMed] [Google Scholar]
- 82.Inda MM, Munoz J, Coullin P, et al. High promoter hypermethylation frequency of p14/ARF in supratentorial PNET but not in medulloblastoma. Histopathology. 2006;48:579–587. doi: 10.1111/j.1365-2559.2006.02374.x. [DOI] [PubMed] [Google Scholar]
- 83.Rizos H, Darmanian AP, Holland EA, Mann GJ, Kefford RF. Mutations in the INK4a/ARF melanoma susceptibility locus functionally impair p14ARF. J Biol Chem. 2001;276:41424–41434. doi: 10.1074/jbc.M105299200. [DOI] [PubMed] [Google Scholar]
- 84.Rizos H, Puig S, Badenas C, et al. A melanoma-associated germline mutation in exon 1beta inactivates p14ARF. Oncogene. 2001;20:5543–5547. doi: 10.1038/sj.onc.1204728. [DOI] [PubMed] [Google Scholar]
- 85.Hewitt C, Lee Wu C, Evans G, et al. Germline mutation of ARF in a melanoma kindred. Hum Mol Genet. 2002;11:1273–1279. doi: 10.1093/hmg/11.11.1273. [DOI] [PubMed] [Google Scholar]
- 86.Randerson-Moor JA, Harland M, Williams S, et al. A germline deletion of p14(ARF) but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum Mol Genet. 2001;10:55–62. doi: 10.1093/hmg/10.1.55. [DOI] [PubMed] [Google Scholar]
- 87.Gazzeri S, Della Valle V, Chaussade L, Brambilla C, Larsen CJ, Brambilla E. The human p19ARF protein encoded by the beta transcript of the p16INK4a gene is frequently lost in small cell lung cancer. Cancer Res. 1998;58:3926–3931. [PubMed] [Google Scholar]