Abstract
Over the past decade, numerous nonviral cationic vectors have been synthesized. They share a high density of positive charges and efficiency for gene transfer in vitro. However, their positively charged surface causes instability in body fluids and cytotoxicity, thereby limiting their efficacy in vivo. Therefore, there is a need for developing alternative molecular structures. We have examined tetrabranched amphiphilic block copolymers consisting of four polyethyleneoxide/polypropyleneoxide blocks centered on an ethylenediamine moiety. Cryo-electron microscopy, ethidium bromide fluorescence and light and X-ray scattering experiments performed on vector–DNA complexes showed that the dense core of the nanosphere consisted of condensed DNA interacting with poloxamine molecules through electrostatic, hydrogen bonding and hydrophobic interactions, with DNA molecules also being exposed at the surface. The supramolecular organization of block copolymer/DNA nanospheres induced the formation of negatively charged particles. These particles were stable in a solution that had a physiological ionic composition and were resistant to decomplexation by heparin. The new nanostructured material, the structure of which clearly contrasted with that of lipoplexes and polyplexes, efficiently transferred reporter and therapeutic genes in skeletal and heart muscle in vivo. Negatively charged supramolecular assemblies hold promise as therapeutic gene carriers for skeletal and heart muscle-related diseases and expression of therapeutic proteins for local or systemic uses.
INTRODUCTION
According to the prevailing paradigm, the principle of nonviral gene delivery lies in the condensation of DNA in the presence of cationic molecules with a high density of positive charges [cationic lipids, polyethylenimine (PEI), polylysine] (1–5). Polyplexes and lipoplexes consisting of highly ordered structures of DNA molecules trapped within positively charged supramolecular assemblies have clearly demonstrated their transfection efficiency in vitro (6). However, aggregation in tissue fluids, toxicity and low in vivo efficiency have thus far hampered their clinical use. Non-ionic amphiphilic polymers were recently found to promote gene delivery in vivo, in tissues such as skeletal and cardiac muscle (7,8). The physicochemical properties of particles formed by non-ionic polymers and DNA are, however, ill-defined (8). This is a concern for in vivo investigation and clinical use, as the latter requires detailed physicochemical characterization. Consequently, new approaches to produce small and highly active assemblies that are homogeneous in size and exhibit colloidal stability are sought. These properties are needed for production of pharmaceutically acceptable formulations aimed at vector-based gene therapy. With this goal in mind, we decided to combine in the same molecule, the properties of cationic vectors and those of non-ionic amphiphilic polymers. We used poloxamine block copolymers, which have a tetrafunctional structure consisting of four polyethyleneoxide/polypropyleneoxide (PEO/PPO) blocks centered on an ethylenediamine moiety. Such block copolymers are largely used in the pharmaceutical sciences. Block copolymers are adsorbed on nanoparticles through binding of their hydrophobic PPO segments while their hydrophilic PEO segments extend outwardly. The steric stabilization thus achieved improves their pharmacokinetics (9). We defined experimental conditions under which DNA molecules collapsed into oligomolecular complexes exhibiting new structural characteristics. These negatively charged nanospheres are able to deliver reporter and therapeutic genes to skeletal and heart muscle cells in vivo.
MATERIALS AND METHODS
Plasmid DNA, poloxamine and formulation
Plasmids encoding reporter genes were described previously (8). The chloramphenicol acetyl transferase (CAT) plasmid was kindly provided by D. Gill (Oxford, UK) and described previously (10). The plasmid encoding human minidystrophin-GFP (11) was kindly provided by J. Tremblay (Université de Laval, Ste Foy, Qc, Canada) and J.T. Vilquin (Inserm U582, Paris, France). The plasmid encoding the secreted human alkaline phosphatase (SeAP) contains the cytomegalovirus (CMV) IE1 promoter and the bovine growth hormone polyadenylation signal. The plasmid encoding the tetracycline inducible murine erythropoietin (EPO) (pTetO-mEPO) has been described previously (12). All plasmids were purified from recombinant Escherichia coli using EndoFree plasmid purification columns (Qiagen).
Poloxamines, which are four-branched block copolymers of PEO and PPO, were generously provided by BASF, and were used as received. Stock solutions (20% w/v) were prepared in water. Formulations of DNA with poloxamine were always prepared by mixing equal volumes of poloxamine stock solution in water and plasmid DNA solution at the desired concentration in 300 mM NaCl or in 2× Tyrode's solution.
Fluorescence studies
Fluorescence measurements were carried out on a Kontron SFM25 spectrofluorimeter. Fluorescence was monitored immediately after adding ethidium bromide (final concentration of 5 μM) to solutions containing 10 μg/ml of DNA and various concentrations of poloxamine.
Electrophoretic and dynamic light scattering
Electrophoretic and dynamic light scattering measurements were made on a Zetasizer 3000HSA (Malvern) at 20°C. The poloxamine 304/DNA complexes were prepared using different DNA and poloxamine 304 concentrations in the Tyrode's solution.
Cryo-transmission electron microscopy (cryo-TEM)
Poloxamine 304/DNA complexes were prepared with 0.3 mg/ml of DNA. A 5 μl sample was deposited onto a holey carbon coated copper grid, the excess was blotted with a filter paper, and the grid was plunged into a liquid ethane bath cooled with liquid nitrogen (Leica EM CPC). Specimens were observed at a temperature of ∼−170°C, using a cryo holder (Gatan), with a FEI Tecnai F20 electron microscope operating at 200 kV. Images were obtained at a nominal magnification of 50 000× under low-dose conditions. Micrographs were recorded on Kodak SO163 films.
X-ray scattering
Poloxamine 304 and poloxamine 304/DNA complexes were prepared in Tyrode's solution or in 150 mM NaCl. Samples were loaded into 1 mm Lindemann type glass capillary. The X-ray scattering experiments were carried out by using the synchrotron radiation source of European Synchrotron Facility (Grenoble, France) on beamline ID2. The experiments were performed at an incident wavelength of 1.45 Å with a sample to detector distance of 3 mm and the scattering vector in the range 0.07 Å−1< q < 0.5 Å−1. Two-dimension SAXS patterns were recorded using an X-ray image intensifier (Thomson) lens coupled to a fast read out CCD (FreloN) camera, and appropriate corrections were carried out (13).
Reporter gene assay
Intramuscular and intracardiac gene transfer procedures were performed as described previously (8). Luciferase, β-galactosidase and green fluorescent protein (GFP) activities were measured as described previously (8). For SeAP quantification, blood was collected from the retro-orbital cavity and the serum, obtained by centrifugation. Mouse serum levels of SeAP were measured by ELISA (Ozyme).
In vivo delivery of therapeutic genes
Doxycycline (Ronaxan, Merial) was dissolved in drinking water to a final concentration of 200 μg/ml in 5% sucrose. Swiss mice were anesthetized with an intraperitoneal injection of 40 mg/kg hypnomidate. Fifty microliters of the poloxamine 304/DNA formulation was injected into shaved tibial anterior muscles at one site, using a microfine syringe (U100, Becton Dickinson). At least five female Swiss mice (8 week) were injected in each experimental group for hematocrit measurement. Hematocrit was determined by microcapillary centrifugation of blood samples. For EPO measurements, blood was collected from the retro-orbital cavity and serum obtained by centrifugation. Mice serum EPO levels were measured as described previously (12). Male mdx mice (8 week) were obtained from Iffa Credo. Mdx mice were anesthetized with xylazine and 50 μl of the formulation with Tyrode's solution containing 50 μg of DNA was injected into shaved tibial anterior muscles. Muscles were dissected, embedded in Tissue-Tek, frozen in isopentane, and cut into sections 10 μm thick. Minidystrophine-GFP expression was analyzed by fluorescence microscopy in transverse sections.
RESULTS AND DISCUSSION
DNA complexation with poloxamine
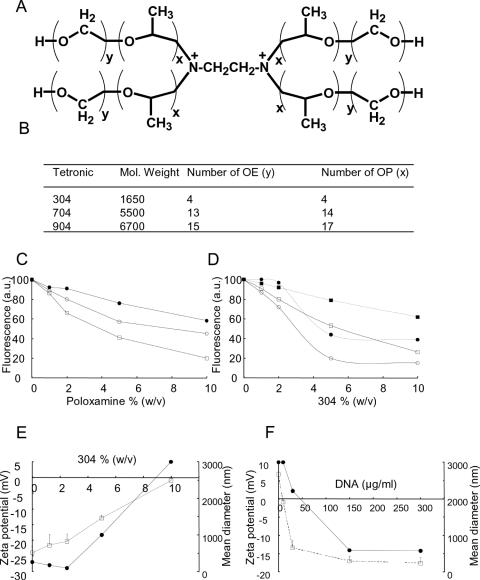
Poloxamines are branched block copolymers consisting of PEO and PPO blocks fixed to an aliphatic diamine (Figure 1A). The presence of the central ethylenediamine moiety renders the molecule positively charged. At physiological pH, there is one positive charge per poloxamine molecule, due to a shift in pKa of the second amine caused by the effect of charge repulsion (14).The basic ethylenediamine moiety has a pKa of 9.4 (14). The physical properties of the poloxamines used in this study are summarized in Figure 1B. DNA was complexed in saline with increasing amounts of poloxamine and exposed to ethidium bromide, which was used as a fluorescent probe as it intercalates between DNA base pairs. As the concentration of poloxamine increased, plasmid DNA molecules became progressively inaccessible to the probe. Among the different poloxamines tested, 304 was found to be the most potent for DNA complexation, possibly because of its low molecular weight (Figure 1C). We investigated how DNA complexation with poloxamine may be affected by interactions with electrolytes as they are found in vivo and may be required to adjust solution tonicity. In a medium that mimicked physiological extracellular ionic conditions (Tyrode's solution containing divalent cations), poloxamine 304/DNA complexes exhibited a more pronounced decrease in fluorescence intensity than in saline. The minimum fluorescence level was reached at 5% poloxamine 304 (Figure 1D). This suggests that the DNA included in the complexes formed in the Tyrode's solution was in a more condensed state.
Figure 1.
Physicochemical properties of poloxamine and poloxamine/DNA complexes. (A) General structure of poloxamines. (B) Physical properties of various poloxamines. (C) Fluorescence measurements to assess DNA complexation with poloxamine 304 (open squares), 704 (open circles) and 904 (filled circles). (D) Fluorescence measurements of poloxamine 304/DNA complexes formed in saline (open squares) and in Tyrode's solution (open circles). Poloxamine 304/DNA formulations with saline (filled squares) and Tyrode's solution (filled circles) were incubated with 100 mU/ml of heparin (dashed line). (E) Zeta potential (open squares) and hydrodynamic diameter (filled circles) of poloxamine 304/DNA complexes formed in Tyrode's solution with 10 μg/ml DNA, as a function of the concentration in poloxamine 304. (F) Zeta potential (open squares) and hydrodynamic diameter (filled circles) of poloxamine 304/DNA complexes formed in Tyrode's solution with 10% poloxamine 304, as a function of DNA concentration.
Interaction of complexes with the extracellular matrix is another important parameter for in vivo gene delivery since components such as negatively charged heparin can destabilize complexes made with highly cationic vectors, causing the early release of DNA molecules from the lipoplexes and polyplexes (15,16). As shown in Figure 1D, fluorescence intensity of poloxamine 304/DNA complexes moderately increased in the presence of heparin suggesting that DNA molecules remained complexed with 304. On the contrary, DNA did not remain complexed with PEI under identical conditions (data not shown) suggesting different charge surface characteristics. Electrophoretic light scattering measurements showed that the zeta potential of poloxamine 304/DNA complexes became less negative as the concentration of poloxamine 304 increased, and reached zero at 10% poloxamine 304 (Figure 1E). Concomitantly, the hydrodynamic diameter of 304/DNA complexes, measured by dynamic light scattering, increased (Figure 1E), likely because neutral complexes tend to aggregate due to attractive Van der Waals forces. Because of the amphiphilic nature of poloxamines, increasing their concentration induces their self-assembly in solution forming large aggregates of lyotropic liquid crystalline mesophases (17). As a result, we could not apply the strategy used with cationic lipids or cationic homopolymers, which consists in increasing the amount of cationic vector to obtain complexes with a good colloidal stability that is needed for in vivo gene transfer. We therefore tested various DNA concentrations with a constant concentration of poloxamine 304, i.e. 10%. Electrophoretic light scattering measurements revealed that poloxamine 304 alone had a positive zeta potential (Figure 1F). When poloxamine 304 was complexed with increasing amounts of DNA compatible with in vivo experimentation (see below), the zeta potential decreased to −18 mV +/− 3 mV. Concomitantly, the hydrodynamic diameter of poloxamine 304/DNA complexes decreased as DNA concentration increased (Figure 1F). Similar results were obtained with 5% poloxamine 304 (data not shown). Ethidium bromide fluorescence was also measured to monitor condensation of high concentrations of DNA mixed with 10% poloxamine 304. Surprisingly, fluorescence intensity remained low irrespective of DNA concentration (data not shown). In contrast, at a given concentration of cationic homopolymer such as PEI, fluorescence intensity increased when the DNA concentration increased. This suggests that PEI/DNA complexes had to be positively charged for all molecules to condense. Inversely, when poloxamine 304 condensed DNA molecules, negatively charged particles were formed. These complexes formed stable colloids at room temperature for several weeks (data not shown) most likely because of repulsive electrostatic forces which are proportional to the square of the zeta potential (18).
Cryo-electron microscopy and X-ray scattering studies of poloxamine 304 and poloxamine 304/DNA complexes
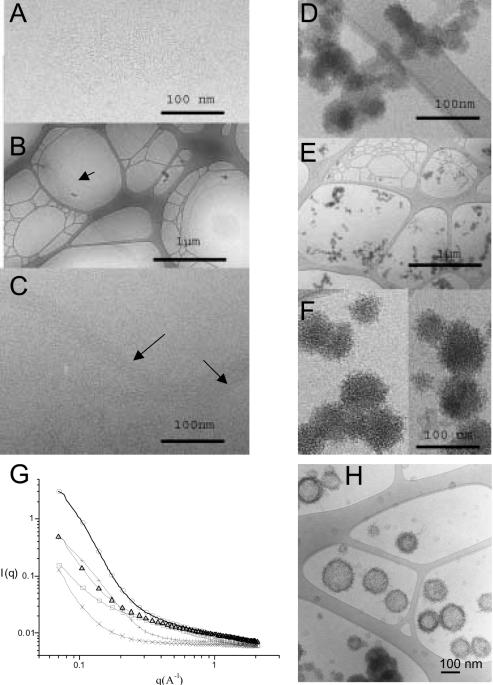
Cryo-electron micrographs showed that plasmid DNA appeared dispersed in 150 mM NaCl (data not shown) whereas poloxamine 304 formed dense stiffs, 25 nm in length (Figure 2A). In 150 mM NaCl, the poloxamine 304/DNA mixture formed a network of bundles, several micrometers in length, suggesting that the presence of poloxamine 304 induced DNA condensation (Figure 2B). At a higher magnification, DNA molecules were linearly condensed and no periodic repeats could be observed between DNA strands (Figure 2C). Molecular interactions between DNA strands were probably mediated through poloxamine 304. In the Tyrode's solution containing divalent cations, poloxamine 304 alone self-assembled into globular nanospheres (of ∼35 nm in diameter) with a smooth surface. These nanospheres were bound together in clusters of several micrometers (Figure 2D). When plasmid DNA in 2× Tyrode's solution and poloxamine 304 in water were combined by equivolumetric mixing, globular structures were also formed although they differed from those formed with poloxamine alone (Figure 2E). These globules were larger (∼60 nm in diameter) and had a rough surface (Figure 2F). In addition, clusters were smaller and did not include as many globules, in agreement with their size determined by dynamic light scattering. These modifications in size and shape of the nanospheres suggested that DNA molecules were condensed inside and at the surface of the globules, in agreement with their negative zeta potential. Altering the ionic environment had a drastic effect on self-assemblies of poloxamine 304, which in turn modulated DNA condensation.
Figure 2.
Cryo-TEM micrographs and X-ray scattering of poloxamine 304 and poloxamine 304/DNA complexes in saline and in Tyrode's solution. (A) Cryo-TEM micrograph of a 10% poloxamine 304 in saline. (B) Cryo-TEM micrograph of 304/DNA complexes with 10% poloxamine 304 in saline. (C) Cryo-TEM micrograph of the same sample at higher magnification. (D) Cryo-TEM micrograph of poloxamine 304 in Tyrode's solution. (E) Cryo-TEM micrograph of a poloxamine 304/DNA complexes with 10% poloxamine 304 in Tyrode's solution. (F) The same sample at a higher magnification. (G) Small-angle X-ray scattering scans of: poloxamine 304 in saline (crosses), in Tyrode's solution (plus sign), of DNA in Tyrode's solution (open squares), of poloxamine 304/DNA complexes in saline (open triangles) and in Tyrode's solution (open circles). (H) Cryo-TEM micrograph of poloxamine 304/DNA complexes in Tyrode's solution interacting with unilamellar vesicles of phosphatidylcholine/phosphatidylglycerol (30:70, w:w).
X-ray scattering data for poloxamine 304 revealed that intensity scattering was higher in Tyrode's solution than in saline, indicating that self-assembly of poloxamine 304 molecules was more efficient in the Tyrode's solution (Figure 2G). In the Tyrode's solution, the presence of both poloxamine 304 and DNA increased scattering intensity by a factor of five as compared to the sum of the scattering intensities for each constituent. This indicated that DNA and poloxamine interacted attractively to form the nanospheres seen in cryo-TEM. A similar, although less spectacular, interaction was also observed in saline. X-ray scattering study of poloxamine/DNA complexes did not reveal a correlation peak within the investigated q range, suggesting that no periodic structure was present irrespective of the medium used. X-ray scan data were corroborated by cryo-TEM results.
To account for the data, it is suggested that the dense core of nanopsheres consisted of condensed DNA interacting with poloxamine molecules through electrostatic, hydrogen bonding and hydrophobic interactions, with DNA molecules also being exposed at the surface. This architecture contrasts with that of classical cationic complexes where DNA molecules are trapped into a positively charged supramolecular assembly (19–23).
Cryo-TEM micrographs of combinations of poloxamine/DNA nanospheres with an artificial lipid membrane showed that the complexes were concentrated at the surface of liposomes with some complexes releasing their DNA (Figure 2H). This indicated that poloxamine/DNA nanospheres were able to fuse with lipid membranes. An attractive hypothesis would be that, in the living cell, DNA molecules associated with the nanospheres may be delivered directly to the cytoplasm following fusion with the plasma and/or endosomal membrane. Therefore, this double mode of cytoplasm entry could avoid the endosomal/lysosomal degradation by enzymes which are activated at low pH. In contrast, it has been fully documented that cationic complexes are internalized through endocytosis (24,25) with only a fraction of DNA molecules escaping lysosomal degradation.
In vivo reporter gene delivery
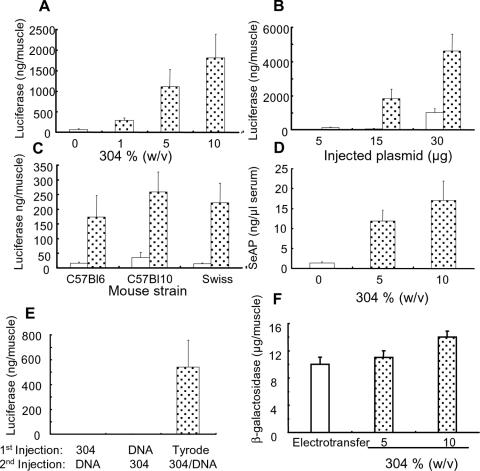
We assessed the potential of poloxamine/DNA nanospheres generated in the Tyrode's solution to transfer genes into the tibial anterior muscle. Seven days after injection of poloxamine/DNA nanospheres, the expression of reporter gene was monitored. The presence of 5 and 10% poloxamine 304 increased luciferase expression by a factor of 17 and 30, respectively, as compared with naked DNA (Figure 3A). A comparable increase in transfection efficiency was observed with the CAT gene (data not shown). Luciferase expression increased as the amount of injected formulated plasmid increased (Figure 3B). Figure 3C shows that luciferase expression increased with poloxamine 304 irrespective of the mouse strain tested (C57Bl6, Swiss, C57Bl10).
Figure 3.
In vivo reporter gene transfer in the mouse tibial anterior muscle. (A) Luciferase activity after intramuscular injection of 15 μg of DNA, naked (open column) or complexed with poloxamine 304 (dotted columns). (B) Expression of luciferase in the muscle as a function of the amount of injected DNA, naked (open columns) or complexed with 10% poloxamine 304 (dotted columns). (C) Luciferase expression after intramuscular injection of 15 μg of DNA, naked (open columns) or complexed with 10% poloxamine 304 (dotted columns), in various mouse strains. (D) Secreted alkaline phosphatase quantification after intramuscular injection of 15 μg of naked plasmid DNA (open column) or complexed with poloxamine 304 (dotted columns). (E) Luciferase expression as a function of the composition of the injected formulation. The luciferase plasmid (15 μg of DNA) was first injected, and 3 h later, a solution of 10% poloxamine 304 was injected into the same tibial anterior muscle or vice versa. In the control experiment, Tyrode's solution alone was injected at first. The poloxamine 304/DNA complexes, containing 15 μg of DNA and 10% poloxamine 304 in Tyrode's solution, was injected 3 h later. Reporter gene expression was monitored 7 days after the intramuscular injection of DNA, either naked or complexed with poloxamine 304. (F) Comparison of the muscle transfection efficiency of poloxamine 304/DNA complexes and electrotransferred naked DNA. Quantitative determination of β-galactosidase gene expression 7 days after electrotransfer of pCMV-βgal (open column) or injection of pCMV-βgal complexed with poloxamine 304 (dotted columns).
Mice were further injected with a plasmid encoding the SeAP reporter gene. Expression in serum was evaluated 7 days post-injection. Formulations containing 5 and 10% (w/v) poloxamine 304 increased SeAP activity in serum by a factor of 12 and 18, respectively, as compared with naked DNA (Figure 3D). In vivo transfection efficiency of poloxamine 304/DNA nanospheres was compared with that of sequential injection of DNA and poloxamine 304. Poloxamine 304/DNA nanopsheres were by far more effective in promoting transfection in the muscle (Figure 3E), suggesting that poloxamine 304 was active when complexed with DNA and did not act as a facilitating agent. Figure 3F shows transfection performance of our nanospheres in comparison with electrotransfer of naked DNA, a potent method for intramuscular transfection. Quantification of β-galactosidase activity showed that the level of transgene expression obtained with poloxamine 304/DNA nanospheres was slightly higher than that achieved by the electrotransfer procedure. Transgene distribution was assessed using β-galactosidase and GFP reporter plasmids. The number of transgene-expressing fibers increased when poloxamine 304/DNA nanospheres were injected. Transfection of ∼40% of the muscle fibers were so obtained (Figure 4B–F). Poloxamine 304/DNA nanospheres also increased transfection efficiency in the zebrafish myotome (Figure 4H–J) as compared with naked DNA (Figure 4G).
Figure 4.
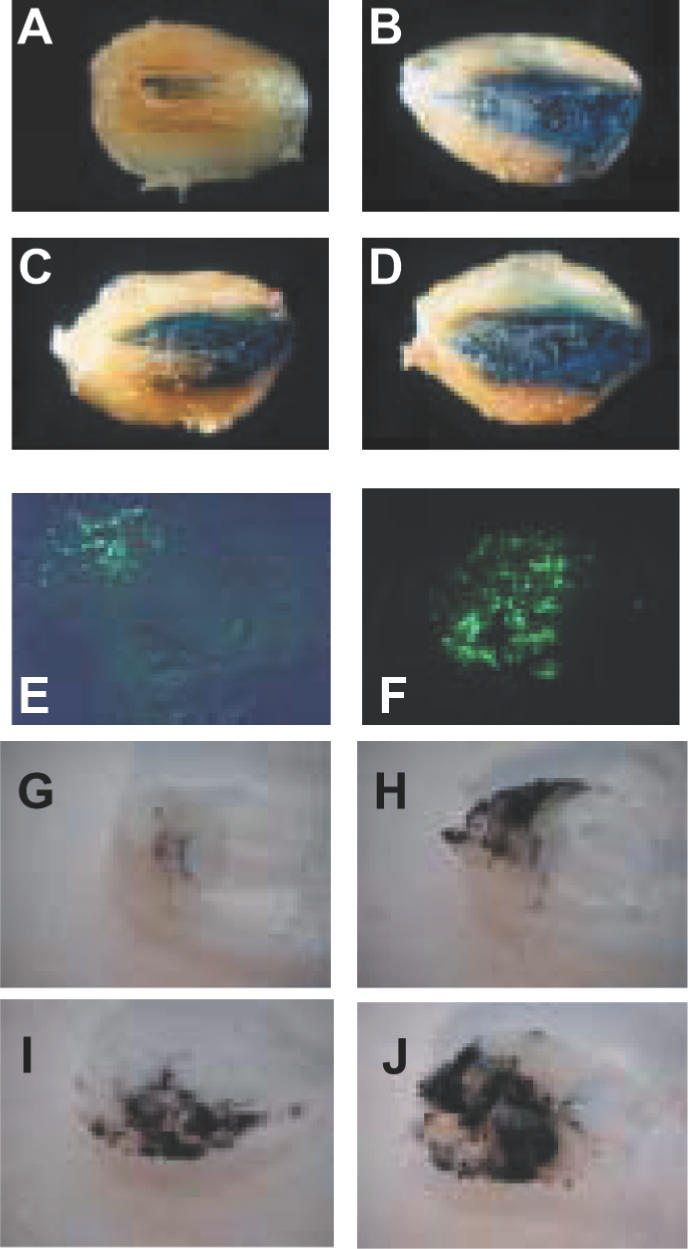
Visualization of reporter gene expression in the mouse tibial anterior muscle and in zebrafish muscle. Photographs of tibial anterior muscles were taken 7 days after intramuscular injection of plasmid DNA encoding the β-galactosidase gene in the absence of poloxamine 304 (A), with 5% (B) or 10% poloxamine 304 (C and D). Tissue sections of mouse tibial anterior muscles, 7 days after injection of naked plasmid DNA encoding GFP (E), and complexes of DNA with 5% poloxamine 304 (F). Photographs of sections of zebrafish myotomes 7 days after injection of plasmid DNA encoding β-galactosidase in the absence of poloxamine 304 (G) or in the presence of 1% (H), 5% (I) and 10% poloxamine 304 (J).
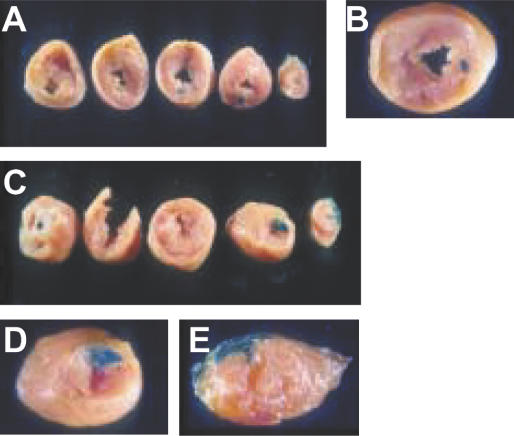
We also investigated transfection efficiency of poloxamine/DNA nanospheres in the rat heart muscle. Naked DNA or poloxamine 304/DNA nanospheres were injected into the free wall of the left ventricle through a thoracotomy. A larger area of tissue expressed β-galactosidase with poloxamine 304/DNA nanospheres (Figure 5C–E) than with naked DNA (Figure 5A and B).
Figure 5.
Gene transfer in rat heart muscle after direct injection into the left ventricle myocardium. Heart sections 3 days after injection of 240 μg of naked plasmid DNA encoding β-galactosidase (A and B). Heart sections 3 days after intramyocardial injection of β-galactosidase with 5% poloxamine 304 (C, D, E).
In vivo delivery of therapeutic genes with poloxamine 304
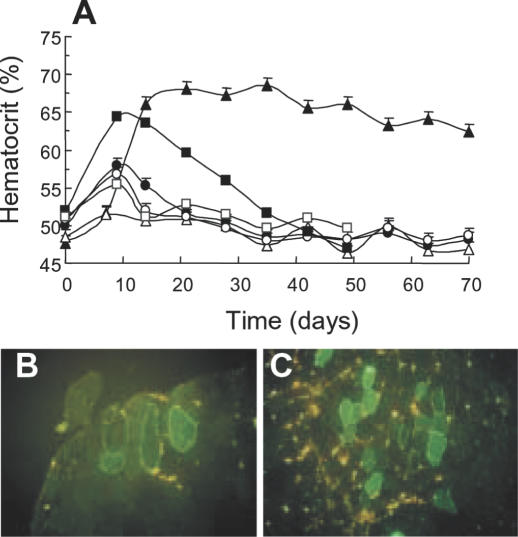
Next, poloxamine/DNA nanospheres were tested for the delivery of therapeutic genes. We investigated the expression of EPO and dystrophin because of their potential relevance in renal disease and Duchenne muscular dystrophy. Mice were intramuscularly injected with a naked plasmid encoding Tet-on inducible murine EPO, or with the plasmid formulated with poloxamine 304. Doxycycline was given in drinking water to half of the animals from day one and the hematocrit was repetitively determined. In the absence of doxycyline, mice injected with naked DNA or poloxamine 304/DNA nanospheres had a slightly, albeit nonsignificant, increased hematocrit (Figure 6A). In the presence of doxycycline, the hematocrit increased significantly at day 14 with 30 μg of naked DNA (Figure 6A). This increase was however transient and values progressively returned to control levels. In contrast, in mice treated with doxycycline that were injected with 10 μg of DNA complexed with poloxamine 304, the hematocrit remained elevated for up to 70 days post-injection (Figure 6A). The transient increase in hematocrit following injection of naked DNA was interpreted as resulting from an immune response to unmethylated CpG dinucleotides present in bacterial DNA. Lowering the amount of DNA injected in muscles with poloxamine 304 possibly caused a milder immune reaction, which resulted in prolonged expression of erythropoietin. One hundred and ten days after injection of the poloxamine 304/DNA nanospheres containing 10 μg of DNA and 5% poloxamine 304, serum EPO was 51 +/− 15 mU/ml in the presence of doxycycline and 3.5 +/− 3.3 mU/ml in its absence (data not shown).
Figure 6.
In vivo delivery of therapeutic genes. (A) Mice hematocrit as a function of time after injection of plasmid DNA encoding inducible murine EPO either naked or complexed with 5% poloxamine 304. Tibial anterior muscles were injected with 10 μg (filled and open circles) and 30 μg (filled and open squares) of naked DNA, and with the poloxamine 304/DNA complexes containing 10 μg of DNA (filled and open triangles). Ten mice were injected in each experiment: five were treated with doxycycline (filled symbols) while the other five were not (empty symbols). Photomicrographs of tissue sections of tibial anterior muscles of mdx mice injected with naked plasmid DNA encoding the human minidystrophin-GFP (B) or the poloxamine 304/DNA complexes (C).
Human minidystrophin-GFP plasmids formulated with or without poloxamine 304 were injected into the tibial anterior muscle of mdx mice. After 7 days, tissue sections showed that the percentage of transfected fibers was much greater with poloxamine 304 (15% of fibers on average) (Figure 6C) than without (Figure 6B).
By preventing non-specific interactions with extracellular matrix components and due to their small size, poloxamine/DNA nanostructured complexes may facilitate diffusion through the whole tissue and promote internalization in the target cell. On the contrary, biodiffusion of naked DNA and highly positively charged complexes may be hampered by the large size and electrostatic interaction with extracellular matrix respectively. These DNA-coated nanostructured complexes can theoretically carry DNA fragments of unlimited size and are compatible with multiple injection schemes. Poloxamine vectors offer advantageous production conditions for pharmaceutically acceptable formulations, which makes them promising new agents for gene therapy.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Clothilde Gourden for her excellent technical expertise, Jacques Tremblay and Jean-Thomas Vilquin for providing us with human minidystrophin-GFP-encoding plasmid, P. Panine for helpful discussion during the synchrotron experiments, and Jean Léger, L. Auvray and P. Davidson for fruitful discussions. This work was supported by the APEX program financed by INSERM, by an ACI program from the Ministère délégué à la recherche et aux nouvelles technologies, and by special grants from the Association Française contre les Myopathies and Vaincre la Mucoviscidose.
REFERENCES
- 1.Felgner P.L., Gadek,T.R., Holm,M., Roman,R., Chan,H.W., Wenz,M., Northrop,J.P., Ringold,G.M. and Danielsen,M. (1987) Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl Acad. Sci. USA, 84, 7413–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao X.A. and Huang,L. (1991) A novel cationic liposome reagent for efficient transfection of mammalian cells. Biochem. Biophys. Res. Commun., 179, 280–285. [DOI] [PubMed] [Google Scholar]
- 3.Behr J.P., Demeneix,B., Loeffler,J.P. and Perez-Mutul,J. (1989) Efficient gene transfer into mammalian primary endocrine cells with lipopolyamine-coated DNA. Proc. Natl Acad. Sci. USA, 86, 6982–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boussif O., Lezoualc'h,F., Zanta,M.A., Mergny,M.D., Scherman,D., Demeneix,B. and Behr,J.P. (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA, 92, 7297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Midoux P., Mendes,C., Legrand,A., Raimond,J., Mayer,R., Monsigny,M. and Roche,A.C. (1993) Specific gene transfer mediated by lactosylated poly-L-lysine into hepatoma cells. Nucleic Acids Res., 21, 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitard B. (2002) Supramolecular assemblies of DNA delivery systems. Somat Cell Mol. Genet., 27, 5–15. [DOI] [PubMed] [Google Scholar]
- 7.Lemieux P., Guerin,N., Paradis,G., Proulx,R., Chistyakova,L., Kabanov,A. and Alakhov,V. (2000) A combination of poloxamers increases gene expression of plasmid DNA in skeletal muscle. Gene Ther., 7, 986–999. [DOI] [PubMed] [Google Scholar]
- 8.Pitard B., Pollard,H., Agbulut,O., Lambert,O., Vilquin,J.T., Cherel,Y., Abadie,J., Samuel,J.L., Rigaud,J.L., Menoret,S., Anegon,I. and Escande,D. (2002) A nonionic amphiphile agent promotes gene delivery in vivo to skeletal and cardiac muscles. Hum. Gene Ther., 13, 1767–1775. [DOI] [PubMed] [Google Scholar]
- 9.Neal J.C., Stolnik,S., Garnett,M.C., Davis,S.S. and Illum,L. (1998) Modification of the copolymers poloxamer 407 and poloxamine 908 can affect the physical and biological properties of surface modified nanospheres. Pharm. Res., 15, 318–324. [DOI] [PubMed] [Google Scholar]
- 10.Pitard B., Oudrhiri,N., Lambert,O., Vivien,E., Masson,C., Wetzer,B., Hauchecorne,M., Scherman,D., Rigaud,J.L., Vigneron,J.P., Lehn,J.M. and Lehn,P. (2001) Sterically stabilized BGTC-based lipoplexes: structural features and gene transfection into the mouse airways in vivo. J. Gene Med., 3, 478–487. [DOI] [PubMed] [Google Scholar]
- 11.Chapdelaine P., Moisset,P.A., Campeau,P., Asselin,I., Vilquin,J.T. and Tremblay,J.P. (2000) Functional EGFP–dystrophin fusion protein for gene therapy vector development. Protein Eng., 13, 611–615. [DOI] [PubMed] [Google Scholar]
- 12.Richard P., Pollard,H., Lanctin,C., Bello-Roufaï,M., Désigaux,L., Escande,D. and Pitard,B. (2004) Inducible production of erythropoietin using intramuscular injection of block copolymer/DNA formulation. J. Gene Med., doi:10.1002/jgm.631. [DOI] [PubMed] [Google Scholar]
- 13.Narayananan T., Diat,O. and Boesecke,P. (2001) SAXS and USAXS on the high brilliance beamline at the ESRF. Nucl. Instrum. Methods Phys. Res. A, 467–468, 1005–1009. [Google Scholar]
- 14.Armstrong J.K., Chowdhry,B.Z., Snowden,M.J., Dong,J. and Leharne,S.A. (2001) The effect of pH and concentration upon aggregation transitions in aqueous solutions of poloxamine T701. Int. J. Pharm., 229, 57–66. [DOI] [PubMed] [Google Scholar]
- 15.Moret I., Esteban Peris,J., Guillem,V.M., Benet,M., Revert,F., Dasi,F., Crespo,A. and Alino,S.F. (2001) Stability of PEI–DNA and DOTAP–DNA complexes: effect of alkaline pH, heparin and serum. J. Control. Release, 76, 169–181. [DOI] [PubMed] [Google Scholar]
- 16.Oupicky D., Howard,K.A., Konak,C., Dash,P.R., Ulbrich,K. and Seymour,L.W. (2000) Steric stabilization of poly-L-lysine/DNA complexes by the covalent attachment of semitelechelic poly[N-(2-hydroxypropyl)methacrylamide]. Bioconjug. Chem., 11, 492–501. [DOI] [PubMed] [Google Scholar]
- 17.Alexandridis P. (1997) Poly(ethylene oxide)/poly(propylene oxide) block copolymer surfactants. Curr. Opin. Colloid Interface Sci., 2, 478–489. [Google Scholar]
- 18.Washington C. (1996) Stability of lipid emulsions for drug delivery. Adv. Drug Delivery Rev., 20, 131–145. [Google Scholar]
- 19.Pitard B., Aguerre,O., Airiau,M., Lachages,A.M., Boukhnikachvili,T., Byk,G., Dubertret,C., Herviou,C., Scherman,D., Mayaux,J.F. and Crouzet,J. (1997) Virus-sized self-assembling lamellar complexes between plasmid DNA and cationic micelles promote gene transfer. Proc. Natl Acad. Sci. USA, 94, 14412–14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitard B., Oudrhiri,N., Vigneron,J.P., Hauchecorne,M., Aguerre,O., Toury,R., Airiau,M., Ramasawmy,R., Scherman,D., Crouzet,J., Lehn,J.M. and Lehn,P. (1999) Structural characteristics of supramolecular assemblies formed by guanidinium-cholesterol reagents for gene transfection. Proc. Natl Acad. Sci. USA, 96, 2621–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lasic D., Strey,H., Stuart,M., Podgornik,R. and Frederik,P.M. (1997) The structure of DNA–liposome complexes. J. Am. Chem. Soc., 119, 832–833. [Google Scholar]
- 22.Rädler J.O., Koltover,I., Salditt,T. and Safinya,C.R. (1997) Structure of DNA-cationic liposome complexes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science, 275, 810–814. [DOI] [PubMed] [Google Scholar]
- 23.Koltover I., Salditt,T., Rädler,J.O. and Safinya,C.R. (1998) An inverted hexagonal phase of cationic liposome–DNA complexes related to DNA release and delivery. Science, 281, 78–81. [DOI] [PubMed] [Google Scholar]
- 24.Zabner J., Fasbender,A.J., Moninger,T., Poellinger,K.A. and Welsh,M.J. (1995) Cellular and molecular barriers to gene transfer by a cationic lipid. J. Biol. Chem., 270, 18997–19007. [DOI] [PubMed] [Google Scholar]
- 25.Labat-Moleur F., Steffan,A.M., Brisson,C., Perron,H., Feugeas,O., Furstenberger,P., Oberling,F., Brambilla,E. and Behr,J.P. (1996) An electron microscopy study into the mechanism of gene transfer with lipopolyamines. Gene Ther., 11, 1010–1017. [PubMed] [Google Scholar]