Abstract
Three members of the genus Borrelia (B.burgdorferi, B.garinii, B.afzelii) cause tick-borne borreliosis. Depending on the Borrelia species involved, the borreliosis differs in its clinical symptoms. Comparative genomics opens up a way to elucidate the underlying differences in Borrelia species. We analysed a low redundancy whole-genome shotgun (WGS) assembly of a B.garinii strain isolated from a patient with neuroborreliosis in comparison to the B.burgdorferi genome. This analysis reveals that most of the chromosome is conserved (92.7% identity on DNA as well as on amino acid level) in the two species, and no chromosomal rearrangement or larger insertions/deletions could be observed. Furthermore, two collinear plasmids (lp54 and cp26) seem to belong to the basic genome inventory of Borrelia species. These three collinear parts of the Borrelia genome encode 861 genes, which are orthologous in the two species examined. The majority of the genetic information of the other plasmids of B.burgdorferii is also present in B.garinii although orthology is not easy to define due to a high redundancy of the plasmid fraction. Yet, we did not find counterparts of the B.burgdorferi plasmids lp36 and lp38 or their respective gene repertoire in the B.garinii genome. Thus, phenotypic differences between the two species could be attributable to the presence or absence of these two plasmids as well as to the potentially positively selected genes.
INTRODUCTION
The genus Borrelia comprises 19 species of which 10 belong to the Borrelia burgdorferi sensu lato complex (1). Only three species of this complex cause the multisystem disorder Lyme borreliosis, B.burgdorferi sensu stricto, B.garinii and B.afzelii. In the United States, B.burgdorferi sensu stricto is the only causative agent for this disease (2), whereas in Europe B.garinii as well as B.afzelii are major contributors to the reported case numbers (3,4).
All these borreliae live in the gastro-intestinal tract of ticks (Ixodes spec.) and are able to infect multiple hosts via tick bite. The host range is thought to be defined by gene variations on a group of redundant plasmids (5–7). Furthermore, the disease patterns observed in humans are dependent on the particular Borrelia species involved. B.garinii is primarily associated with neuroborreliosis (3), B.afzelii with acrodermatitis chronica athrophicans (a chronic skin disease) (8), whereas B.burgdorferi sensu stricto was found to be prevalent in Lyme arthritis (9), which, however, was not confirmed by two other studies (10,11).
The nuclear genomes of all B.burgdorferi sensu lato species consist of one linear chromosome and varying amounts of several linear and circular plasmids (12). The chromosomes are highly similar (13), but plasmids show a wealth of diversity and can be lost during culture of the bacteria (14,15). The genomic sequence of the B.burgdorferi B31 chromosome was determined in 1997 (16). The sequence analysis of B.burgdorferi sensu stricto showed that the chromosome has a length of 0.94 Mb harbouring all genes for basic cellular functions. The plasmids often share sequence motifs and segments, and seem to contain a large number of fragmented genes, since several sequence motifs occur in predicted non-coding as well as coding regions. This fact and the lack of orthologs in other species made it difficult to define genes (17). Although the plasmid fraction of the genome seems to be responsible for host range selection and pathogenicity (6,18), large parts of it are likely to be dispensable for viability in culture and are highly variable, even in a single species (19).
The Borrelia species belong to the spirochetes, a group of bacteria that have long, helically coiled cells. To date, the genomes of the following spirochete species are known in addition to Borrelia burgdorferi: Treponema denticola, Treponema pallidum and Leptospira interrogans (16,20–22). Their genome sizes range from 1.6 to 4.3 Mb, with B.burgdorferi possessing the smallest genome. The B.burgdorferi sequence was recently used for microarray experiments to identify similar sequences in B.hermsii (23). This analysis revealed that at least 81% of the chromosome and 41% of the plasmid sequences of B.burgdorferi are present in this distantly related species. Yet, hybridization experiments can only detect highly similar sequences shared between organisms. On the other hand, a comparative analysis of sequences is able to detect not only low similarities that would not give hybridization signals, but provides also access to sequence differences of the organisms compared. Closely related species reveal species-specific differences and evolutionary selection pressures on genes. At the same time, a comparative sequence analysis provides the means for a better annotation as was shown with several Saccharomyces species (24). The comparison of more distantly related species helps to describe core sets of proteins within a specific evolutionary branch (25). The goal of the present comparative study was (i) to define the orthologous gene set, which presumably is the basic set of borreliosis causing Borreliae; (ii) to identify the stable and variable parts in the genomes of Borrelia; (iii) to determine, which genome part is dispensable without loosing pathogenicity; (iv) to improve the the B.burgdorferi annotation by the B.garinii orthology information. Furthermore, gene groups exposed to different evolutionary changes can be defined. Since positive selection is an indicator for environmental adaptations of the pathogen, genes evolving in such a manner are highly suspicious to be involved in pathogenicity. Many of the B.burgdorferi genes are so far described as only hypothetical. Thus, a comparative analysis would also help to define true orthologous pairs and exclude false positive predictions.
Therefore, we decided to sequence and analyse another species from the B.burgdorferi sensu lato complex, B.garinii. Previous studies had revealed that selected genes of both species are highly similar with >90% sequence identity on DNA level. Moreover, there were hints that the chromosomes of the two species are collinear (26). This relatively high similarity enabled us to apply a low redundant sequencing strategy. Thus, we were able to generate a complete analysis of the chromosome and two conserved plasmids of B.garinii. The diverse plasmid fraction of the genome could also be defined and analysed although no clear-cut assignment to B.burgdorferi plasmids was possible due to redundancies and rearrangements.
MATERIALS AND METHODS
The B.garinii strain PBi (OspA serotype 4), a cerebrospinal fluid (CSF) isolate from a German patient with neuroborreliosis, was used for sequence analysis (27). OspA-serotype 4 strains are enriched in CSF, but they have been isolated only exceptionally from ticks (27). A low passage of strain PBi (12th subculture) was cultured in MVP-medium as described (28). Passage 12 is still infectious for gerbils, infectivity was lost between passage 30 and 60. DNA was extracted using the Genomic DNA Bufferset (Quiagen GmbH, Hilden) and Genomic tip 500/6 and 100/6 (Quiagen, GmbH, Hilden). A genomic library with a target insert size of 1.5 kb using total DNA was constructed as described previously (29). From this library, 5740 clones were sequenced from both ends resulting in an estimated coverage of the whole genome of three times. According to the similarity of the obtained sequences to B.burgdorferi counterparts they were binned into a plasmid and a chromosome group using BLAST (30). The chromosomal sequences were assembled using the assembler GAP4 (31) utilizing the finished chromosome of B.burgdorferi (16,17) as a backbone. Using the backbone sequence as a ruler and orientation measure, we defined primer pairs for PCR reactions to close the gaps. For convenient primer design, we have written a Perl script for the automatic definition of primers in a Staden package project with the program ‘primer3’ (http://www-genome.wi.mit.edu/genome_software/other/primer3.html) as kernel (R.Lehmann, unpublished data). Nucleotide differences between the B.garinii and the B.burgdorferi chromosome were calculated automatically if the Phred score was 20 or better at the differing base. Bases below this score were inspected by eye to ensure proper difference calculation.
Initial gene finding was performed using GeneMarkS (http://opal.biology.gatech.edu/GeneMark) (32). Orthology to B.burgdorferi counterparts was determined by aligning the best bidirectional hit (BBH) to each predicted protein. Only proteins located at the same position within the different genomes and with <10% length difference were considered as orthologs. Orphan genes of both organisms contained no functional domains according to an InterPro analysis (http://www.ebi.ac.uk/interpro/). Transmembrane domains were predicted using TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). The GenBank gene descriptions of the B.burgdorferi genome (NC_001318, NC_000948 to NC_000957, NC_001849 to NC_001857, NC_001903, NC_001904, NC_004971) were used for the annotation of the corresponding B.garinii coding sequences.
To perform an independent cross-check of the reliability of this annotation approach, all potential protein coding sequences (CDS; potential start codon to stop codon without length threshold) of B.garinii were used for BLASTP searches against the GenBank database. Whenever a gene is referred to as ‘hypothetical’, no match in any database could be found. A ‘conserved hypothetical gene’ is a gene, which can be detected with sufficient similarity (p < 10−10) in other genera.
The alignment between the B.garinii and B.burgdorferi collinear chromosomes and plasmids was generated using the program stretcher, which is part of the EMBOSS package (http://www.emboss.org).
The B.garinii sequences were deposited in GenBank with the accession numbers CP000013, CP000014, CP000015 and AY722917 to AY722953. The B.garinii genome data as well as the results of the comparative analysis of B.garinii with B.burgdorferi are also available from the Spirochetes Genome Browser at http://sgb.imb-jena.de/. The browser is based on our genome annotation and analysis system GenColors (manuscript in preparation).
RESULTS
The DNA for the libraries was obtained from the total DNA content of B.garinii strain PBi. Thus, besides the chromosome, the plasmids of this strain should also be represented at least in part in the shotgun data. All sequences from the whole-genome library were binned according to their similarities to the B.burgdorferi genome parts (i.e. chromosome and plasmids): 37.8% of all B.garinii reads were derived from plasmids (Table 1). This value is comparable to that of the plasmid fraction of B.burgdorferi (40%).
Table 1. Comparison of the B.garinii low redundancy WGS assembly to the B.burgdorferi sequence.
| B.garinii | B.burgdorferi | ||||
|---|---|---|---|---|---|
| Number | % | Coverage depth | Length | ||
| Reads | 7562 | 100.0 | |||
| Chromosome | 4704 | 62.2 | 3.38 | 904 246 | 910 724 |
| Coding | 840 399 | 861 829 | |||
| lp54 | 464 | 6.1 | 5.11 | 55 560 | 53 561 |
| cp26 | 181 | 2.4 | 3.65 | 27 108 | 26 498 |
| Plasmid fragments >2 kb | 2018 | 26.7 | 4.92 | 239 965 | 567 176 |
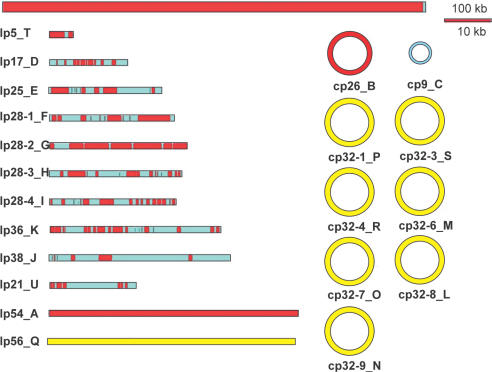
Altogether the assembly comprises 1.227 Mb of B.garinii sequence. Three contigs completely cover the counterparts of the corresponding B.burgdorferii chromosome and plasmids lp54 and cp26. Additional 37 contigs >2 kb amounting to 239 kb were obtained for the remaining plasmid fraction of B.garinii (Figure 1). Clear-cut and error-free assignment of these plasmid contigs to defined B.burgdorferi plasmids was not possible, further underlining the variability of the plasmid complement in Borrelia species. The whole chromosome and two plasmids (lp54,A and cp26, B) are completely represented in B.garinii. Eight plasmids of B.burgdorferi are highly similar (cp32: L, M, N, O, P, R, S; lp56: Q). These seem to be also completely represented by B.garinii plasmid contigs although neither an exact assignment of individual contigs to a specific B.burgdorferi plasmid nor the calculation of their copy number is possible. The remaining plasmid contigs of B.garinii show also similarities to B.burgdorferi plasmids, but some regions are either unique to B.burgdorferi or have low DNA similarities. In four contigs, we observed similarities to different plasmids of B.burgdorferi indicating breakage/fusion points between different plasmids.
Figure 1.
View of the B.burgdorferi genome indicating similarities to sequences of the B.garinii genome. The calculation of the similarities was done using BLAST. Threshold for identity was 75% on a length of 40 bases. Portions of the B.burgdorferi genome with matches are depicted in red, unmatched in blue. The complex consisting of eight nearly identical plasmids is drawn in yellow. Here, no exact orthologous regions could be defined since sequences as well as CDSs are redundant. The scale for the plasmids is 10× magnified compared to the chromosome.
Chromosome
In our low redundancy project, the average coverage of the chromosome assembly is 3.38 (Table 1). The initial assembly of the B.garinii chromosome had 260 gaps (comprising sequencing and clone gaps). After applying gap closure procedures, we obtained one contig covering almost (99.5%) the complete linear B.burgdorferi chromosome. Despite the low redundancy of the sequence reads >80% of the chromosome is endowed with a error frequency of <10−4. The overall expected error rate is 0.26% (Supplementary Figure S1). The comparative analysis shows an unbroken collinearity between the B.burgdorferi and B.garinii chromosomes. The only parts of the B.burgdorferi chromosome with no counterpart in B.garinii reside in both telomeric regions with a size of 168 and 8458 bp, respectively. Since ends of linear chromosomes are not clonable without further manipulation, the missing bases on the left end are probably due to a cloning bias. It was previously shown that the right end of the chromosome in B.burgdorferi exhibits length variations in defined steps in different strains (33). The shorter right end of the B.garinii chromosome is comparable to one form of these stepwise variable lengths of the B.burgdorferi chromosomes.
Substitutions, insertions and deletions
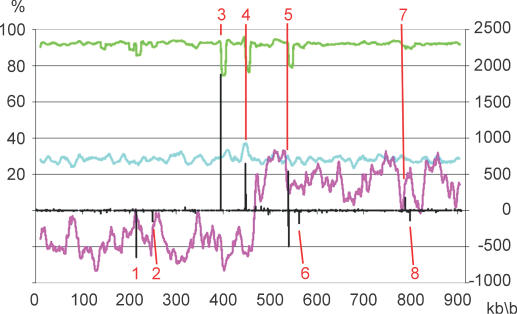
Base substitutions are a measure for evolutionary distance between organisms. The overall identity of the B.garinii with the B.burgdorferi chromosome is 92.7%. A calculation based on the shared CDS on amino acid level gives the same result indicating an equal distribution of substitutions over the chromosome irrespective of information content. The decrease of similarity between the two chromosomes below 80% in three regions around the origin of replication as can be seen in Figure 2 is apparently caused by larger insertions and deletions (indels; Figure 2, number 4–6). In total, we found 66 482 single base substitutions (Table 2). Transitions and transversions are almost equally distributed in the genic and intergenic regions.
Figure 2.
A sketch of the B.garinii chromosome compared to the collinear B.burgdorferi counterpart. Base identity of B.garinii versus B.burgdorferi is shown as green line, GC content as light blue line (both left scale) and GC skew as purple line. All values were calculated using a sliding window of 10 kb with a step width of 1 kb. Positions of all indels (deletions—negative peaks, insertions—positive peaks) in the alignment of the two analysed chromosomes are shown as black bars (right scale). Indels with a distance <10 bp to each other were defined as one single indel. All larger indels are indexed with numbers in red, exact positions are given in Supplementary Table S1. A cluster of five insertions appears in this figure as one peak (number 4), in a similar manner two deletions are located near position 540 000 (number 5 together with an insertion). The deletion of 8458 bases at the telomeric end is not shown.
Table 2. Frequency of single base substitutions in the collinear genomic elements of B.burgdorferi and B.garinii.
| Single base substitution | Total | Genic regions | Extragenic regions |
|---|---|---|---|
| Chromosome | |||
| Sum | 66 482 (7.36%) | 62 072 (7.35%) | 4410 (7.50%) |
| Transversions | 14 890 (1.65%) | 13 784 (1.63%) | 1106 (1.88%) |
| Transitions | 51 592 (5.71%) | 48 288 (5.71%) | 3304 (5.62%) |
| cp26 | |||
| Sum | 2 405 (8.87%) | 2 001 (8.81%) | 404 (9.20%) |
| Transversions | 708 (2.61%) | 569 (2.50%) | 139 (3.16%) |
| Transitions | 1 697 (6.26%) | 1 432 (6.30%) | 265 (6.03%) |
| lp54 | |||
| Sum | 8 364 (15.06%) | 6 088 (15.23%) | 2 276 (14.63%) |
| Transversions | 3 398 (6.12%) | 2 398 (6.00%) | 1000 (6.43%) |
| Transitions | 4 966 (8.94%) | 3 690 (9.23%) | 1276 (8.20%) |
Besides the shorter right end of the chromosome, we found eight insertions and six deletions with a size >100 bp (Figure 2, numbers 1–8; Supplementary Table S1) in the B.garinii chromosome as major structural differences relative to the chromosome of B.burgdorferi.
The largest observed insertion with a size of 1878 bp is caused by a duplication of a region containing the bmpA gene and part of the bmpB gene (see below) resulting in a tandem repeat of these genes (Figure 2, number 3). A series of five insertions is separated by short orthologous sequences of at most 470 bases (Figure 2, number 4; Supplementary Table S1). This cluster of insertions is located in a region containing two tRNA genes (tRNA-Ile-1, tRNA-Ala-1) and expands only intergenic regions.
Indel region 5 consists of an insertion of 538 bases (Figure 2, peak 3) followed by two deletions of 211 and 498 bases, respectively, which are separated by 133 bases. In this indel region 5 resides the gene encoding inositol monophosphatase (BB0524) in B.burgdorferi. This gene is only partly represented (59 of 284 amino acids) in B.garinii. The eighth insertion (Figure 2, number 7) is located in an intergenic region. According to the observed indel regions, two ‘hotspots’ for rearrangements on the Borrelia chromosome could be defined: indel region 4 and indel region 5. Most interestingly, these two regions are located in close vicinity to the origin of replication of the chromosome at position 475 kb.
All deletions >100 bases including the missing chromosomal ends comprise 10 448 bases, all insertions 4099 bases. Thus, the B.garinii chromosome is by 0.7% shorter than that of B.burgdorferi (Table 2).
Genes
A comparative annotation of the B.garinii chromosome was performed using the previously published B.burgdorferi gene prediction and annotation (GenBank NC_001318.1) (16). In parallel, we used GeneMarkS for ab initio gene predictions. This program was not able to detect 36 of the original gene predictions on the B.burgdorferi chromosome (Supplementary Table S3). Two of these non-verified coding sequences are fused to neighbouring coding sequences in B.garinii (BB0410, BB0510). The failure of GeneMarkS to identify BB0412 is possibly a false negative result, as the program predicted the ortholog in B.garinii. Additional ten potential genes of the remaining 814 genes annotated on the chromosome of B.burgdorferi are not predicted on the chromosome of B.garinii (Supplementary Table S4). Interestingly, these genes are annotated in B.burgdorferi only as predicted coding region without any other supportive evidence like similarities to other genes. Twelve predicted B.burgdorferi genes are fused in B.garinii (BB0078 + BB0079, BB0356 + BB0357, BB0410 + BB0411, BB0510 + BB0511, BB0521 + BB0522, BB0710 + BB0711). Eight annotated genes show extensive length divergence and overlap only partly their B.garinii counterpart, seven of which are altered due to differing open reading frames (ORFs) on an otherwise orthologous genomic sequence (BB0475, BB0524, BB0532, BB0546, BB0591, BB0749, BB0758). The lmp1 gene (BG0212) is affected by the largest deletion in B.garinii (Figure 2, number 1) and thereby shortened by 648 bases at the 5′ end. In summary, 786 GenBank annotated genes of B.burgdorferi are supported by GeneMarkS predictions in both Borrelia species and thus most likely represent the orthologous set of chromosomal genes. The length of 452 orthologs is unaltered, 255 genes are longer in B.burgdorferi and 79 genes are longer in B.garinii, but since these length differences are only small, the core of the deduced amino acid sequence is not affected.
BB0086 is split in B.garinii. Two genes are affected by a large duplication in B.garinii leading to a second copy of bmpA (BB0382) and a partial copy of bmpB (BB0383; Figure 2, number 3). Due to nonsense mutations, this partial copy is represented in the predicted gene set by four small ORFs.
In addition to the 807 predicted B.garinii genes with homologous sequences (including split, fused and other altered gene structures) in annotated CDS of B.burgdorferi, GeneMarkS predicts 33 further genes. These genes are comparably small (<60 amino acids) and most likely represent false positive predictions. This is further underlined by the fact that four of these potential genes lie within rRNA and tRNA gene regions and additional four predictions are apparently derived from the truncated copy of bmpB. The additional 39 genes on the B.burgdorferi chromosome, which are predicted by GeneMarkS, may also be false positive predictions.
On the DNA level, no predicted protein-coding gene of B.garinii is identical to its ortholog in B.burgdorferi whereas 20 tRNA genes (out of a total of 33 tRNA genes) are identical to their orthologous counterparts. Interestingly, the mutations seem to affect tRNA genes not randomly, since most sequence changes occur in non-unique tRNA genes (11 of 13). All four copies of tRNA-Leu, all three copies of tRNA-Ser, two of the three tRNA-Arg copies, and the second copy of tRNA-Lys and tRNA-Thr, respectively, are mutated.
Due to the high similarity of the chromosomes, the statistics of the codon usage shows only a slight difference between the two species. For example, in B.burgdorferi a higher preference for TTG as a start codon than in B.garinii predicted genes could be observed (Supplementary Table S2).
On the protein level, only 11 of all B.garinii genes are not altered in comparison to B.burgdorferi. This includes the ribosomal proteins rpsU, rpsL, rpmG, rpsJ, rpsS, rpmF, a putative subunit K of an ATPase, the flagellar motor switch protein fliG-2, the phosphocarrier protein ptsH-2, and the chemotaxis-related proteins cheX and cheY-3.
Additional 25 genes are affected by conservative exchanges with amino acids having the same chemical properties, thus increasing the number of highly conserved proteins to 38; not surprisingly 18 genes of this expanded group code for ribosomal proteins.
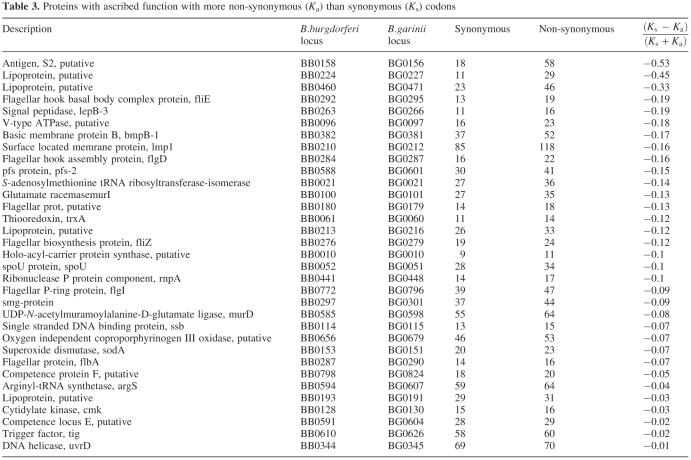
As an indication for positive selection, 94 (11.7%) of all orthologous genes and genes with similar sequences contain more non-synonymous than synonymous exchanges. Of these, 61 have no functional assignments. Interestingly, a higher than average proportion of the deduced amino acid sequences is predicted to contain transmembrane domains (39% compared to 26% for all proteins, Supplementary Table S8). The remaining 33 predicted genes are listed in Table 3. Many of these proteins seem to be located, according to their function, on the surface of the cell.
Table 3. Proteins with ascribed function with more non-synonymous (Ka) than synonymous (Ks) codons.
Plasmids
Different strains of the same Borrelia species can carry different sets of plasmids. The differing plasmid repertoire of the cells can be partly a function of the living conditions (34). Additionally, strains can loose parts of their equipment due to a lack of selection pressure (35). Since the primary assembly of the chromosomal reads without additional gap closure sequences resulted in 91% coverage of the chromosome, we may conclude that also the plasmids are represented in the same range of coverage. Using the whole-genome shotgun data, it was possible to assemble two individual plasmids of the B.garinii PBi strain completely (Table 1). These two plasmids are highly similar and collinear to the linear plasmid lp54 and the circular plasmid cp26 of B.burgdorferi B31. The nearly two times higher coverage of one of these plasmids (lp54) compared to that of the chromosome indicates that it should be present in about two copies per cell. Compared to the chromosome, we find an equal number of base substitutions (8.9%) on the cp26 plasmid. Interestingly, with 15% substitutions are twice as frequent on plasmid lp54. Most remarkably, the transition:transversion ratio on lp54 is 3:2, whereas that of the chromosome as well as that of cp26 is approximately 4:1 (Table 2). The coding capacity of both plasmids is comparable to their counterparts in B.burgdorferi. Only three B.garinii lp54 genes predicted by GenMark are orphans, whereas the majority of predicted genes (49 of 74) have orthologs in the B.burgdorferi plasmid: 22 of the predicted genes match as pairs to different parts of 11 B.burgdorferi genes indicating nonsense mutations leading to split CDSs in B.garinii. On the other hand, 14 predicted B.burgdorferi genes have no counterparts on the B.garinii plasmid (Supplementary Table S5). None of these genes has an ascribed function. Interestingly, the B.burgdorferi lp54 gene family (BBA68 to BBA73) appears to be almost completely conserved in B.garinii, only BBA72 being split into two predicted genes. The analysis of the coding capacity of cp26 showed that all 26 predicted B.garinii genes have orthologs in B.burgdorferi, only three (BBB15, BBB20, BBB21) of the B.burgdorferi predicted genes are orphans (Supplementary Table S5). Interestingly, two cp26 encoded genes are subjected to a rapid positive selection: ospC and BBB08. OspC is well characterized as outer surface protein, whereas BBB08 so far has no assigned function. On the other hand, only 17 of the 55 lp54 encoded proteins may be subjected to a neutral evolution or purifying selection.
The assignment of clusters of orthologous groups (COG) (36) to the predicted proteins is clearly different between the chromosome and the plasmids. Whereas 81.6% of the chromosome-encoded orthologous proteins can be assigned to a COG, only 53.9% (cp26) and 26.5% (lp54) can be categorized this way (Supplementary Table S6).
All other B.garinii plasmids are represented in our assembly as 37 contigs >2 kb comprising 239 kb. We here refer to these plasmid parts as variable plasmid segments (VPS). As it is known from the assembly of the B.burgdorferi plasmids, there are redundant segments distributed over several plasmids (37,38). The same holds true for the B.garinii VPS. Some redundant regions containing polymorphisms could separately be assembled. Yet, the read coverage of some contigs is higher than that of the chromosome and cp26. Thus, it is very likely that these portions of the VPS represent paralogous sequences. Therefore, they cannot be assembled properly into individual plasmids. Accordingly, due to this non-unique nature of many segments in the plasmids, a clear 1:1 assignment to defined plasmids of B.burgdorferi is not possible.
A GeneMarkS gene prediction revealed 338 complete and truncated potential protein-coding genes on the VPS: 284 of these predicted genes have matches to predicted genes in the B.burgdorferi genome on protein level. Many, mainly small genes (117) show partial matches, but 167 predicted genes exhibit similar lengths in both genomes. One of these genes is a true ortholog to BB0844, which is encoded on the chromosome in B.burgdorferi. All other predicted genes are related to plasmid-encoded genes.
To get an overview of our B.garinii VPS assembly, we performed a BLAST search on nucleotide level of all plasmid-derived contigs against a database of B.burgdorferi plasmids. This BLAST search revealed that 70% (167 kb) of the VPS are similar enough to B.burgdorferi plasmid sections to be detected (Figure 1). The remaining sequences (73 kb) have no detectable similarity to B.burgdorferi plasmids on the DNA level. Yet, if we search for similarities on protein level, we find matches (40–70% identity on amino acid level) for all contigs to putative B.burgdorferi plasmid-encoded proteins. The contig with the lowest similarity to B.burgdorferi sequences is contig AY722928, which encodes a vls locus involved in antigenic variation in the mammalian host (39). This locus is located on lp28-1 in B.burgdorferi. Thus, all VPS sequences seem to be represented in B.burgdorferi plasmids.
We then asked, which part of the coding capacity of the B.burgdorferi plasmids is present in B.garinii. Since small predicted genes are often false positives, for a reliable comparison of the gene sets, we took into account only B.burgdorferi plasmid-derived proteins >100 amino acids, and searched for their counterparts in the whole B.garinii shotgun data. Only one protein each from plasmids lp5 (T), lp54 (Q), lp21 (U), cp32-3 (S), lp25 (E), lp28-1 (F), lp28-2 (G), lp28-3 (H) and lp28-4 (I) had no counterpart. The failure to detect these proteins in the B.garinii genome could be due to missing shotgun data. Interestingly, from plasmids lp38 (J) 15 of 21 and from plasmid lp36 (K) 9 of 24 proteins were not represented in the shotgun data (Supplementary Table S7). A more detailed inspection revealed that the predicted protein-coding regions from these two plasmids that have matches to the shotgun data belong to protein families. Members of these protein families are encoded also on different plasmids. These results taken together indicate that B.garinii PBi lacks the counterparts of plasmids lp38 and lp36.
Since the copy number of plasmid segments can affect the phenotype of Borreliae (40), we also analysed the data in this respect. According to the BLAST hits against B.burgdorferi proteins >100 amino acids plasmids lp28-1 (F), lp28-3 (H), lp28-4 (I), lp17 (D), lp25 (E) and lp5 (T) are present in one copy per cell. Plasmid lp28-2 is represented with two independent segments in our assembly. Thus, it should exist as two slightly different copies in B.garinii. For the proteins from the highly redundant cp32 and cp56 plasmids, we observed between three and four copies each. Furthermore, since parts of these segments are identical, the assembly resulted in parts of these segments in a higher coverage than average. We thus estimate that this plasmid group is at least present in five copies. The plasmid cp9 encodes similar proteins as the cp32 plasmids, albeit with much lower similarities. Thus, we are not able to determine whether a counterpart of this particular plasmid belongs to the B.garinii genome.
DISCUSSION
Closely related pathogenic species can cause different symptoms or even a different disease based on unique species-specific features. To reveal the molecular basis of such differences, one can examine the genomic repertoire of two closely related species. In cases where previous studies revealed not only a high similarity on DNA level but also almost complete collinearity of the chromosomes or large segments thereof, a direct comparative analysis without a completely finished genome is feasible (41,42). The limitation of this method lies in the inability to resolve highly rearranged and repetitive structures of the genome. Here, we report on an in-depth exploration of the B.garinii genome in comparison to that of B.burgdorferi. The analysis revealed a complete collinearity of the chromosome as well as of two plasmids in the compared species. The other genome parts seem to be subjected to rapid sequence changes as well as rearrangements and duplications. Thus, Borreliae genomes seem to consist of a remarkably invariant part mainly responsible for survival in ticks and a fluctuating part responsible for pathogenicity and disease symptoms in humans.
Borrelia core gene repertoire annotation on the invariant collinear genome fraction
An ab initio prediction of genes is flawed by the uncertainty as to what constitutes a real gene in a given organism (43). In recent years, it was successfully shown that a comparative genomics approach could improve if not largely replace an annotation from scratch (44,45). If the relationship between the two compared species is close enough, most if not all genes should have an ortholog in the sister species. Thus, the use of orthology information is the best approach to discern between true genes and false positive predictions or species-specific adaptations. On the stable fraction of the genome (chromosome and plasmids lp54 and cp26), we could easily define, which of the predicted genes in both organisms are orthologous gene pairs. Some of the orthologous pairs found may not be true genes especially if they are short. But the high conservation would suggest at least a regulatory function of these chromosomal regions. In total, we found 861 orthologous gene pairs compared to 955 (B.burgdorferi) and 932 (B.garinii) predicted genes on these conserved genomic elements. Since the other plasmids seem to be dispensable for viability, this set of orthologous protein-coding gene pairs is very likely the basic repertoire of Borrelia burgdorferi sensu lato species. This is supported by the fact that for example the proteins OspA, OspB and OspC, which seem to be required for survival in the tick midguts (46), are encoded on the two conserved plasmids.
COG categories show interspecies relationships, i.e. only proteins more common than for one genus can be categorized this way (36). The fraction of categorizable proteins decreases from the chromosome to cp26 and then to lp54 considerably, whereas the fraction of positively selected genes increases. Recently, it was shown that B.burgdorferi is not able to live without cp26 (47). Our analysis further supports the view that cp26 is an essential genome part of Borrelia species. Both the presence of two members of a family of genes (ospA and ospB) known to be needed in tick midguts and the conserved collinearity of the plasmid leads us to the conclusion that lp54 very likely plays also a pivotal role in survival in ticks. Since most of the encoded proteins are subjected to positive selection, we hypothesize that lp54 is a major player in host response evasion.
Selection pressure
The analysis of the ratio of non-synonymous to synonymous substitutions shows that the overwhelming majority of functionally described proteins of the chromosome (475/513; 92.6%) are under neutral evolution or purifying selection. The remaining 33 proteins (Table 3) show positive selection to different degrees. As expected, we find many surface-associated proteins in this list, which may be involved in the escape from the host response. On the other hand, of the 295 proteins described only as predicted or hypothetical, 56 (19%) seem to be under positive selection. This is a larger fraction than that in the subset with known function. Many of the proteins with functional assignment as well as the hypothetical proteins seem to be located at the surface of the cell and presumably generate a strain variability to fool the immune response from ticks as well as from humans. Thus, this subset of hypothetical proteins would be a rewarding target for further functional studies.
Variable plasmid complement of B.garinii
Only an isolation and analysis of each single plasmid would allow the resolution of the entire plasmid fraction of B.garinii. But since the whole-genome shotgun data should represent most parts of the plasmids (see Results), we are able to calculate similarities between contigs and plasmids to uncover the relationship of B.garinii to B.burgdorferi plasmids, and to reveal differences in plasmid content between the two species.
The noncollinear plasmids are not only rearranged but also the encoded proteins are not as conserved as in the collinear genome parts. Yet, two-thirds of the B.garinii VPS sequences are similar enough on DNA level to match B.burgdorferi counterparts. For the remaining sequences, no DNA similarities could be found. Nevertheless, our ability to detect similarities on the protein level shows that these plasmid parts are more likely subject to an accelerated evolution than unique B.garinii genome constituents: 167 of the 338 predicted genes have counterparts of similar length in the B.burgdorferi genome. Despite their high divergence, they may thus be ascribed as proteins with orthologous functions. BB0844 is encoded on the right-end extension of the B.burgdorferi chromosome and its ortholog on a plasmid in B.garinii. This may point to a mechanism, which enables variable Borrelia chromosome lengths by exchanging parts between chromosome and distinct plasmid segments.
Based on comparisons of the coding capacity of the VPS with the B.burgdorferi plasmids, we are able to define, which plasmids are presumably present in B.garinii and which not. With this analysis, we could show that two plasmids (lp38, J and lp36, K) are missing from the B.garinii genome. This is especially important since lp38 encodes a member of the osp family of proteins (ospD), a protein family widely studied for its role in pathogenicity and survival in the hosts.
It was shown that interplasmidal duplications and rearrangements are able to change the virulence phenotype of Borrelia species (40). Thus, it is of high value to know, which plasmids or segments are represented more than once in the genome. Generally, duplication and diversification events seem to affect the same plasmids in the two species. We also see an amplification of the cp32 plasmid sequences, although there may be a few copies less in B.garinii than in B.burgdorferii. Most other plasmid sequences are represented only as one copy in each species. Yet, in B.garinii plasmid lp28-2 sequences underwent also duplication and diversification. Thus, while the main difference in the protein family sets lies in the presence or absence of plasmids lp36 and lp38, additional diversity is achieved not only by mutation of single genes but also by a duplication of lp28-2, and possibly modification of the copy number of cp32 plasmid sequences.
Evolution and species definition
In previous studies (48–50), a large genetic distance between and within Borrelia species was observed on the basis of highly variable genes and genomic regions. Most of the genes examined are located on the plasmids, which are far less conserved than the chromosome. In contrast, we found a strong conservation of similarity and collinearity between B.garinii and B.burgdorferi not only of the chromosome but also of two plasmids. Interestingly, the amino acid identity of chromosomally encoded proteins is not higher than the conservation of the whole chromosome on DNA level. Thus, despite positive selection observed for specific proteins (Table 3), the chromosome on the whole is subjected to a neutral evolution.
Since plasmid repertoire variability is observed also in closely related strains causing similar disease patterns, the species definition is based only on the chromosome. Proteins encoded on the chromosome may also slightly influence the disease pattern. As discussed before, the survival in ticks may be mediated by the two collinear plasmids. Thus, pathogenicity in vertebrate hosts could be mainly dependent on the VPS. Future work has to determine, which plasmids or plasmid parts are lost during loss of pathogenicity. Since both Borrelia species have amplified mainly the same plasmids or segments thereof, it is conceivable that it is not enough to keep one member of each plasmid-encoded protein for maintenance of pathogenicity. Rather a larger number of paralogous proteins encoded by redundant plasmids could be required to successfully infect vertebrates.
Possibly the genes under positive selection are also causative for the symptom differences of the various Borrelia species. Thus, both the variable plasmid part of the genome and the positively selected genes represent prime targets for further functional studies.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
DDBJ/EMBL/GenBank accession nos+ CP000013—CP000015, AY722917—AY722953
REFERENCES
- 1.Wang G., van Dam,A.P., Schwartz,I. and Dankert,J. (1999) Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev., 12, 633–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marti Ras N., Postic,D., Foretz,M. and Baranton,G. (1997) Borrelia burgdorferi sensu stricto, a bacterial species ‘made in the U.S.A.’? Int. J. Syst. Bacteriol., 47, 1112–1117. [DOI] [PubMed] [Google Scholar]
- 3.Wilske B., Busch,U., Eiffert,H., Fingerle,V., Pfister,H.W., Rossler,D. and Preac-Mursic,V. (1996) Diversity of OspA and OspC among cerebrospinal fluid isolates of Borrelia burgdorferi sensu lato from patients with neuroborreliosis in Germany. Med. Microbiol. Immunol. (Berl.), 184, 195–201. [DOI] [PubMed] [Google Scholar]
- 4.Saint Girons I., Gern,L., Gray,J.S., Guy,E.C., Korenberg,E., Nuttall,P.A., Rijpkema,S.G., Schonberg,A., Stanek,G. and Postic,D. (1998) Identification of Borrelia burgdorferi sensu lato species in Europe. Zentralbl. Bakteriol., 287, 190–195. [DOI] [PubMed] [Google Scholar]
- 5.Stevenson B. and Miller,J.C. (2003) Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol., 57, 309–324. [DOI] [PubMed] [Google Scholar]
- 6.Purser J.E., Lawrenz,M.B., Caimano,M.J., Howell,J.K., Radolf,J.D. and Norris,S.J. (2003) A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol., 48, 753–764. [DOI] [PubMed] [Google Scholar]
- 7.Lagal V., Postic,D. and Baranton,G. (2002) Molecular diversity of the ospC gene in Borrelia. Impact on phylogeny, epidemiology and pathology. Wien. Klin. Wochenschr., 114, 562–567. [PubMed] [Google Scholar]
- 8.Canica M.M., Nato,F., du Merle,L., Mazie,J.C., Baranton,G. and Postic,D. (1993) Monoclonal antibodies for identification of Borrelia afzelii sp. nov. associated with late cutaneous manifestations of Lyme borreliosis. Scand. J. Infect. Dis., 25, 441–448. [DOI] [PubMed] [Google Scholar]
- 9.Lunemann J.D., Zarmas,S., Priem,S., Franz,J., Zschenderlein,R., Aberer,E., Klein,R., Schouls,L., Burmester,G.R. and Krause,A. (2001) Rapid typing of Borrelia burgdorferi sensu lato species in specimens from patients with different manifestations of Lyme borreliosis. J. Clin. Microbiol., 39, 1130–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eiffert H., Karsten,A., Thomssen,R. and Christen,H.J. (1998) Characterization of Borrelia burgdorferi strains in Lyme arthritis. Scand. J. Infect. Dis., 30, 265–268. [DOI] [PubMed] [Google Scholar]
- 11.Vasiliu V., Herzer,P., Rossler,D., Lehnert,G. and Wilske,B. (1998) Heterogeneity of Borrelia burgdorferi sensu lato demonstrated by an ospA-type-specific PCR in synovial fluid from patients with Lyme arthritis. Med. Microbiol. Immunol. (Berl.), 187, 97–102. [DOI] [PubMed] [Google Scholar]
- 12.Xu Y. and Johnson,R.C. (1995) Analysis and comparison of plasmid profiles of Borrelia burgdorferi sensu lato strains. J. Clin. Microbiol., 33, 2679–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casjens S., Delange,M., Ley,H.L.,III, Rosa,P. and Huang,W.M. (1995) Linear chromosomes of Lyme disease agent spirochetes: genetic diversity and conservation of gene order. J. Bacteriol., 177, 2769–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwan T.G., Burgdorfer,W. and Garon,C.F. (1988) Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect. Immun., 56, 1831–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busch U., Will,G., Hizo-Teufel,C., Wilske,B. and Preac-Mursic,V. (1997) Long-term in vitro cultivation of Borrelia burgdorferi sensu lato strains: influence on plasmid patterns, genome stability and expression of proteins. Res. Microbiol., 148, 109–118. [DOI] [PubMed] [Google Scholar]
- 16.Fraser C.M., Casjens,S., Huang,W.M., Sutton,G.G., Clayton,R., Lathigra,R., White,O., Ketchum,K.A., Dodson,R., Hickey,E.K. et al. (1997) Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature, 390, 580–586. [DOI] [PubMed] [Google Scholar]
- 17.Casjens S., Palmer,N., van Vugt,R., Huang,W.M., Stevenson,B., Rosa,P., Lathigra,R., Sutton,G., Peterson,J., Dodson,R.J. et al. (2000) A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol., 35, 490–516. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y., Kodner,C., Coleman,L. and Johnson,R.C. (1996) Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect. Immun., 64, 3870–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elias A.F., Stewart,P.E., Grimm,D., Caimano,M.J., Eggers,C.H., Tilly,K., Bono,J.L., Akins,D.R., Radolf,J.D., Schwan,T.G. et al. (2002) Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun., 70, 2139–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren S.X., Fu,G., Jiang,X.G., Zeng,R., Miao,Y.G., Xu,H., Zhang,Y.X., Xiong,H., Lu,G., Lu,L.F. et al. (2003) Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature, 422, 888–893. [DOI] [PubMed] [Google Scholar]
- 21.Seshadri R., Myers,G.S., Tettelin,H., Eisen,J.A., Heidelberg,J.F., Dodson,R.J., Davidsen,T.M., DeBoy,R.T., Fouts,D.E., Haft,D.H. et al. (2004) Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc. Natl Acad. Sci. USA, 101, 5646–5651. Epub 2004 Apr 5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser C.M., Norris,S.J., Weinstock,G.M., White,O., Sutton,G.G., Dodson,R., Gwinn,M., Hickey,E.K., Clayton,R., Ketchum,K.A. et al. (1998) Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science, 281, 375–388. [DOI] [PubMed] [Google Scholar]
- 23.Zhong J. and Barbour,A.G. (2004) Cross-species hybridization of a Borrelia burgdorferi DNA array reveals infection- and culture-associated genes of the unsequenced genome of the relapsing fever agent Borrelia hermsii. Mol. Microbiol., 51, 729–748. [DOI] [PubMed] [Google Scholar]
- 24.Kellis M., Patterson,N., Endrizzi,M., Birren,B. and Lander,E.S. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature, 423, 241–254. [DOI] [PubMed] [Google Scholar]
- 25.Hardison R.C. (2003) Comparative genomics. PLoS Biol., 1, E58. Epub 2003 Nov 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ojaimi C., Davidson,B.E., Saint Girons,I. and Old,I.G. (1994) Conservation of gene arrangement and an unusual organization of rRNA genes in the linear chromosomes of the Lyme disease spirochaetes Borrelia burgdorferi, B.garinii and B.afzelii. Microbiology, 140, 2931–2940. [DOI] [PubMed] [Google Scholar]
- 27.Wilske B., Preac-Mursic,V., Gobel,U.B., Graf,B., Jauris,S., Soutschek,E., Schwab,E. and Zumstein,G. (1993) An OspA serotyping system for Borrelia burgdorferi based on reactivity with monoclonal antibodies and OspA sequence analysis. J. Clin. Microbiol., 31, 340–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preac-Mursic V., Wilske,B. and Reinhardt,S. (1991) Culture of Borrelia burgdorferi on six solid media. Eur. J. Clin. Microbiol. Infect. Dis., 10, 1076–1079. [DOI] [PubMed] [Google Scholar]
- 29.Roe B.A. (2004) Shotgun library construction for DNA sequencing. Methods Mol. Biol., 255, 171–187. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonfield J.K., Smith,K. and Staden,R. (1995) A new DNA sequence assembly program. Nucleic Acids Res., 23, 4992–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Besemer J., Lomsadze,A. and Borodovsky,M. (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res., 29, 2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casjens S., Murphy,M., DeLange,M., Sampson,L., van Vugt,R. and Huang,W.M. (1997) Telomeres of the linear chromosomes of Lyme disease spirochaetes: nucleotide sequence and possible exchange with linear plasmid telomeres. Mol. Microbiol., 26, 581–596. [DOI] [PubMed] [Google Scholar]
- 34.Iyer R., Kalu,O., Purser,J., Norris,S., Stevenson,B. and Schwartz,I. (2003) Linear and circular plasmid content in Borrelia burgdorferi clinical isolates. Infect. Immun., 71, 3699–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimm D., Elias,A.F., Tilly,K. and Rosa,P.A. (2003) Plasmid stability during in vitro propagation of Borrelia burgdorferi assessed at a clonal level. Infect. Immun., 71, 3138–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natale D.A., Galperin,M.Y., Tatusov,R.L. and Koonin,E.V. (2000) Using the COG database to improve gene recognition in complete genomes. Genetica, 108, 9–17. [DOI] [PubMed] [Google Scholar]
- 37.Stevenson B., Zuckert,W.R. and Akins,D.R. (2000) Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J. Mol. Microbiol. Biotechnol., 2, 411–422. [PubMed] [Google Scholar]
- 38.Zuckert W.R. and Meyer,J. (1996) Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J. Bacteriol., 178, 2287–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J.R., Hardham,J.M., Barbour,A.G. and Norris,S.J. (1997) Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell, 89, 275–285. [DOI] [PubMed] [Google Scholar]
- 40.Penningon P.M., Cadavid,D., Bunikis,J., Norris,S.J. and Barbour,A.G. (1999) Extensive interplasmidic duplications change the virulence phenotype of the relapsing fever agent Borrelia turicatae. Mol. Microbiol., 34, 1120–1132. [DOI] [PubMed] [Google Scholar]
- 41.Kirkness E.F., Bafna,V., Halpern,A.L., Levy,S., Remington,K., Rusch,D.B., Delcher,A.L., Pop,M., Wang,W., Fraser,C.M. et al. (2003) The dog genome: survey sequencing and comparative analysis. Science, 301, 1898–1903. [DOI] [PubMed] [Google Scholar]
- 42.Anzai T., Shiina,T., Kimura,N., Yanagiya,K., Kohara,S., Shigenari,A., Yamagata,T., Kulski,J.K., Naruse,T.K., Fujimori,Y. et al. (2003) Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc. Natl Acad. Sci. USA, 100, 7708–7713. Epub 2003 Jun 7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aggarwal G. and Ramaswamy,R. (2002) Ab initio gene identification: prokaryote genome annotation with GeneScan and GLIMMER. J. Biosci., 27, 7–14. [DOI] [PubMed] [Google Scholar]
- 44.Nowrousian M., Wurtz,C., Poggeler,S. and Kuck,U. (2004) Comparative sequence analysis of Sordaria macrospora and Neurospora crassa as a means to improve genome annotation. Fungal Genet. Biol., 41, 285–292. [DOI] [PubMed] [Google Scholar]
- 45.Dubchak I. and Frazer,K. (2003) Multi-species sequence comparison: the next frontier in genome annotation. Genome Biol., 4, 122. Epub 2003 Nov 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang X.F., Pal,U., Alani,S.M., Fikrig,E. and Norgard,M.V. (2004) Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med., 199, 641–648. Epub 2004 Feb 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Byram R., Stewart,P.E. and Rosa,P. (2004) The essential nature of the ubiquitous 26-kilobase circular replicon of Borrelia burgdorferi. J. Bacteriol., 186, 3561–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farlow J., Postic,D., Smith,K.L., Jay,Z., Baranton,G. and Keim,P. (2002) Strain typing of Borrelia burgdorferi, Borrelia afzelii, and Borrelia garinii by using multiple-locus variable-number tandem repeat analysis. J. Clin. Microbiol., 40, 4612–4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bunikis J., Garpmo,U., Tsao,J., Berglund,J., Fish,D. and Barbour,A.G. (2004) Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology, 150, 1741–1755. [DOI] [PubMed] [Google Scholar]
- 50.Xu Y., Bruno,J.F. and Luft,B.J. (2003) Detection of genetic diversity in linear plasmids 28-3 and 36 in Borrelia burgdorferi sensu stricto isolates by subtractive hybridization. Microb. Pathog., 35, 269–278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.