Abstract
Autophagy is a highly conserved protein degradation pathway from yeasts to humans that is essential for removing protein aggregates and misfolded proteins in healthy cells. Recently, autophagy-related genes polymorphisms have been implicated in several autoimmune diseases including systemic lupus erythematosus, rheumatoid arthritis, psoriasis, and multiple sclerosis. Numerous studies reveal autophagy and autophagy-related proteins also participate in immune regulation. Conditional deletions of autophagy-related proteins in mice have rendered protection from experimental autoimmune encephalomyelitis, and TNF-mediated joint destruction in animal models of multiple sclerosis and experimental arthritis respectively. As autophagy is strongly implicated in immune functions such as removal of intracellular bacteria, inflammatory cytokine secretion, antigen presentation, and lymphocyte development, in this review we summarized current understanding of the roles of autophagy and autophagy proteins in autoimmune diseases.
1. Introduction
1.1 Autophagy pathways
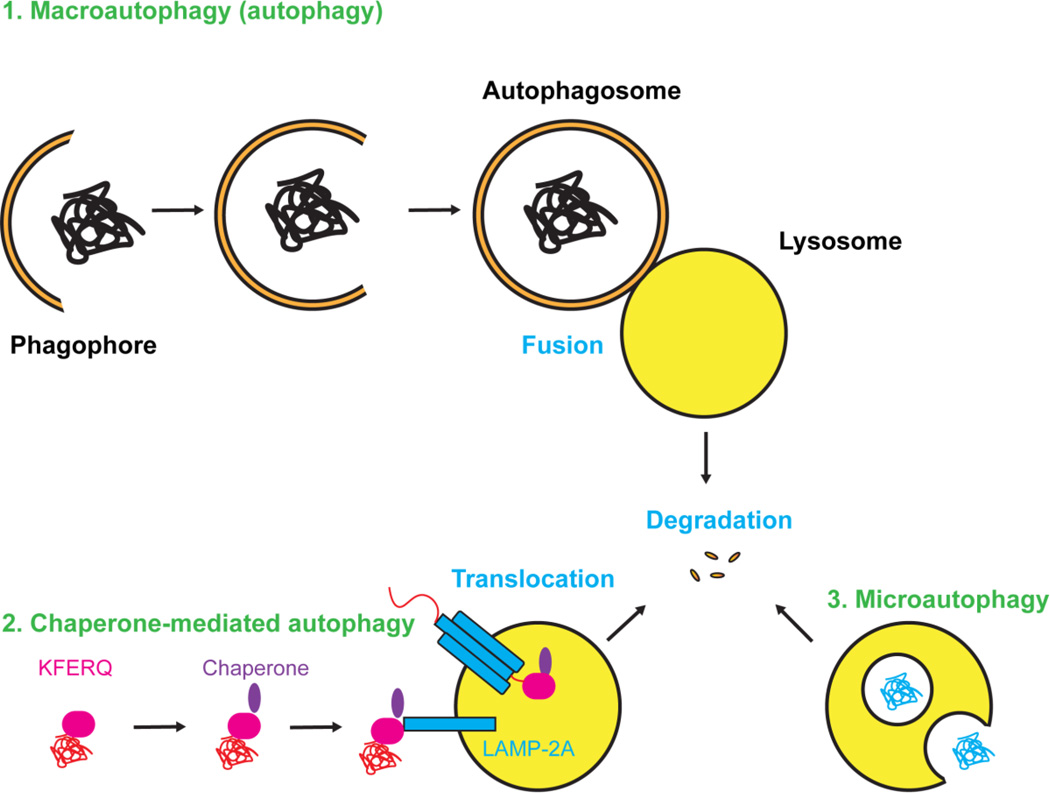
Autophagy is the only known conserved protein degradation pathway other than the ubiquitin-proteasome system (UPS). There are three major types of autophagy: 1) macroautophagy (referred as autophagy in general), 2) chaperone-mediated autophagy (CMA), and 3) microautophagy (Fig. 1).
Figure 1. Illustration of the three types of autophagy.
1) Macroautophagy is generally referred to as autophagy. In the process, protein aggregates and misfolded proteins are recruited by the phagophore and then enclosed in double-membrane vesicles named autophagosomes that later fuse with lysosomes for degradation. 2) In chaperone-mediated autophagy, proteins contain KFERQ degradation signal are recruited by chaperone and bind to LAMP-2A on lysosomes. Proteins then translocate from cytosol through LAMP-2A multimers into lysosomes for degradation. 3) Microautophagy directly engulfs cytoplasmic components for degradation.
1.2 Macroautophagy
The most studied autophagy pathway is macroautophagy that is generally referred as autophagy. Because the autophagy pathway is highly conserved, studies in yeast genetics advance our knowledge tremendously in the molecular process of autophagy. This process involves phagophore formation, autophagosome formation, and fusion of autophagosomes and lysosomes to form autolysosomes for protein degradation. Phagophore is an isolation membrane, which may derive from the endoplasmic reticulum (ER) or mitochondria. Phagophore can recruit and enclose cytoplasmic components selectively or non-selectively to form a double layer membrane vesicle called autophagosome. Autophagosome later fuses with the lysosome and forms the autolysosome where degradative enzymes break down cytoplasmic components.
Similar to UPS, ubiquitin also plays essential roles in regulating autophagy, including activating autophagy-related (ATG) proteins and labeling targeted cargos for degradation [1]. ATG7 is one of the ubiquitin-activating enzymes, also know as E1 enzymes that initialized the process [2]. E2 ATG10 is responsible for the conjugation of ATG5-ATG12 to ATG16, which then facilitate conjugation of phosphatidylethanolamine (PE) to LC3 together with E2 ATG3 [3]. PE-conjugated LC3, also known as LC3-II tightly bound to autophagosomal membranes, therefore, is used as autophagic marker protein [4–6].
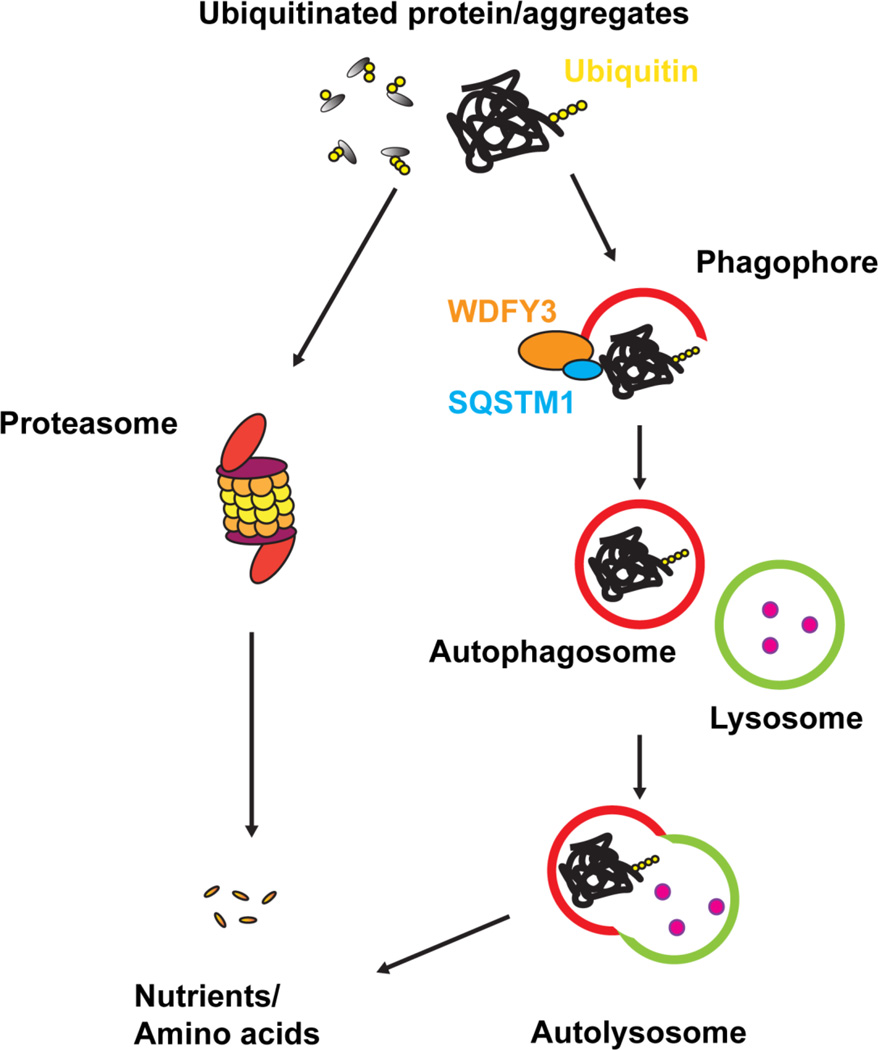
Autophagy has been long considered to be a non-selective process. However, recent studies demonstrate that autophagy also regulates highly selective degradation processes such as clearing damaged mitochondria (mitophagy) and clearing ubiquitinated protein aggregates (aggrephagy)(Fig. 2)[7–10]. Deficient in mitophagy-related proteins such as PTEN-induced putative kinase 1 (PINK1), Parkin, optineurin (OPTN) and Nix leads to impaired clearance of damaged mitochondria [11, 12]. Deficient in other autophagy adaptor proteins such as sequestosome-1 (SQSTM1), neighbor of Brca1 gene (NBR1), WDFY3 or HDAC6 leads to impaired clearance of misfolded proteins and protein aggregates [13–15].
Figure 2. Illustration of ubiquitinated proteins undergoes two major degradation pathways.
Ubiquitinated-labeled misfolded proteins or protein aggregates can be degraded via ubiquitin-proteasome system (UPS) and/or autophagy. UPS system cannot process large proteins or protein aggregates due to its limitation of size. In contrast, macroautophagy can enclose large proteins and aggregates in autophagosomes (0.5–1.5µm), which later fuse with lysosomes for degradation. Autophagy adaptor proteins such as SQSTM1 and WDFY3 can recruit ubiquitinated-labeled misfolded proteins or protein aggregates into phagophore and form autophagosomes. The autophagosomes then fuse with lysosomes that contain degradative enzymes and form autolysosomes and degrade proteins.
1.3 Chaperone-mediated autophagy
Chaperone-mediated autophagy (CMA) is another type of autophagy that does not involve the formation of autophagosomes [16]. CMA involves several steps including 1) substrate recognition and lysosomal targeting, 2) substrate binding and unfolding, 3) substrate translocation and substrate degradation [17]. Firstly, cargos containing short degradation signal sequence related to KFERQ are recruited by a chaperone complex including Heat shock-cognate protein of 70KDa (Hsc70) in the cytosol [18]. Secondly, Lysosome-associated membrane protein type 2A (LAMP-2A) on the lysosomes targets and binds to the chaperone complex, which brings the targeted proteins close to lysosomes. Thirdly, the interaction between LAMP-2A and chaperone complex further induces LAMP-2A oligomerization to facilitate translocation of targeted proteins from the cytosol into lysosomes for degradation [19]. Recently, modulation of deregulated CMA by a phosphopeptide has been shown significantly reduced autoimmune pathologies in patients and an animal autoimmune disease model [20], but further studies are required to elucidate CMA mechanisms in autoimmune diseases.
1.4 Microautophagy
Microautophagy involves direct lysosomal engulfment of cytoplasmic cargo without forming autophagosomes, which is essential for cell survival when cells are under stress such as nutrient starvation [21, 22]. Microautophagy can be divided into five sequential steps: 1) microautophagic invagination and autophagic tubes, 2) vesicle formation, 3) vesicle expansion, 4) vesicle scission and 5) vesicle degradation [23]. In microautophagy, lipids are essential for maintaining invagination and forming autophagic tubes, which are distinct from other types of autophagy [24].
Recently, microautophagy has been shown to regulate synaptic protein turnover in neurons and thus defects in microautophagy may result in accumulation of dysfunctional proteins and cause neurodegenerative disorders [25]. Although studies in yeast and Drosophila models have advanced our knowledge of microautophagy, the physiological function of microautophagy in mammalian cells remains poorly understood.
2. Autophagy in the immune system
Autophagy plays four principle roles in the immune system including 1) removal of intracellular pathogens, 2) secretory pathway, 3) lymphocyte development, and 4) pro-inflammatory signaling [26, 27].
2.1 Autophagy for removal of intracellular pathogens
There are two routes to eliminate intracellular pathogens through the autophagy pathway. The first route is termed xenophagy [28] that involves the engulfment of intercellular pathogens in double-membrane autophagosomes. The second route is termed LC3-associated phagocytosis (LAP) and is characterized by the enclosing of pathogens in single-membrane phagosomes decorated with LC3 [29, 30]. Some pathogen removal via autophagy requires additional receptors such as toll-like receptors (TLRs) [31]. The vesicles containing intracellular pathogens then fuse with lysosomes to form autolysosomes or autophagolysosomes [32] for degradation and elimination of intercellular pathogens.
2.2 Autophagy in the secretory pathway
The process of phagocytosis and the secretory pathway share a lot of common functions including vesicle trafficking and membrane fusion. Therefore, it’s no surprise that the autophagy pathway/autophagy proteins that play roles in phagocytosis also participate in secretory pathways. For instance, mice deficient in autophagy-related protein 5 (ATG5fl/fl LysM Cre+) present high level of IL-1α secreted by macrophages in vitro and in vivo, which lead to excessive inflammatory responses [33]. Furthermore, inhibition of autophagy in antigen-presenting cells leads to elevated IL-1β secretion upon TLRs stimulation in vitro and induction of autophagy along with LPS stimulation reduced IL-1β secretion in vivo [34]. Inhibition of autophagy in antigen-presenting cells also leads to elevated IL-23 secretion as a consequential event of increased IL-1 β level [35]. Recently, autophagic regulation of mitochondrial reactive oxygen species (ROS) has been shown to control the secretion of another pro-inflammatory cytokine, macrophage migration inhibitory factor (MIF) [36], which aligns with previous studies and suggest defects in autophagy leads to increased pro-inflammatory cytokines secretion.
2.3 Autophagy in lymphocyte development
Lymphocytes including T cells and B cells are important for adaptive immunity. Proper activation of lymphocytes is critical for lymphocyte development, and defective activation may result in autoimmunity. One major T cell activation signal is antigen presentation via the major histocompatibility complex (MHC) molecules I, II that reside on the cell surface to display antigens. MHC class I are found on all nucleated cells and MHC class II are found in antigen presenting cells (APCs) including macrophages, dendritic cells and B cells. To enable proper presentation of antigens, peptides derived from intracellular or extracellular proteins need to be digested or processed via degradation pathways including the ubiquitin-proteasome system and autophagy.
Autophagy can enhance MHC class I presentation of viral antigens in macrophages during infection [37] and also promote MHC class II presentation of viral antigens [38, 39]. Furthermore, other reports have elegantly demonstrated that autophagy is required for generation of major histocompatibility complex (MHC) class II antigen-specific CD4 (+) T cell responses in dendritic cells [40, 41]. Similarly, autophagy deficiency in thymic epithelial cells (TECs) causes altered MHC class II presentation of MHC peptide ligands and tissue-restricted antigens, which contributes to the generation of autoreactive CD4 (+) T cell repertoire [42].
B cells can differentiate into plasma cells that are responsible for generating autoantibodies and are critical for autoimmunity. It was previously shown that ATG5 is required for B cell survival during development and for the maintenance of B cell subset (B-1a) in the periphery [43] and plasma cells require autophagy for sustainable immunoglobulin production [44]. In fact, autophagy-deficient plasma cells secrete more antibodies accompanied with higher apoptosis rate compared to wild type plasma cells in vitro, suggesting that autophagy is specifically required for plasma cell homeostasis and long-lived humoral immunity. [44]. Defective or overactive autophagy modulates B cell development and function and therefore contributes to autoimmunity
2.4 Autophagy in pro-inflammatory signaling
Recent evidence supports crosstalk between autophagy/autophagy-related proteins in nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) activation. For instance, T cell receptor (TCR) mediated NF-κB activation is modulated by B-cell lymphoma/leukemia 10 (BCL10) in association with the autophagy adaptor p62/SQSTM1 [45]. In macrophages, SQSTM1/p62-dependent clearance of damaged mitochondria modulates NLRP3-inflammasome activation; ablation of SQSTM1/p62 leads to increased activation of inflammasome and overproduction of IL-1β [46]. Although this mechanism limits excessive IL-1β dependent inflammation, other studies have shown that autophagy enhances NF-κB activity in specific tissue macrophages by sequestering A20 to boost antifungal immunity [47]. This evidence suggests that autophagy can modulate survival and pro-inflammatory signaling via many pathways including NF-κB activation in many cell types. Since autophagy plays multiple roles in the immune system, disturbances in autophagic activity are likely to affect the development of autoimmunity. In fact, a plethora of evidence from genome-wide association studies and basic research highlight the autophagy roles in autoimmune diseases and are summarized in (Table 1.).
Table 1.
Summary of effect of inhibiting autophagy in cellular pathways and outcomes of different autoimmune diseases.
| Inhibition of Autophagy |
Outcomes | References | |
|---|---|---|---|
| Multiple sclerosis | Reduce autoreactive T cells, disrupted antigen presentation, defect neutrophils degranulation |
↓ disease severity | [48], [49], [50] |
| Systemic Lupus Erythematosus |
Partly inhibits plasma cells differentiation |
↓ disease severity | [51] |
| Rheumatoid Arthritis |
Reduced inflammatory cytokines secretion, reduced osteoclast function |
↓ disease severity ↓ bone destruction |
[52], [53] |
| Psoriasis/Psoriatic arthritis |
Increased inflammatory cytokine secretion |
↑ disease severity | [54], [55] |
| Inflammatory bowel diseases |
Disrupted exocytosis of antimicrobial peptides of Paneth cells, increased inflammatory cytokine |
↑ disease severity | [56], [57] |
3. Autophagy/Autophagy-related proteins in autoimmune diseases
3.1 Multiple sclerosis
Multiple sclerosis (MS) is a common autoimmune disease that caused inflammation and demyelination in the human central nervous system [58]. Since autophagy plays multiple roles in the immune system, extensive studies have investigated the role of autophagy in multiple sclerosis. A link between autophagy and multiple sclerosis is provided by the observed elevated autophagy-related protein, ATG5, expression in autoreactive T cells isolated from multiple sclerosis patients and mice with experimental autoimmune encephalomyelitis (EAE), [59]. The study further demonstrates that autophagy promotes T cell survival by degradation of apoptosis proteins in EAE model, and inhibition of autophagy in CD4 T cells using Beclin-1 conditional knockout mice (Beclin-1fl/fl CD4 Cre+) leads to protective phenotypes in EAE model [48]. In myeloid cells, disrupted antigen presentation in dendritic cells is observed in ATG7 conditional knockout mice (ATG7fl/fl CD11c Cre+) that again lead to reduced disease severity in EAE model [49]. Moreover, autophagy deficiency in neutrophils also reduced disease severity in EAE model due to defective degranulation [50]. Taken together, inhibition of autophagy leads to ameliorated disease severity in EAE model by regulating survival and activation of autoreactive T cells and reduced inflammatory cytokine secretion from neutrophils. The implication of autophagy in MS remains to be elucidated.
3.2 Systemic Lupus Erythematosus
Systemic Lupus Erythematosus (SLE) is an autoimmune disease that is characterized by acute and chronic inflammation of various tissues of the body including skin, joints, heart, kidneys and/or nervous system [60]. Several cellular and immune system components are disturbed in SLE, such as clearance of apoptotic cells, abnormal B and T cell signaling, autoantibody secretion and deregulated cytokine secretion. The cause of the SLE is currently unknown, but both genetic and environmental factors contribute to disease development [61]. Genome-wide association studies link autophagy-related gene 5 (ATG5) with SLE in both Chinese [62] and European [63] populations suggesting that defects in the autophagy pathway may contribute to SLE pathogenesis. A follow-up study suggested that ATG5 single nucleotide polymorphism (SNP) rs573775 implicated a role of the mutation with aberrant IL-10 cytokine secretion and higher risk for SLE [64]. Deregulation of autophagy has also been observed in T cells derived from SLE patients and confirmed in animal models of the disease [65], [66]. Moreover, activation of autophagy in B cell differentiation is observed in a lupus mouse model (NZB/W) and SLE patients. Inhibition of autophagy by inhibitors or transgenic mice partly inhibits plasma cells differentiation that suggests autophagy may regulate the survival rate of autoreactive B Cells and plasma cell differentiation in SLE [51].
Increased methylation patterns of histone deacetylase-6 (HDAC6), an ubiquitin-binding deacetylase critical for aggrephagy and mitophagy [67], compare to healthy controls are also observed in SLE patients [68]. Further animal studies reveal increased HDAC6 expression in T cells and B cells derived from NZB/W mice [69], and inhibition of HDAC6 reduces lupus pathogenesis in NZB/W mice, which suggest a potential role of HDAC6-dependent selective autophagy in SLE pathogenesis. Therapeutic treatments inhibiting autophagy pathway or autophagy-related proteins may improve SLE clinical outcomes by reducing autoreactive lymphocytes differentiation and their associated functions.
3.3 Rheumatoid Arthritis
Rheumatoid arthritis (RA) is a chronic inflammatory autoimmune disease that exhibits various clinical manifestations including synovial inflammation and bone loss. Immune cells such as Th17 cells, B cells, macrophages, neutrophils, mast cells and fibroblast-like synoviocytes are critical for inducing and maintaining synovial inflammation in RA pathology [70, 71]. This chronic inflammation leads to secretion of a plethora of pro-inflammatory cytokines and RANKL, which are primarily responsible for the activation of osteoclasts and the subsequent bone destruction. Autophagy has been associated with RA fibroblast-like synoviocytes (RA-FLS) survival, which is a major source of pro-inflammatory cytokines and RANKL. Specifically in in vitro experiments RA-FLS treated with a proteasome inhibitor (MG-132) prevented cell death whereas in contrast, treatment with an endoplasmic reticulum (ER) stress inducer, (thapsigargin) led to the formation of ubiquitinated protein aggregates and cell death via a mechanism involving autophagy proteins SQSTM1 and WDFY3 [72]. Other reports have shown that inhibition of autophagy-related protein HDAC6 using Tubastitin A reduces the inflammatory cytokine secretion from macrophages and FLS, and ameliorated arthritis disease severity in collagen antibody-induced arthritis (CAIA) and collagen-induced arthritis (CIA) in mouse models [73, 74]. Collectively, these data demonstrate a prominent role of autophagy in synovial inflammation both in vitro and in vivo.
Autophagy also modulates osteoclast-mediated bone destruction in rheumatoid arthritis. Increased expressions of autophagy-related protein Beclin1 and ATG7 are observed in osteoclasts of rheumatoid arthritis patients and inhibition of autophagy using ATG7fl/fl LysM Cre+ transgenic mice show reduced bone destruction in TNF-mediated arthritis [52]. The protective effect in TNF-mediated bone destruction resulted from the inhibition of autophagy may be in part due to impaired secretion of inflammatory cytokines IL-1 and IL-6 that affects osteoclastogenesis [52]. In addition, autophagy regulates the osteoclast ruffled border formation, (specialized organelle that facilitates bone resorption) which is evidenced by reduced bone resorption in both in vitro and in vivo assays in ATG5 deficient mice (ATG5fl/fl LysM Cre+) [53]. Furthermore, mutations in autophagy-related protein SQSTM1 impair osteoclast differentiation and are associated with Paget’s disease of bone [75]. Disrupted RANKL-induced osteoclastogenesis is observed in SQSTM1 deficient mice where SQSTM1 forms complex with TNF receptor associated factor 6 (TRAF6) and leads to NF-κB activation [76]. Deletion of SQSTM1 ubiquitin-binding domain (UBD) leads to increased osteoclast differentiation and function suggesting that SQSTM1 may regulate osteoclastogenesis via multiple pathways [77]. Interaction of autophagy-related proteins SQSTM1 and WDFY3 has been observed in human osteoclasts [78], and we recently showed that WDFY3 deficient mice (WDFY3fl/fl LysM Cre+) show enhanced osteoclastogenesis and RANKL-mediated bone resorption in vitro and in vivo assays via TRAF6 dependent activation of NF-κB [79]. Another autophagy-related protein, optineurin (OPTN) also negatively regulates osteoclastogenesis by modulating NF-κB and IFN-β signaling [80]. Furthermore, inhibition of autophagy using chloroquine prevents TRAF3 degradation and inhibits osteoclast differentiation in vitro and in vivo [81]. In summary, autophagy influences RA pathologies in two major ways; synovial inflammation and bone destruction. As inhibition of autophagy may ameliorate RA disease pathologies by modulating inflammation and bone destruction at synovial joints a detailed understanding of autophagy mechanisms in RA are needed to develop effective treatments.
3.4 Psoriasis/Psoriatic arthritis
Psoriasis is a chronic skin autoimmune disease where the skin undergoes abnormally excessive proliferation of keratinocytes, which also contribute to skin inflammation with increased secretion of pro-inflammatory cytokines. [82]. About 6–42% of psoriasis patients also have psoriatic arthritis [83]. Increased epidermal expression of the autophagy-related protein, SQSTM1, has been observed in psoriatic skin [54]. Autophagy negatively regulates TLR2/6 mediated NF-κB activation, SQSTM1 expression, and cytokine secretion in human keratinocytes that are critical to skin inflammation as observed in psoriasis/psoriatic arthritis [54]. Indeed other studies have shown that mutation of psoriasis risk gene AP1S3 that leads to impaired autophagy, and accumulation of SQSTM1, results in up-regulation of IL-36 in keratinocytes and causes skin inflammation [84]. Furthermore, increased expression of autophagy-related protein ATG16L1 is observed in dendritic cells derived from psoriatic arthritis patients compared to healthy controls that suggests autophagy involvement in psoriatic arthritis pathogenesis [85]. Inhibition of autophagy by chloroquine may aggravate psoriasis by increased IL-23 secretion from myeloid cells, which also leads to an induction of Th17 cells [55]. Taken together, inhibition of autophagy results in exacerbating skin inflammation in psoriasis and psoriatic arthritis. Modulation of autophagy may be a therapeutic approach for psoriasis/psoriatic arthritis, which merits further studies.
3.5 Inflammatory bowel diseases
Crohn’s disease and ulcerative colitis are the two common forms of inflammatory bowel disease (IBD) that are characterized as autoimmune diseases [86]. A human genome-wide association study identified autophagy-related genes ATG16L1 and immunity related GTPase M (IRGM) for Crohn’s disease and implicated autophagy in disease pathogenesis [87]. Deletion polymorphism upstream of IRGM alters IRGM expression, leads to defect autophagy and associates with Crohn's disease [88]. Deletion of autophagy-related proteins such as ATG16L1 and ATG5 leads to disrupted exocytosis of antimicrobial peptides of Paneth cells, which are essential for mucosal immunity [56]. Furthermore, deletion of ATG16L1 also leads to increased IL-1β production in macrophages that may also contribute to Crohn’s disease pathogenesis [57]. ATG16L1 interacts with nucleotide-binding oligomerization domain-containing protein 2 (NOD2) to degrade intracellular bacteria via autophagy pathway which is also deregulated in Crohn’s disease [89]. In summary, modulation of autophagy/autophagy-related proteins contributes to inflammatory bowel diseases. Restoring the functional autophagy-related proteins may be a great therapeutic approach for treating inflammatory bowel diseases in the future.
4. Conclusions
Autophagy is a conserved cellular degradation pathway. Recent evidence ranging from genome-wide association studies to basic in vivo and in vitro research have linked the autophagy pathways and/or autophagy-related proteins to autoimmunity. Crosstalk between autophagy and immune system includes removal of intracellular pathogens, secretory pathway, lymphocytes development, and pro-inflammatory signaling. Using transgenic animals, to model human diseases, important roles of autophagy in autoimmunity have been uncovered. Although in certain studies presented in this review inhibition of autophagy ameliorates diseases including multiple sclerosis, systemic lupus erythematosus, and rheumatoid arthritis in other cases it seems to exacerbate diseases such as psoriasis, psoriatic arthritis and Crohn’s disease. Since the autophagy pathway and autophagy-related proteins are highly conserved in many cell types the variation between effectors and transducers in different cells/tissues (bone-osteoclasts, skin-keratinocytes etc.) affected in the autoimmune diseases discussed and crosstalk of multiple pathways may be the underlying cause for this effect. For instance, SQSTM1 can regulate NF-κB activation via forming a complex with TRAF6 but also can degrade NF-κB via selective autophagy. In such cases the availability of effectors and transducers within a given cell may dictate the outcome. Distinguishing the autophagy-related protein's roles in autophagy or other cellular mechanisms in autoimmune disease pathologies remains to be a challenge [90]. A detailed understanding of autophagy is paramount, for the development of treatments for autoimmune diseases in the near future.
Highlights.
Autophagy is a highly conserved protein degradation pathway essential for removing protein aggregates and misfolded proteins in healthy cells.
Autophagy pathways are strongly implicated in immune functions such as removal of intracellular bacteria, inflammatory cytokine secretion, antigen presentation, and lymphocyte development.
Autophagy-related genes polymorphisms have been implicated in several autoimmune diseases including systemic lupus erythematosus, rheumatoid arthritis, psoriasis, and multiple sclerosis.
Acknowledgments
This work was partly supported by NIH/NIAMS-R01AR062173 and SHC 85700 grants to IEA, and by the UC Davis, graduate group in immunology fellowship to DW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None of the authors has any potential financial conflict of interest related to this manuscript.
References
- 1.Lamark T, Johansen T. Autophagy: links with the proteasome. Curr Opin Cell Biol. 2010;22:192–198. doi: 10.1016/j.ceb.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 3.Hanada T, Ohsumi Y. Structure-function relationship of Atg12, a ubiquitin-like modifier essential for autophagy. Autophagy. 2005;1:110–118. doi: 10.4161/auto.1.2.1858. [DOI] [PubMed] [Google Scholar]
- 4.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO journal. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. Journal of cell science. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 6.Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. The Journal of biological chemistry. 2006;281:3017–3024. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 7.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Molecular cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 8.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 9.Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell death and differentiation. 2013;20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrane J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO reports. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. Journal of cell science. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 14.Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Overvatn A, Stenmark H, Bjorkoy G, Simonsen A, Johansen T. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–344. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 15.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, Krainc D, Brech A, Stenmark H, Simonsen A, Yamamoto A. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Molecular cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 17.Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 19.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperonemediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macri C, Wang F, Tasset I, Schall N, Page N, Briand JP, Cuervo AM, Muller S. Modulation of deregulated chaperone-mediated autophagy by a phosphopeptide. Autophagy. 2015;11:472–486. doi: 10.1080/15548627.2015.1017179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 22.Mizushima N. Autophagy: process and function. Genes & development. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 23.Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller O, Sattler T, Flotenmeyer M, Schwarz H, Plattner H, Mayer A. Autophagic tubes: vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. The Journal of cell biology. 2000;151:519–528. doi: 10.1083/jcb.151.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uytterhoeven V, Lauwers E, Maes I, Miskiewicz K, Melo MN, Swerts J, Kuenen S, Wittocx R, Corthout N, Marrink SJ, Munck S, Verstreken P. Hsc70-4 Deforms Membranes to Promote Synaptic Protein Turnover by Endosomal Microautophagy. Neuron. 2015;88:735–748. doi: 10.1016/j.neuron.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nature reviews. Immunology. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, Glogauer M, Grinstein S, Brumell JH. Activation of antibacterial autophagy by NADPH oxidases. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6226–6231. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 31.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. The EMBO journal. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klionsky DJ, Eskelinen EL, Deretic V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes... wait, I'm confused. Autophagy. 2014;10:549–551. doi: 10.4161/auto.28448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castillo EF, Dekonenko A, Arko-Mensah J, Mandell MA, Dupont N, Jiang S, Delgado-Vargas M, Timmins GS, Bhattacharya D, Yang H, Hutt J, Lyons CR, Dobos KM, Deretic V. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3168–3176. doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, Kornfeld H, Fitzgerald KA, Lavelle EC. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. The Journal of biological chemistry. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peral de Castro C, Jones SA, Ni Cheallaigh C, Hearnden CA, Williams L, Winter J, Lavelle EC, Mills KH, Harris J. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol. 2012;189:4144–4153. doi: 10.4049/jimmunol.1201946. [DOI] [PubMed] [Google Scholar]
- 36.Lee JP, Foote A, Fan H, Peral de Castro C, Lang T, Jones SA, Gavrilescu N, Mills KH, Leech M, Morand EF, Harris J. Loss of autophagy enhances MIF/macrophage migration inhibitory factor release by macrophages. Autophagy. 2016;12:907–916. doi: 10.1080/15548627.2016.1164358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippe R, Desjardins M. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 39.Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Muller M, Kreymborg K, Altenberend F, Brandenburg J, Kalbacher H, Brock R, Driessen C, Rammensee HG, Stevanovic S. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, Mizushima N, Grinstein S, Iwasaki A. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–239. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 42.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 43.Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, Virgin HWt. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 44.Pengo N, Scolari M, Oliva L, Milan E, Mainoldi F, Raimondi A, Fagioli C, Merlini A, Mariani E, Pasqualetto E, Orfanelli U, Ponzoni M, Sitia R, Casola S, Cenci S. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013;14:298–305. doi: 10.1038/ni.2524. [DOI] [PubMed] [Google Scholar]
- 45.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity. 2012;36:947–958. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, He F, Boassa D, Perkins G, Ali SR, McGeough MD, Ellisman MH, Seki E, Gustafsson AB, Hoffman HM, Diaz-Meco MT, Moscat J, Karin M. NF-kappaB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell. 2016;164:896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanayama M, Inoue M, Danzaki K, Hammer G, He YW, Shinohara ML. Autophagy enhances NFkappaB activity in specific tissue macrophages by sequestering A20 to boost antifungal immunity. Nature communications. 2015;6:5779. doi: 10.1038/ncomms6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovacs JR, Li C, Yang Q, Li G, Garcia IG, Ju S, Roodman DG, Windle JJ, Zhang X, Lu B. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell death and differentiation. 2012;19:144–152. doi: 10.1038/cdd.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhattacharya A, Parillon X, Zeng S, Han S, Eissa NT. Deficiency of autophagy in dendritic cells protects against experimental autoimmune encephalomyelitis. The Journal of biological chemistry. 2014;289:26525–26532. doi: 10.1074/jbc.M114.575860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhattacharya A, Wei Q, Shin JN, Abdel Fattah E, Bonilla DL, Xiang Q, Eissa NT. Autophagy Is Required for Neutrophil-Mediated Inflammation. Cell reports. 2015;12:1731–1739. doi: 10.1016/j.celrep.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 51.Clarke AJ, Ellinghaus U, Cortini A, Stranks A, Simon AK, Botto M, Vyse TJ. Autophagy is activated in systemic lupus erythematosus and required for plasmablast development. Annals of the rheumatic diseases. 2015;74:912–920. doi: 10.1136/annrheumdis-2013-204343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin NY, Beyer C, Giessl A, Kireva T, Scholtysek C, Uderhardt S, Munoz LE, Dees C, Distler A, Wirtz S, Kronke G, Spencer B, Distler O, Schett G, Distler JH. Autophagy regulates TNFalpha-mediated joint destruction in experimental arthritis. Annals of the rheumatic diseases. 2013;72:761–768. doi: 10.1136/annrheumdis-2012-201671. [DOI] [PubMed] [Google Scholar]
- 53.DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, Takahata Y, Klumperman J, Tooze SA, Teitelbaum SL, Virgin HW. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Developmental cell. 2011;21:966–974. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee HM, Shin DM, Yuk JM, Shi G, Choi DK, Lee SH, Huang SM, Kim JM, Kim CD, Lee JH, Jo EK. Autophagy negatively regulates keratinocyte inflammatory responses via scaffolding protein p62/SQSTM1. J Immunol. 2011;186:1248–1258. doi: 10.4049/jimmunol.1001954. [DOI] [PubMed] [Google Scholar]
- 55.Said A, Bock S, Lajqi T, Muller G, Weindl G. Chloroquine promotes IL-17 production by CD4+ T cells via p38-dependent IL-23 release by monocyte-derived Langerhans-like cells. J Immunol. 2014;193:6135–6143. doi: 10.4049/jimmunol.1303276. [DOI] [PubMed] [Google Scholar]
- 56.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HWt. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxininduced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 58.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372:1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 59.Alirezaei M, Fox HS, Flynn CT, Moore CS, Hebb AL, Frausto RF, Bhan V, Kiosses WB, Whitton JL, Robertson GS, Crocker SJ. Elevated ATG5 expression in autoimmune demyelination and multiple sclerosis. Autophagy. 2009;5:152–158. doi: 10.4161/auto.5.2.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lisnevskaia L, Murphy G, Isenberg D. Systemic lupus erythematosus. Lancet. 2014;384:1878–1888. doi: 10.1016/S0140-6736(14)60128-8. [DOI] [PubMed] [Google Scholar]
- 61.Gualtierotti R, Biggioggero M, Penatti AE, Meroni PL. Updating on the pathogenesis of systemic lupus erythematosus. Autoimmun Rev. 2010;10:3–7. doi: 10.1016/j.autrev.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 62.Zhou XJ, Lu XL, Lv JC, Yang HZ, Qin LX, Zhao MH, Su Y, Li ZG, Zhang H. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Annals of the rheumatic diseases. 2011;70:1330–1337. doi: 10.1136/ard.2010.140111. [DOI] [PubMed] [Google Scholar]
- 63.International Consortium for Systemic Lupus Erythematosus G, Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, James JA, Merrill JT, Gilkeson GS, Seldin MF, Yin H, Baechler EC, Li QZ, Wakeland EK, Bruner GR, Kaufman KM, Kelly JA. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nature genetics. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lopez P, Alonso-Perez E, Rodriguez-Carrio J, Suarez A. Influence of Atg5 mutation in SLE depends on functional IL-10 genotype. PloS one. 2013;8:e78756. doi: 10.1371/journal.pone.0078756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alessandri C, Barbati C, Vacirca D, Piscopo P, Confaloni A, Sanchez M, Maselli A, Colasanti T, Conti F, Truglia S, Perl A, Valesini G, Malorni W, Ortona E, Pierdominici M. T lymphocytes from patients with systemic lupus erythematosus are resistant to induction of autophagy. FASEB J. 2012;26:4722–4732. doi: 10.1096/fj.12-206060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gros F, Arnold J, Page N, Decossas M, Korganow AS, Martin T, Muller S. Macroautophagy is deregulated in murine and human lupus T lymphocytes. Autophagy. 2012;8:1113–1123. doi: 10.4161/auto.20275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, Taylor JP, Cuervo AM, Yao TP. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. The EMBO journal. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang TJ, Lin YZ, Liu CC, Lin CH, Li RN, Wu CC, Ou TT, Tsai WC, Yen JH. Methylation and gene expression of histone deacetylases 6 in systemic lupus erythematosus. Int J Rheum Dis. 2015 doi: 10.1111/1756-185X.12783. [DOI] [PubMed] [Google Scholar]
- 69.Regna NL, Vieson MD, Gojmerac AM, Luo XM, Caudell DL, Reilly CM. HDAC expression and activity is upregulated in diseased lupus-prone mice. Int Immunopharmacol. 2015;29:494–503. doi: 10.1016/j.intimp.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423:356–361. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 71.Bottini N, Firestein GS. Duality of fibroblast-like synoviocytes in RA: passive responders and imprinted aggressors. Nature reviews. Rheumatology. 2013;9:24–33. doi: 10.1038/nrrheum.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kato M, Ospelt C, Gay RE, Gay S, Klein K. Dual role of autophagy in stressinduced cell death in rheumatoid arthritis synovial fibroblasts. Arthritis Rheumatol. 2014;66:40–48. doi: 10.1002/art.38190. [DOI] [PubMed] [Google Scholar]
- 73.Lee J, Hong EC, Jeong H, Hwang JW, Kim H, Bae EK, Ahn JK, Choi YL, Han J, Cha HS, Koh EM. A novel histone deacetylase 6-selective inhibitor suppresses synovial inflammation and joint destruction in a collagen antibody-induced arthritis mouse model. Int J Rheum Dis. 2015;18:514–523. doi: 10.1111/1756-185X.12501. [DOI] [PubMed] [Google Scholar]
- 74.Vishwakarma S, Iyer LR, Muley M, Singh PK, Shastry A, Saxena A, Kulathingal J, Vijaykanth G, Raghul J, Rajesh N, Rathinasamy S, Kachhadia V, Kilambi N, Rajgopal S, Balasubramanian G, Narayanan S. Tubastatin, a selective histone deacetylase 6 inhibitor shows anti-inflammatory and anti-rheumatic effects. Int Immunopharmacol. 2013;16:72–78. doi: 10.1016/j.intimp.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 75.Johnson-Pais TL, Wisdom JH, Weldon KS, Cody JD, Hansen MF, Singer FR, Leach RJ. Three novel mutations in SQSTM1 identified in familial Paget's disease of bone. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2003;18:1748–1753. doi: 10.1359/jbmr.2003.18.10.1748. [DOI] [PubMed] [Google Scholar]
- 76.Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, Moscat J, Diaz-Meco MT. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Developmental cell. 2004;6:303–309. doi: 10.1016/s1534-5807(03)00403-9. [DOI] [PubMed] [Google Scholar]
- 77.Yip KH, Feng H, Pavlos NJ, Zheng MH, Xu J. p62 ubiquitin binding-associated domain mediated the receptor activator of nuclear factor-kappaB ligand-induced osteoclast formation: a new insight into the pathogenesis of Paget's disease of bone. Am J Pathol. 2006;169:503–514. doi: 10.2353/ajpath.2006.050960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hocking LJ, Mellis DJ, McCabe PS, Helfrich MH, Rogers MJ. Functional interaction between sequestosome-1/p62 and autophagy-linked FYVE-containing protein WDFY3 in human osteoclasts. Biochemical and biophysical research communications. 2010;402:543–548. doi: 10.1016/j.bbrc.2010.10.076. [DOI] [PubMed] [Google Scholar]
- 79.Wu DJ, Gu R, Sarin R, Zavodovskaya R, Chen CP, Christiansen BA, Adamopoulos IE. Autophagy-linked FYVE containing protein WDFY3 interacts with TRAF6 and modulates RANKL-induced osteoclastogenesis. J Autoimmun. 2016 doi: 10.1016/j.jaut.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Obaid R, Wani SE, Azfer A, Hurd T, Jones R, Cohen P, Ralston SH, Albagha OM. Optineurin Negatively Regulates Osteoclast Differentiation by Modulating NF-kappaB and Interferon Signaling: Implications for Paget's Disease. Cell reports. 2015;13:1096–1102. doi: 10.1016/j.celrep.2015.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiu Y, Xu H, Zhao C, Li J, Morita Y, Yao Z, Xing L, Boyce BF. Chloroquine reduces osteoclastogenesis in murine osteoporosis by preventing TRAF3 degradation. The Journal of clinical investigation. 2014;124:297–310. doi: 10.1172/JCI66947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 83.Gladman DD, Antoni C, Mease P, Clegg DO, Nash P. Psoriatic arthritis: epidemiology, clinical features, course, and outcome. Annals of the rheumatic diseases. 2005;64(Suppl 2):ii14–ii17. doi: 10.1136/ard.2004.032482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mahil SK, Twelves S, Farkas K, Setta-Kaffetzi N, Burden AD, Gach JE, Irvine AD, Kepiro L, Mockenhaupt M, Oon HH, Pinner J, Ranki A, Seyger MM, Soler-Palacin P, Storan ER, Tan ES, Valeyrie-Allanore L, Young HS, Trembath RC, Choon SE, Szell M, Bata-Csorgo Z, Smith CH, Di Meglio P, Barker JN, Capon F. AP1S3 Mutations Cause Skin Autoinflammation by Disrupting Keratinocyte Autophagy and Up-Regulating IL-36 Production. J Invest Dermatol. 2016 doi: 10.1016/j.jid.2016.06.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wenink MH, Santegoets KC, Butcher J, van Bon L, Lamers-Karnebeek FG, van den Berg WB, van Riel PL, McInnes IB, Radstake TR. Impaired dendritic cell proinflammatory cytokine production in psoriatic arthritis. Arthritis and rheumatism. 2011;63:3313–3322. doi: 10.1002/art.30577. [DOI] [PubMed] [Google Scholar]
- 86.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 87.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nature genetics. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, Duerr RH, Silverberg MS, Taylor KD, Rioux JD, Altshuler D, Daly MJ, Xavier RJ. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nature genetics. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology. 2010;139:1630–1641. doi: 10.1053/j.gastro.2010.07.006. 1641 e1631-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO reports. 2013;14:143–151. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]