Abstract
Twenty Fmoc-protected trinucleotide phosphoramidites representing a complete set of codons for the natural amino acids were chemically synthesized for the first time. A pool of these reagents was incorporated into oligonucleotides at substoichiometric levels to generate two libraries of variants that randomly carry either few or many codon replacements on a region encoding nine amino acids of the bacterial enzyme TEM-1 β-lactamase. Assembly of the libraries was performed in a completely automated mode through a simple modification of ordinary protocols. This technology eliminates codon redundancy, stop codons and enables complete exploration of sequence space for single, double and triple mutations throughout a protein region spanning several residues. Sequence analysis of many non-selected clones revealed a good incorporation of the trinucleotides, producing combinations of mutations quite different from those obtained using conventional degenerate oligonucleotides. Ceftazidime-selection experiments yielded several never before reported variants containing novel amino acid combinations in the β-lactamase omega loop region.
INTRODUCTION
In the last years, many methods have been developed to accelerate artificial modification of proteins, either by enzymatic mutagenesis of the genes that encode them or by amplification from mutant primers. Enzymatic methods are mainly based on error-prone PCR (1) and/or in vitro recombination (2) generating quasirandom single base changes throughout a gene sequence. Unfortunately, a large number of amino acid replacements are not adequately represented in these approaches, such as those requiring 2–3 base pair changes per codon (3). This is the major drawback of methods that generate random point mutations. A similar effect is seen with libraries of spiked oligos, where each wild-type nucleotide is contaminated with the other three bases during oligonucleotide assembly (4,5). However, some reports have demonstrated the necessity to perform amino acid changes by replacements of two or three bases on the wild-type codons to optimize a target protein function (6). In order to cover all sequence space in a protein region, some schemes of saturation mutagenesis have been described to produce pools of degenerate oligonucleotides. ‘NNG/C’ approach is a widely used and commercially available method (7), which generates a mixture of 31 sense codons that encode the twenty amino acids and also one non-sense codon. This method is saturating by nature and the exploration of amino acid replacements on several positions requires numerous experiments and oligonucleotides (8). Additionally, the frequency of mutants generated with NNG/C is biased towards those amino acids encoded by redundant codons (9). Codon redundancy has been circumvented by the use of DMTr-protected trinucleotide phosphoramidites (DMTr represents the 4,4′-dimethoxytrityl protecting group) that encode all twenty amino acids (10) or a subset of them (11,12) again in schemes that are suited for regional saturation. These regional saturation methods are ill-suited for the exploration of variants displaying few amino acid replacements within segments of genes, in contrast to the spiked and enzymatic methods. For instance, if a small region comprising five amino acids is completely randomized with a mixture containing 20 DMTr-trinucleotides, a total number of 3.2 × 106 variants will be generated, where the single—95 variants—and double—3610 variants—mutants together represent only a 0.1% of the population. The situation gets worse as the region grows.
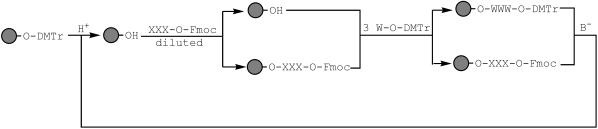
Library enrichment with variants carrying few codon replacements can be performed by combining Fmoc-protected trinucleotide phosphoramidites (Fmoc represents the 9-fluorenylmethoxycarbonyl group) and ordinary DMTr-protected monomers during oligonucleotide synthesis (Figure 4), such as was previously demonstrated with codons TTT and GCT (13). The success of the method relies in the chemical orthogonality of both protecting groups: whereas DMTr is labile to acid and stable to alkali, Fmoc is the converse. Mutant codons are introduced by substoichiometric incorporation of the Fmoc-trinucleotides to the growing oligonucleotide chains, whereas wild-type codons are assembled through DMTr-monomers.
Figure 4.
Fmoc-trinucleotide approach for the creation of codon-based amino acid substitutions in a non-saturating mode. The starting sphere represents a DMTr-protected oligonucleotide growing in a solid support. Synthesis proceeds in common 3′ to 5′ direction. In the first step, the DMTr group is removed by acid treatment (H+). A part of the growing oligo reacts with a diluted solution of a single Fmoc-trinucleotide phosphoramidite (XXX-O-Fmoc), or a mixture, without performing capping, oxidation and detritylation. The remaining chains are saturated with three sequential ordinary couplings (W-O-DMTr) to assemble the wild-type codon to be partially replaced. Removal of both protecting groups by sequential acid and base (B−) treatments prepares the oligo for a new cycle of mutagenesis.
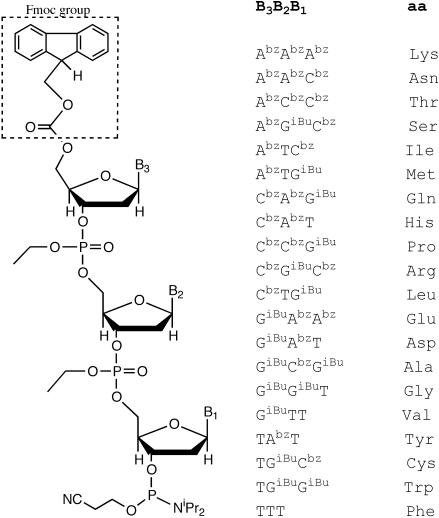
The goal of this report is to demonstrate the applicability of this mutagenesis technology for all amino acids, through the chemical synthesis of 20 Fmoc-trinucleotide phosphoramidites (Figure 2). A pool containing 19 codons (the 20th codon, GAA, representing Gly was mistakenly omitted) was incorporated into oligonucleotides to create two libraries of variants carrying different ratios of amino acid replacements in a region spanning nine residues of the bacterial enzyme TEM-1 β-lactamase. The sequence of several clones obtained in experiments with and without ceftazidime selection was used to analyze the distribution and frequency of mutations, as well as the sequence requirements in the region 170–178 of TEM-1 to generate a specificity change.
Figure 2.
Fmoc-trinucleotide phosphoramidites synthesized in this work. Abz: N6-benzoyl adenine, Cbz: N4-benzoyl cytosine, GiBu: N2-isobutyryl guanine, T: thymine.
MATERIALS AND METHODS
Chemistry
The chemical reagents used for oligonucleotide assembly were purchased from different suppliers. 1H-tetrazole and 4,4′-dimethoxytrityl chloride (DMTr-Cl) were acquired from Chemgenes. Ordinary 4,4′-dimethoxytrityl-β-cyanoethyl phosphoramidites of dAbz, dCbz, dGiBu and dT were from Glen Research. Iodine, pyridine, acetic anhydride (Ac2O), N-methylimidazole (NMI), trichloroacetic acid (TCA), ammonium hydroxide (NH4OH), 9-fluorenylmethoxycarbonyl chloride (Fmoc-Cl), 1,8-diazabicyclo(5.4.0)undec-7-ene (DBU), thiophenol, triethylamine and 2-cyanoethyl diisopropylchlorophosphoramidite were from Aldrich. Dichloromethane and tetrahydrofurane (THF) were from Burdick & Jackson, while anhydrous acetonitrile was from American Bioanalytical.
Chemical reactions in solution-phase were followed by thin layer chromatography (TLC) on aluminum-backed silica gel 60 F254 sheets (Merck). Intermediate phosphoramidites were analyzed on ethyl acetate/dichloromethane/triethylamine (45:45:10 v/v), while all the other reactions were analyzed on chloroform:methanol 9:1. 5′-Fmoc-monomers were synthesized and recovered by the method described by Lehmann et al. (14). 3′-DMTr-monomers were prepared by our reported procedure (13). The phosphitylating agent chloro(diisopropylamino)-ethoxyphosphine (EtOPClNiPr2) used in the synthesis of Fmoc-trimers was prepared according to Atkinson and Smith, using ethoxydichlorophosphite instead of methoxydiclorophosphite (15).
HPLC analyses were performed on a System Gold chromatograph and an ultrasphere C18 analytical column (4.6 × 250 mm) both from Beckman, with detection at 260 nm. Elution of deprotected tetramers was performed by a linear acetonitrile gradient from 5 to 15% in 0.1 M triethylammonium acetate, pH 7.2, over 20 min at a flow rate of 1 ml/min. Fmoc-trinucleotide phosphoramidites were analyzed under similar conditions but using a linear acetonitrile gradient from 65 to 85%.
Chemical synthesis
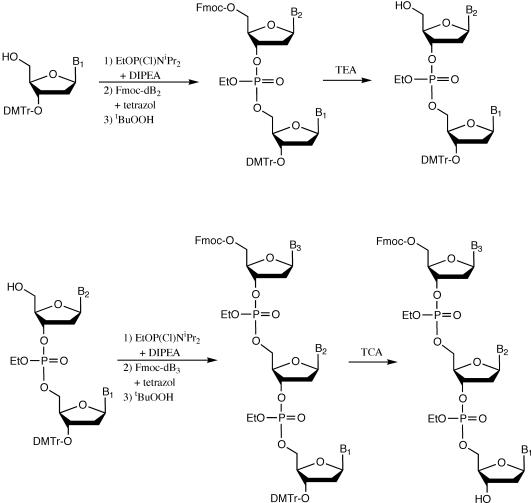
Synthesis of Fmoc-trinucleotides, precursor synthons of the target Fmoc-trinucleotide phosphoramidites, proceeded in the 3′→5′ direction in a completely protected way as shown in Figure 1. The procedures for synthesis of the Fmoc-TTT trimer and its corresponding phosphoramidite are described as an example. The other 19 Fmoc-trinucleotide phosphoramidites AAA, AAC, ACC, AGC, ATC, ATG, CAG, CAT, CCG, CGC, CTG, GAA, GAT, GCG, GGT, GTT, TAT, TGC and TGG were prepared using the same procedure. For characterization and functionality, each Fmoc-trinucleotide phosphoramidite was coupled to a deoxynucleoside-bound support. The resulting tetramers and standards synthesized by ordinary chemistry were fully deprotected and co-injected in HPLC.
Figure 1.
Synthetic pathway used to assemble the Fmoc-trinucleotides. Abbreviations of reagents are indicated in Materials and Methods. B1, B2 and B3 represent Abz: N6-benzoyl adenine, Cbz: N4-benzoyl cytosine, GiBu: N2-isobutyryl guanine or T: thymine. Reagent abbreviations are indicated in Materials and Methods.
3′-O-4,4′-dimethoxytrityl(thymidine-3′-yl-ethylphosphate-5′-yl-thymidine) (3′DMTr-TT dimer): 3′-O-(4,4′-dimethoxytrityl)thymidine (1.53 g, 3.3 mmol) was dried by co-evaporation with anhydrous THF (10 ml × 2) and re-dissolved in 25 ml of the same solvent. N,N-diisopropylethylamine (0.68 ml, 3.9 mmol) and the phosphitylating agent (0.84 ml, 3.9 mmol) were added with independent disposable syringes under nitrogen atmosphere and magnetic stirring. After 30 min of reaction, TLC analysis showed a complete conversion of the nucleoside to its phosphoramidite, and this reaction mixture was transferred by teflon tubing to another flask containing a mixture of 5′-O-(9-fluorenylmethoxycarbonyl)thymidine (5′Fmoc-dT, 2.2 g, 4 mmol) and tetrazol (1.05 g, 15 mmol) previously co-evaporated with THF (2 × 10 ml). After 60 min of the second reaction, the TLC analysis showed a complete disappearance of the intermediate phosphoramidite (limiting reagent) and an excess of tert-butyl hydroperoxide (5 ml) was added to oxidize the recently formed phosphite-triester bond to the more stable phosphate-triester. After 1 h of reaction, the mixture was concentrated to dryness, dissolved in dichloromethane (100 ml) and sequentially washed with a saturated solution of sodium bicarbonate (2 × 20 ml) and brine (2 × 20 ml). The organic phase was dried over anhydrous sodium sulfate, filtered and concentrated until formation of foam. The fully protected dimer was submitted to a quick chromatographic purification to remove the excess of 5′Fmoc-dT, and then the Fmoc group was removed at room temperature with a solution of pyridine/TEA/water 3:1:1 (20 ml) during 15 min. The mixture was evaporated, concentrated to foam and re-submitted to chromatographic purification to eliminate any non-polar impurity that would have eluted in the first purification. The expected product 3′DMTr-TT dimer eluted with 3% methanol in dichloromethane. The pure compound was recovered as a white powder after precipitation over n-hexane and vacuum drying.
5′-O-(9-fluorenylmethoxycarbonyl)thymidine-3′-yl-ethylphosphate-5′-yl-thymidine-3′-yl-ethylphosphate-5′-yl-thymidine (5′Fmoc-TTT trimer): This compound was synthesized as 3′DMTr-TT dimer, excepting last steps. Once the fully protected trimer was semi-purified, it was submitted to detritylation with 3% TCA in dichloromethane (200 ml) at 0°C during 10 min, and was immediately neutralized with a saturated solution of sodium bicarbonate. The mixture was washed as before and concentrated to foam. The Fmoc-TTT trimer was purified by flash column chromatography using silica gel 60 H (Merck, 5–40 μm) as the stationary phase, eluting with 4% methanol in dichloromethane. The pure compound was recovered as a white powder after precipitation over n-hexane and vacuum drying.
5′-O-(9-fluorenylmethoxycarbonyl)thymidine-3′-yl-ethylphosphate-5′-yl-thymidine-3′-yl-ethylphosphate-5′-yl-thymidine-3′-yl-O-ethyl-N,N-diisopropylaminophosphoramidite (Fmoc-TTT amidite): The Fmoc-TTT trimer (1.19 g, 1.5 mmol) was dried by co-evaporation with anhydrous THF (10 ml × 2) and was re-dissolved in 15 ml of the same solvent. N,N-diisopropylethylamine (0.43 ml, 2.5 mmol) and 2-cyanoethyl diisopropylchlorophosphoramidite (0.47 ml, 2.25 mmol) were added with independent disposable syringes under nitrogen atmosphere and magnetic stirring. After 60 min of reaction, TLC analysis showed a complete transformation of the starting trimer to a more hydrophobic product corresponding to its phosphoramidite. The reaction was quenched with a saturated solution of sodium bicarbonate (5 ml), diluted with dichloromethane and washed as before. The crude compound was purified over silicagel 60, using 20% pyridine in dichloromethane as eluting solution. Finally, it was recovered as a white powder after precipitation over n-hexane and vacuum drying.
Assembly of oligonucleotide libraries
Two oligonucleotide libraries carrying codon substitutions were assembled in completely automated mode using an ASM-800 DNA synthesizer (Biosset, Novosibirsk, Russia), in a single synthesis column at 0.2 μmol scale. Note that this process can be implemented in any commercial DNA synthesizer equipped with an X-vial designed for especially phosphoramidites and performing manual Fmoc deprotection (13). The synthesis protocol is supplied as Supplementary Material.
The synthesizer was loaded with ordinary reagents for oligonucleotide synthesis. Detritylating reagent: 3% TCA in dichloromethane; cap A: 10% acetic anhydride in THF; cap B: THF:pyridine:N-methylimidazole 8:1:1; oxidizing reagent: 0.02 M iodine in THF:pyridine:water 8:1:1; DMTr-β-cyanoethylamidites of dAbz, dCbz, dGiBu and dT at 0.1 M concentration in anhydrous acetonitrile were loaded in vials A, C, G and T, respectively. The fifth vial, m5, for amidites was loaded with either of two mutagenic solutions, containing an equimolar pool of 19 Fmoc-trimer amidites. Total concentration of the first mutagenic solution was 6 mM, and the second one was 24 mM. The 6 mM solution was used to prepare an oligonucleotide library aiming at a few substituted codons on a region comprising nine codons; this library was termed Low Mutagenesis Rate (LMR). Consequently, the 24 mM solution was used to prepare a second oligonucleotide library whose variants should contain many codon replacements; this library was termed High Mutagenesis Rate (HMR). Finally, bottle R1 was loaded with 0.1 M DBU in acetonitrile for removal of the Fmoc group incorporated with the mutant codons. Figure S1, supplied as Supplementary Material, shows the DNA synthesizer implemented for assembly of the libraries.
The program for automated assembly of mutagenic oligos was devised to perform three types of synthesis cycles. (i) A cycle to perform ordinary coupling of DMTr-monomers, represented in the DNA sequence with letters a, c, g and t. It sequentially contains detritylation, coupling, capping and oxidation steps. (ii) A cycle to incorporate the Fmoc-trinucleotide phosphoramidites and the 3′ base of the wild-type codon to be partially replaced. It is represented in the DNA sequence with bold letters a, c, g and t. It contains the steps of detritylation, Fmoc removal, substoichiometric coupling of the growing oligo with the mutagenic mixture, saturation of remaining growing oligos with the 3′ monomer of the wild-type codon, capping and oxidation. (iii) A cycle to assemble the first wild-type base flanking the 5′ terminal of the mutagenized region. It is represented with underlined letters a, c, g or t and contains the steps of detritylation, removal of Fmoc group, saturation of growing oligos with the wild-type monomer, capping and oxidation.
The DNA sequence was programmed as follows and separated as codons in the mutagenized region for more clarity: 5′-accg gagctc aat gaa gcc ata cca aac gac gag cgt gataccacgatgcctgc-3′. This sequence also contains a SacI restriction site gagctc for cloning purposes.
Once the mutagenic oligos were assembled, the solid support containing them was treated with 0.5 ml of thiophenol/triethylamine (3:1 v/v) during 2 h at 55°C to remove the internucleotidic ethyl groups introduced with mutant codons. Supernatants were removed by filtration and the supports were subjected to an overnight treatment with concentrated ammonium hydroxide to remove all remaining protecting groups and recover the oligonucleotide in solution. Libraries LMR and HMR were purified on 15% polyacrylamide gel containing 8 M urea, and recovered in deionized water after n-butanol desalting.
The mutagenesis rate—taken as the fraction of trimer incorporation—per wild-type codon was determined from the absorbance difference between the DMTr cation released from the previous nucleotide to the mutagenized codon and the DMTr-monomer phosphoramidite incorporated after addition of the trimers. For libraries LMR and HMR, the average mutagenesis rate was 0.21 and 0.69, respectively.
Biological assessment
Kanamycin (KAN) was bought from Sigma Chemical Co. Restriction endonucleases, T4 DNA ligase, Klenow DNA polymerase and deoxynucleoside-triphosphates (dNTPs) were purchased from New England Biolabs, and used according to standard protocols. Plasmid pT4BlaSac (16) was the parent plasmid of libraries LMR and HMR. It contains the TEM-1 β-lactamase gene (including its original promoter), a KAN-resistance gene, and the ColE1 and f1 origins of DNA replication within 3.2 kb. This plasmid contains the restriction sites SacI (496-gagctc-501) and PstI (541-ctgcag-546) within the blaTEM-1 gene. E.coli XL1-Blue [recA1, endA1, gyrA96, thi-1, hsdR17, supE44, relA1, lac, {F′ proAB, lacIq lacZΔM15, Tn 10(tetr)}] was obtained from Stratagene, Inc. and used for cloning and plasmid production.
Cloning of mutant cassettes
Duplexes of libraries LMR and HMR were generated by extension of the partially complementary primer 5′-tgccattgcaggcatcgtggtatc-3′, using the Klenow fragment (3′→5′ exo−) of DNA polymerase I as described earlier (13,17). Complementary primer was designed with a PstI restriction site to generate mutant cassettes containing SacI/PstI sites at the ends. The products were double digested with both restriction enzymes and cloned into the pT4BlaSac plasmid. The resultant recombinant plasmids were electroporated into XL1-Blue cells, and a part of the electroporation mixture was plated on LB agar containing either KAN at 25 μg/ml or on selective plates containing KAN plus ceftazidime at 0.5 μg/ml. The plates were incubated overnight at 37°C, and several clones per library were randomly sequenced to analyze the frequency and distribution of mutations.
Antibiotic susceptibility
Five microliters of a diluted (10−5-fold) overnight LB culture of XL1-blue harboring either each ceftazidime-resistant plasmid or the wild-type plasmid was spotted onto LB kanamycin plates containing increasing concentrations of ampicillin or ceftazidime. The minimum concentration of antibiotic completely inhibiting the growth of cells was taken as the MIC.
RESULTS
Chemical synthesis
Out of 61 sense codons that make up the genetic code, we selected 20 trimers—avoiding those poorly expressed in E.coli—to be synthesized as Fmoc-trinucleotide phosphoramidites. The Fmoc-trinucleotide phosphoramidites and the encoded amino acids are shown in Figure 2. Notice the kind of protection for the internucleotidic phosphates; in contrast to the protecting groups—methyl (10,18), 2-cyanoethyl (11) and 2-chlorophenyl (12,19)—commonly used in the preparation of dinucleotide and trinucleotide phosphoramidites, we decided to use the ethyl group (12,16,19,20). The reason is that methyl, 2-cyanoethyl and 2-chlorophenyl are too labile to nucleophiles, labile to alkali or few stable as phosphitylating reagent. In contrast, the ethyl group is very stable to base, is removed only with powerful nucleophiles and gives rise to very stable phosphitylating reagents (16). Thus, no special care must be taken during the reaction process and intermediate workup on the preparation of the precursor synthons (Figure 1). Synthesis of Fmoc-trinucleotides was accomplished in a 3′ to 5′ direction, first synthesizing dinucleotides whose 3′ hydroxyl was protected with the DMTr group and then preparing the trinucleotides whose 5′ hydroxyl was protected with the Fmoc group. Each dinucleotide and trinucleotide was synthesized in two stages. In the first stage, fully protected dinucleotide and trinucleotide were obtained through a ‘one pot’ reaction where activation of the 3′ nucleoside, coupling with the 5′ nucleoside and oxidation of the internucleotidic phosphite, took place without isolation of any intermediate material. After this stage, the fully protected product—either dinucleotide or trinucleotide—was purified by chromatography to remove the excess of 5′Fmoc-monomer and avoid contamination of the final product. In the second stage for dinucleotides, the Fmoc group was removed by alkali treatment and re-purified by flash column-chromatography to remove any non-polar material that would have co-eluted in the first purification. In the second stage for trinucleotides, the DMTr group was removed by acid treatment and re-purified to ensure the exclusive presence of the expected material.
As shown in Figure 2, we decided to prepare 2-cyanoethyl phosphoramidites instead of ethyl phosphoramidites of the Fmoc-trinucleotides to enable their chromatographic purification. 2-cyanoethyl phosphoramidites are less reactive and therefore more stable to purification conditions than alkyl phosphoramidites (21). Purity of Fmoc-trinucleotide phosphoramidites was determined by reverse-phase HPLC, and it was higher than 80% in all cases. H-phosphonate was the major subproduct found in each sample as a result of the intrinsic environmental humidity. This subproduct reduces the purity of the expected phosphoramidites, although it is innocuous for the coupling reaction. When synthesizing monomer, dimer and trimer phosphoramidites bearing DMTr protection, H-phosphonates are mainly avoided by the use of high triethylamine proportions during workup and purification, however for Fmoc-containing compounds this reagent is deleterious to stability.
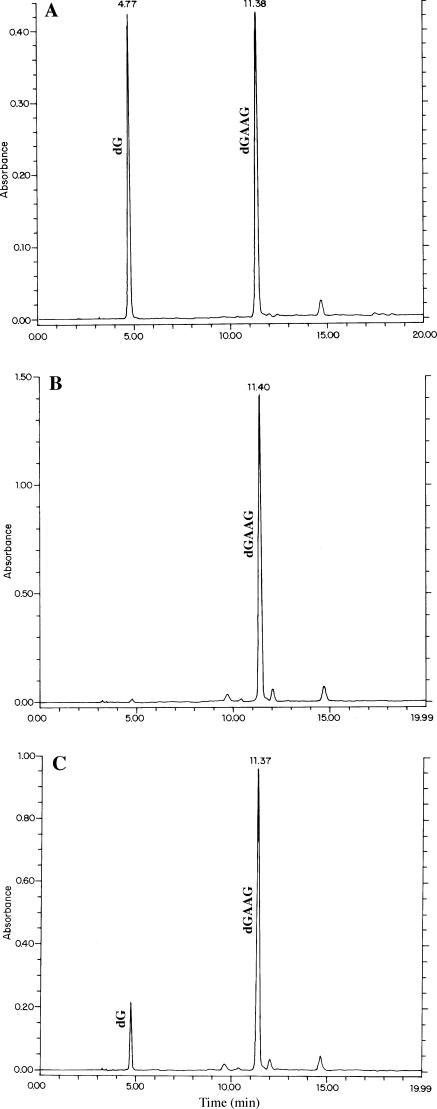
Testing of each Fmoc-trinucleotide phosphoramidite for solid-phase synthesis was performed by addition to a deoxynucleoside-bound support. As controls, the resulting tetramers and standards—assembled by ordinary methods—were fully deprotected and HPLC co-injected, giving rise to only one peak corresponding to the expected tetramer, as exemplified in Figure 3 for the missing Fmoc-GAA phosphoramidite. Additional confirmation of sequence for the other trimers was obtained after their incorporation into oligonucleotide libraries and sequencing several clones (see below).
Figure 3.
HPLC analysis of the fully deprotected tetranucleotide GAAG. (A) Assembled by incorporation of the Fmoc-trinucleotide phosphoramidite GAA to dG-bound support. (B) Standard assembled by ordinary mononucleotide chemistry. (C) Co-injection of samples shown in (A) and (B).
Mutagenesis
In a previous paper (13), using two trinucleotides independently, we demonstrated that a combination of Fmoc-trinucleotide phosphoramidites and ordinary DMTr-monomer phosphoramidites during oligonucleotide synthesis affords semi-automation of a novel codon-based mutagenesis method (Figure 4). In this strategy, mutant codons are assembled with Fmoc-protected trinucleotide phosphoramidites whereas wild-type codons are assembled with DMTr-protected monomers. This approach goes beyond most methods of mutagenesis based on the use of DMTr-trinucleotides (10–12,19) and/or NNG/C degenerate oligonucleotides (7), as it is able to create oligonucleotide variants carrying few codon replacements when a region comprising several codons is explored. Furthermore, we demonstrated that the ‘Fmoc-trinucleotide approach’ follows a combinatorial codon replacement pattern that obeys a binomial distribution (3). By varying the mutagenesis rate, libraries ranging from one to several amino acid replacements per gene can be preferentially generated.
To extend the Fmoc-trinucleotide approach to the other amino acids and demonstrate the utility of the trinucleotides synthesized here, we decided to randomize nine codons that encode amino acids 170–178 of the bacterial enzyme TEM-1 β-lactamase. This region was selected because it is close to the natural PstI restriction site located in the TEM-1 β-lactamase gen (blaTEM-1) and to a putative SacI site. PstI is 13 bp downstream of the codon for Arg-178. The SacI restriction site was created by a single base substitution 501G>T in blaTEM-1, located two codons upstream from that of Asn-170. The blaTEM-1 gene containing both restriction sites was cloned into the plasmid pT4Bla to use it in subsequent cloning experiments.
To better assess the method, we assembled two oligonucleotide libraries at different mutagenesis rates. Theoretically, one library should produce an average of two amino acid replacements per variant and the other an average of six amino acid replacements per variant. The assembled sequence was accggagctc-AAT/XXX-GAA/XXX-GCC/XXX-ATA/XXX-CCA/XXX-AAC/XXX-GAC/XXX-GAG/XXX-CGT/XXX-gataccacgatgcctgc, where lower-case letters represent the 5′ and 3′ flanking regions of the mutated segment. The codons submitted to partial substitution are denoted by capital letters whereas XXX represents the mixture of trinucleotides incorporated along the assembly. The first library—termed Low Mutagenesis Rate or LMR—was mutated through the repetitive incorporation of an equimolar 6 mM solution composed of 19 trimers (the trimer GAA was mistakenly omitted), giving an average mutagenesis rate of 0.21. For the second library—termed High Mutagenesis Rate or HMR—a 24 mM mixture was employed, giving an average mutagenesis rate of 0.69. As an improvement to the original reported method, libraries LMR and HMR were assembled in a completely automated mode using an appropriate DNA synthesizer implemented with additional ancillary bottles (see Supplementary Material for the equipment and program used).
Oligonucleotides were de-ethylated in their support of synthesis before complete deprotection with ammonium hydroxide was performed. This two-step deprotection avoids cleavage of the sugar-phosphate backbone at the ethyl-phosphotriester positions. Oligonucleotides were purified by denaturing polyacrylamide gel and converted into mutant DNA duplexes by annealing and extending a partially complementary primer that contains the PstI restriction site. DNA extension was performed under mild conditions, using the large fragment (Klenow) of DNA polymerase I. Mutant duplexes were digested with restriction enzymes SacI/PstI and cloned as cassettes in the blaTEM-1 gene located in the pT4BlaSac plasmid. We prefer to clone mutagenic libraries as cassettes because incorporation by PCR may result on biases for those mutants that are more similar to the wild-type sequence (22). The cloning process left intact the kanamycin-resistance gene, and therefore plasmid DNA from several clones selected only on this antibiotic was isolated and sequenced to analyze the diversity of codons incorporated into the oligonucleotides. The sequencing results for both libraries are given in Table 1.
Table 1. Sequence of random clones isolated from libraries LMR and HMR without ceftazidime selection.
p represents the mutagenesis rate. Mutant codons are indicated in bold letters. Single nucleotide deletions are marked as asterisks. Unexpected mutant codons are highlighted as gray rectangles.
Once the successful incorporation of each trimer was confirmed, libraries LMR and HMR were subjected to ceftazidime-selection experiments to find out those residues in the region 170–178 of TEM-1 that are able to confer it a specificity change from ampicillin to ceftazidime. To avoid the recovery of either poor ceftazidime-hydrolyzing mutants or wild-type clones, plates containing 0.5 μg/ml of ceftazidime were employed for the selection. This concentration represents a 6.6-fold excess of the maximum concentration at which cells harboring wt TEM-1 β-lactamase are still able to grow.
Several colonies from both libraries grew, their plasmids were recovered and re-transformed to ensure that the resistant effect is caused by a mutant β-lactamase and not by another mechanism of resistance, as has been extensively addressed (23). After isolation of the re-transformed plasmids and DNA sequencing, the results were concentrated in Table 2.
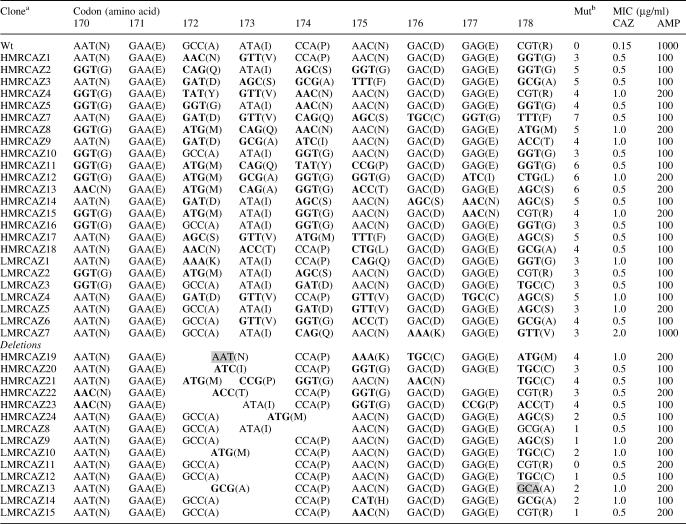
Table 2. Sequence of ceftazidime-resistant mutants generated with the oligonucleotide libraries LMR and HMR.
aThe acronym HMR and LMR for the ceftazidime-resistant clones indicates the library where they came from.
bMut indicates the number of substituted amino acids per variant. Unexpected mutant codons are highlighted as gray rectangles.
DISCUSSION
A careful analysis of the sequences reported in Tables 1 and 2 for non-selected and ceftazidime-selected clones respectively, allows a series of important remarks to be made with regard to the pool of Fmoc-trinucleotide phosphoramidites and their application on the creation of protein libraries, as well as their successful application in changing the substrate specificity of TEM-1 β-lactamase.
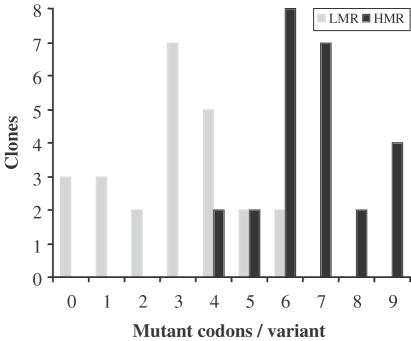
Under non-selecting conditions (Table 1), most mutants found in library LMR carry few codon replacements, an average of 2.9 per variant, whereas mutants in library HMR carry many substitutions, averaging 6.7 per variant. The results agree well with the theoretically expected results of 1.9 and 6.2 average codons per variant, respectively, as calculated by a binomial distribution, using the experimental mutagenesis rates determined during the oligonucleotide assembly (0.21 for library LMR and 0.69 for HMR). The small difference found between the expected and the experimental results clearly indicates the mutagenesis rates are being lightly underestimated, although it does not represent a serious problem. Figure 5 illustrates that the Fmoc-trinucleotide approach incorporates mutant codons in a combinatorial mode, obeying the binomial distribution. From these results, it is concluded that an appropriate selection of the mutagenesis rate can preferentially produce mutants bearing any desired multiplicity.
Figure 5.
Frequency of mutations generated in libraries LMR and HMR without ceftazidime-selection.
The mutations were distributed throughout the region, interspersing wild-type and mutant codons as expected (e.g. mutants LMR1, HMR1, etc.). To our knowledge, only the resin-splitting (24) method and the Fmoc-based approaches (13,16,17) are able to generate these types of variants. However the resin-splitting method is labor-intensive, impractical to automate, deleterious to the overall yield and extremely difficult to control at low mutagenesis rates, whereas the method we are reporting is completely suited for automation and avoids codon redundancy and stop codons.
Under non-selecting conditions, all 19 mutant codons occurred in library HMR, and TAT was the only missing codon in library LMR (Table 3). Although equimolar solutions of the Fmoc-trinucleotide phosphoramidites were used, an uneven representation of codons was obtained, considering both libraries (Table 3). Based on this representation, trimers could be grouped into three categories: ‘highly reactive’, those that occurred >15 times such as AGC, ATC, ATG and GGT; ‘reactive’, those that occurred between 10 and 15 times such as AAA, AAC, ACC, CAG, CTG, GAT, GCG, GTT, TGC, TGG and TTT; and ‘poorly reactive’, those that occurred <10 times, such as CAT, CCG, CGC and TAT. These results correlate with the difference in reactivity found in Fmoc-dinucleotide (16) and DMTr-trinucleotide phosphoramidites previously reported (10,11). Because of this reactivity difference, the concentration of the trimers might be empirically adjusted to generate equal representation of each amino acid.
Table 3. Incorporation of Fmoc-trinucleotide phosphoramidites in libraries LMR and HMR without ceftazidime selection.
| Codon | LMR occurrence | HMR occurrence | Total occurrence |
|---|---|---|---|
| AAA | 3 | 11 | 14 |
| AAC | 2 | 12 | 14 |
| ACC | 1 | 12 | 13 |
| AGC | 7 | 11 | 18 |
| ATC | 6 | 10 | 16 |
| ATG | 4 | 15 | 19 |
| CAG | 4 | 8 | 12 |
| CAT | 2 | 6 | 8 |
| CCG | 1 | 3 | 4 |
| CGC | 4 | 3 | 7 |
| CTG | 3 | 8 | 11 |
| GAT | 4 | 7 | 11 |
| GCG | 2 | 11 | 13 |
| GGT | 5 | 15 | 20 |
| GTT | 4 | 11 | 15 |
| TAT | 0 | 8 | 8 |
| TGC | 2 | 9 | 11 |
| TGG | 5 | 5 | 10 |
| TTT | 4 | 6 | 10 |
Codon GAA was mistakenly omitted from both libraries.
Some 6 of the 24 sequenced clones of library LMR (LMR18-23) and 4 of the 25 clones of library HMR (HMR21-24) displayed single nucleotide deletions. Because all deletions were found within wild-type codons, and wild-type codons were assembled using ordinary DMTr-monomer chemistry, these point deletions might have arisen from incomplete capping during the assembly. Deletions could be reduced increasing either the capping time per cycle of synthesis or the concentration of the capping reagent. Although frameshift mutations reduce the number of useful proteins, the rate generated by the Fmoc-trinucleotide approach is minimal as compared to other mutagenesis methods (25).
Of the 235 mutant codons expressed in both libraries grown under non-selecting conditions, only 5 were unpredicted. This is a low ratio of unexpected mutations, generated perhaps by modifications of bases as a consequence of the numerous chemical treatments to which each trinucleotide is exposed during all the process. For instance, codon CGT of clone LMR19 could have arisen from deamination of the 3′ terminal base of codon CGC.
Whereas library HMR yielded mutants averaging 6 and 7 codon replacements under non-selecting conditions, under ceftzidime selection this average drops to 4 codons. This result clearly reinforces the hypothesis that many amino acid substitutions in a short protein region are commonly deleterious for a target function, hence justifying our proposed non-saturating mutagenesis method.
On both experiments with and without ceftazidime selection, library LMR yielded mutants preferentially bearing three codon replacements. This result correlates well with the hypothesis referred in the previous paragraph. Because the frequency of multiple mutants in library LMR is low, the frequency of triple mutants under ceftazidime selection remains the same.
Palzkill and co-workers (26,27) have extensively studied the identification of residues responsible to alter the substrate specificity in the N- and C-terminal regions of the TEM-1 β-lactamase active site omega loop by randomizing three or four contiguous residues at saturation. The main conclusion of their studies is that several substitutions in both regions are responsible to alter the ceftazidime hydrolytic activity without any specific pattern. In the present work, we found that no substitution, among the members of the functional variant group (Table 2), is allowed at position Glu-171. Positions Asp-176 and Glu-177 were also rarely substituted; only 5 out of 40 functional variants allowed a substitution at position 176. Glycine was the only amino acid able to substitute for the asparagine at position 170, whereas residues Ala-172, Ile-173, Pro-174, Asn-175 and Arg-178 were freely substituted. Our results agree with the previous proposals that removal of some intramolecular interactions in the omega loop (in this case, the salt bridge formed between Asp-176 and Arg-178) could allow more room in the active site for a better fit of the bulky ceftazidime side-chain (26). However, no extra disruptions inside the omega loop region (as the removal of the ionic bond between positions 171 and 179) were allowed, perhaps, because this could produce very unstable variants that are unable to pass the selection step. The different amino acid sequence requirements at some positions between our variants and Palzkill's, could be simply due to a poorer protein expression level in our system as compared to the one used by Palzkill and co-workers. This difference could result in a better selection dynamic range in their system as compared to ours. We suggest that due to the poor expression level of our variants, the selection step is picking up variants not too damaged in the activity or in the stability of the protein. This will explain the restricted variability at position 170 for a residue (glycine) naturally observed to substitute for the asparagine at this position (28), and will also explain that a simple repositioning of the positive charge from residue 178 to residue 176 (variant LMRCAZ7) could produce a variant able to reach wild-type resistance levels for ampicillin together with a broader substrate specificity. We expect this variant might satisfy the hydrogen bonding capabilities of the lysine side-chain by forming a new salt bridge (perhaps with the neighboring acidic residue; a rotation of the Lys-178 side-chain to the protein surface will be needed) or by keeping the contact already made by the former aspartate residue with a water molecule in the inside of the protein core helping to neutralize the new positive charge.
An important number of functional mutants (36% of the total number of variants) showed combinations of substitutions and single codon deletions. The deletions were mostly concentrated at positions 172 or 173, and the substitutions mainly targeted at position 178. From a chemical point of view, those deletions (mutants HMRCAZ19, HMRCAZ20, HMRCAZ21, HMRCAZ22, HMRCAZ24, LMRCAZ10 and LMRCAZ13) where two wild-type codons were replaced by a mutant trimer are easily explained by incomplete removal of the Fmoc group. Hence a trimer added in one cycle, but not deprotected, avoided incorporation of the following trimer or wt codon. Successful Fmoc removal in the next cycle fixed the single deletion. However, those mutations bearing a deletion between two wild-type codons (mutants LMRCAZ8, LMRCAZ9, LMRCAZ11, LMRCAZ12, LMRCAZ14 and LMRCAZ15) can only be explained by deacetylation of truncated sequences as a consequence of the alkali DBU treatment. Encouraged by this finding and another previous reported deletion where residues Glu-240 and Arg-241 were substituted by alanine, have made us to propose a new method of protein evolution based exclusively on codon deletions randomly distributed in some parts of an enzyme active site (29).
In conclusion, we have synthesized a complete set of Fmoc-protected trinucleotide phosphoramidites by a straightforward procedure, in which the key step was the ‘one pot’ preparation of fully protected dimers and trimers whose internucleotidic phosphates were protected with the ethyl group. Also, we have demonstrated that these Fmoc-protected trinucleotide phosphoramidites can be combined with DMTr-protected monomers, during oligonucleotide synthesis, to afford unique libraries in which mutant and wild-type codons are interspersed throughout a long gene segment, in contrast to ordinary degenerate oligonucleotides. Finally, using these compounds for the process of mutagenesis, we achieved a specific change from ampicillin—the original substrate—towards ceftazidime, a third generation cephalosporine in the TEM-1 β-lactamase used as test case.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Eugenio López, Santiago Becerra, Leandro Ordóñez, Timoteo Olamendi and René Hernández for excellent technical assistance. We are also indebted to Dr Hugh Mackie from Glen Research for careful revision of the manuscript, and to Dr Alejandro Garciarrubio for providing us the minimal list of trimers required to complete a set of codons and a set of anticodons (to be described somewhere else). J.O. acknowledges financial support by DGAPA-UNAM (grant IN-214803).
REFERENCES
- 1.Cadwell R.C. and Joyce,G.F. (1992) Randomization of genes by PCR mutagenesis. PCR Methods Appl., 2, 28–33. [DOI] [PubMed] [Google Scholar]
- 2.Stemmer W.P. (1994) DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl Acad. Sci. USA, 91, 10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sirotkin K. (1986) Advantages to mutagenesis techniques generating populations containing the complete spectrum of single codon changes. J. Theor. Biol., 123, 261–279. [DOI] [PubMed] [Google Scholar]
- 4.Hutchison C.A. III, Nordeen,S.K., Vogt,K. and Edgell,M.H. (1986) A complete library of point substitution mutations in the glucocorticoid response element of mouse mammary tumor virus. Proc. Natl Acad. Sci. USA, 83, 710–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derbyshire K.M., Salvo,J.J. and Grindley,N.D.F. (1986) A simple and efficient procedure for saturation mutagenesis using mixed oligodeoxynucleotides. Gene, 46, 145–152. [DOI] [PubMed] [Google Scholar]
- 6.Miyazaki K. and Arnold,F.H. (1999) Exploring nonnatural evolutionary pathways by saturation mutagenesis: rapid improvement of protein function. J. Mol. Evol., 49, 716–720. [DOI] [PubMed] [Google Scholar]
- 7.del Rio G., Osuna,J. and Soberón,X. (1994) Combinatorial libraries of proteins: analysis of efficiency of mutagenesis techniques. Biotechniques, 17, 1132–1139. [PubMed] [Google Scholar]
- 8.Huang W., Petrosino,J., Hirsch,M., Shenkin,P.S. and Palzkill,T. (1996) Amino acid sequence determinants of beta-lactamase structure and activity. J. Mol. Biol., 258, 688–703. [DOI] [PubMed] [Google Scholar]
- 9.Hughes M.D., Nagel,D.A., Santos,A.F., Sutherland,A.J. and Hine,A.V. (2003) Removing the redundancy from randomised gene libraries. J. Mol. Biol., 331, 973–979. [DOI] [PubMed] [Google Scholar]
- 10.Virnekas B., Ge,L., Pluckthun,A., Schneider,K.C., Wellnhofer,G. and Moroney,S.E. (1994) Trinucleotide phosphoramidites: ideal reagents for the synthesis of mixed oligonucleotides for random mutagenesis. Nucleic Acids Res., 22, 5600–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyttle M.H., Napolitano,E.W., Calio,B.L. and Kauvar,L.M. (1995) Mutagenesis using trinucleotide beta-cyanoethyl phosphoramidites. Biotechniques, 19, 274–281. [PubMed] [Google Scholar]
- 12.Ono A., Matsuda,A., Zhao,J. and Santi,D.V. (1995) The synthesis of blocked triplet-phosphoramidites and their use in mutagenesis. Nucleic Acids Res., 23, 4677–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaytán P., Yáñez,J., Sanchez,F., Mackie,H. and Soberón,X. (1998) Combination of DMT-mononucleotide and Fmoc-trinucleotide phosphoramidites in oligonucleotide synthesis affords an automatable codon-level mutagenesis method. Chem. Biol., 5, 519–527. [DOI] [PubMed] [Google Scholar]
- 14.Lehmann C., Xu,Y.Z., Christodoulou,C., Tan,Z.K. and Gait,M.J. (1989) Solid-phase synthesis of oligoribonucleotides using 9-fluorenylmethoxycarbonyl (Fmoc) for 5′-hydroxyl protection. Nucleic Acids Res., 17, 2379–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atkinson T. and Smith,M. (1984) Solid-phase synthesis of oligodeoxyribonucleotides by the phosphite-triester method. In Gait,M.J. (ed.), Oligonucleotide Synthesis: A Practical Approach. IRL Press, Oxford, pp. 35–81. [Google Scholar]
- 16.Gaytán P., Osuna,J. and Soberón,X. (2002) Novel ceftazidime-resistance beta-lactamases generated by a codon-based mutagenesis method and selection. Nucleic Acids Res., 30, e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaytán P., Yáñez,J., Sanchez,F. and Soberón,X. (2001) Orthogonal combinatorial mutagenesis: a codon-level combinatorial mutagenesis method useful for low multiplicity and amino acid-scanning protocols. Nucleic Acids Res., 29, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar G. and Poonian,M.S. (1984) Improvements in oligodeoxyribonucleotide synthesis: methyl N,N-dialkylphosphoramidite dimer units for solid support phosphite methodology. J. Org. Chem., 49, 4905–4912. [Google Scholar]
- 19.Kayushin A.L., Korosteleva,M.D., Miroshnikov,A.I., Kosch,W., Zubov,D. and Piel,N. (1996) A convenient approach to the synthesis of trinucleotide phosphoramidites—synthons for the generation of oligonucleotide/peptide libraries. Nucleic Acids Res., 24, 3748–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallo K.A., Shao,K.L., Phillips,L.R., Regan,J.B., Koziolkiewicz,M., Uznanski,B., Stec,W.J. and Zon,G. (1986) Alkyl phosphotriester modified oligodeoxyribonucleotides. V. Synthesis and absolute configuration of Rp and Sp diastereomers of an ethyl phosphotriester (Et) modified EcoRI recognition sequence, d[GGAA(Et)TTCC]. A synthetic approach to regio- and stereospecific ethylation-interference studies. Nucleic Acids Res., 14, 7405–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinha N.D., Biernat,J., McManus,J. and Koster,H. (1984) Polymer support oligonucleotide synthesis XVIII: use of beta-cyanoethyl-N,N-dialkylamino-/N-morpholino phosphoramidite of deoxynucleosides for the synthesis of DNA fragments simplifying deprotection and isolation of the final product. Nucleic Acids Res., 12, 4539–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward B. and Juehne,T. (1998) Combinatorial library diversity: probability assessment of library populations. Nucleic Acids Res., 26, 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frere J.M., Joris,B., Granier,B., Matagne,A., Jacob,F. and Bourguignon-Bellefroid,C. (1991) Diversity of the mechanisms of resistance to beta-lactam antibiotics. Res. Microbiol., 142, 705–710. [DOI] [PubMed] [Google Scholar]
- 24.Glaser S.M., Yelton,D.E. and Huse,W.D. (1992) Antibody engineering by codon-based mutagenesis in a filamentous phage vector system. J. Immunol., 149, 3903–3913. [PubMed] [Google Scholar]
- 25.Murakami H., Hohsaka,T. and Sisido,M. (2002) Random insertion and deletion of arbitrary number of bases for codon-based random mutation of DNAs. Nat. Biotechnol., 20, 76–81. [DOI] [PubMed] [Google Scholar]
- 26.Petrosino J.F. and Palzkill,T. (1996) Systematic mutagenesis of the active site omega loop of TEM-1 beta-lactamase. J. Bacteriol., 178, 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palzkill T., Le,Q.Q., Venkatachalam,K.V., LaRocco,M. and Ocera,H. (1994) Evolution of antibiotic resistance: several different amino acid substitutions in an active site loop alter the substrate profile of beta-lactamase. Mol. Microbiol., 12, 217–229. [DOI] [PubMed] [Google Scholar]
- 28.Ambler R.P., Coulson,A.F., Frere,J.M., Ghuysen,J.M., Joris,B., Forsman,M., Levesque,R.C., Tiraby,G. and Waley,S.G. (1991) A standard numbering scheme for the class A beta-lactamases. Biochem. J., 276, 269–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osuna J., Yanez,J., Soberon,Y. and Gaytán,P. (2004) Protein evolution by codon-based random deletions. Nucleic Acids Res., 32, e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.