Abstract
As a growing number of complementary transcripts, susceptible to exert various regulatory functions, are being found in eukaryotes, high throughput analytical methods are needed to investigate their expression in multiple biological samples. Serial Analysis of Gene Expression (SAGE), based on the enumeration of directionally reliable short cDNA sequences (tags), is capable of revealing antisense transcripts. We initially detected them by observing tags that mapped on to the reverse complement of known mRNAs. The presence of such tags in individual SAGE libraries suggested that SAGE datasets contain latent information on antisense transcripts. We raised a collection of virtual tags for mining these data. Tag pairs were assembled by searching for complementarities between 24-nt long sequences centered on the potential SAGE-anchoring sites of well-annotated human expressed sequences. An analysis of their presence in a large collection of published SAGE libraries revealed transcripts expressed at high levels from both strands of two adjacent, oppositely oriented, transcription units. In other cases, the respective transcripts of such cis-oriented genes displayed a mutually exclusive expression pattern or were co-expressed in a small number of libraries. Other tag pairs revealed overlapping transcripts of trans-encoded unique genes. Finally, we isolated a group of tags shared by multiple transcripts. Most of them mapped on to retroelements, essentially represented in humans by Alu sequences inserted in opposite orientations in the 3′UTR of otherwise different mRNAs. Registering these tags in separate files makes possible computational searches focused on unique sense–antisense pairs. The method developed in the present work shows that SAGE datasets constitute a major resource of rapidly investigating with high sensitivity the expression of antisense transcripts, so that a single tag may be detected in one library when screening a large number of biological samples.
INTRODUCTION
Antisense transcripts, implicated in various regulatory mechanisms, were initially described in individual gene studies (1). With the progress of genome annotation, an increasing number of gene pairs, potentially capable of encoding complementary transcripts, were reported in eukaryotes. Specifically designed computational search led to the detection of nearly 1600 cis-encoded gene pair candidates in the human genome (2–4). Progress in genome annotation is expected to increase this number (5). In the mouse genome, 2481 pairs of overlapping genes were predicted using the FANTOM2 cDNA set (6). In order to ascertain the natural occurrence of functional double-stranded RNAs, their co-expression must be investigated at the transcriptome level. More than 4 million expressed sequence tags (ESTs) have been sequenced in a wide diversity of cDNA libraries. Nevertheless, these data can hardly be used directly to study the expression of functionally significant complementary sequences (7). Many ESTs were not directionally cloned and even well-known mRNA sequences were registered from both strands of cloned cDNAs (8). There is therefore a need for new experimental datasets.
Large-scale expression profiling tools such as microarrays have been used to analyze the co-expression of potentially complementary transcripts (4,9) and a method dedicated to the direct identification of new overlapping mRNAs has recently been developed (10). Alternatively, Serial Analysis of Sage Expression (SAGE) offers specific advantages (11). SAGE is a high throughput method based on the extraction, amplification and sequencing of 14-nt cDNA tags (11). Directionally reliable tags are generated from a well-defined restriction site, usually CATG, at the 3′ end of each transcript. Following on from this anchoring site, they differ by a 10-nt sequence sufficient in most cases to identify corresponding mRNAs. Tags are randomly assembled as ditags to limit PCR quantitative biases, so as to amplify tag populations which reproduce the original mRNA distribution. Sequencing thousands of concatenated tags and enumerating the frequency of occurrence provide quantitative gene expression profiles. SAGE data are acquired independently of any previous knowledge of gene sequences and may even reveal new transcripts. This heuristic interest is reinforced by the fact that cumulative SAGE data can easily be merged, allowing large-scale comparisons between independently raised libraries, so that any tag sequence can be traced back in the whole collection of published libraries.
Tags matching mRNAs in antisense orientation were observed when SAGE was applied to Plasmodium falciparum (12,13). This phenomenon might initially be considered as a peculiar feature of the malarial parasite but it was also reported in Caenorhabditis elegans (14). When analyzing our human SAGE libraries (15), we found also such ‘reverse’ tags, the presence of which could not be explained merely by experimental artifacts. The alternative hypothesis was that they originate from natural antisense transcripts (NATs) with the corollary that large SAGE datasets, which are now being assembled (16), contain latent information on sense–antisense pairs. Based on this working hypothesis, we developed a strategy for the sensitive detection of antisense transcripts and studies of their expression by large-scale screening of SAGE data.
MATERIALS AND METHODS
RNA sources, SAGE libraries and detection of antisense transcripts
Total RNA was extracted with Trizol (Invitrogen, Cergy-Pontoise, France) from untreated U937 cells and from U937 cells treated for 48 h with Vitamin D and retinoids as previously described (15). Reticulocytes were purified as described (17,18) from fresh blood samples of 10 healthy adult volunteers after informed consent. The reticulocyte library was built as described for U937 cells, using Sau3A1 as anchoring enzyme. Before polymerase chain reaction (PCR), first-strand cDNAs were synthesized with Superscript II reverse transcriptase (Invitrogen) with primers that specifically hybridize to either antisense transcripts (AT) or sense transcripts (ST). All primers annealed within coding regions of the genes, resulting in 119–302-bp PCR products. RPL9 (NM_000661) (AT): 5′-CATGATCAAGGGTGTTACACTGG-3′, (ST): 5′-CCGCTGAATTTGAAACAAGC-3′, RPL27A (NM_000990) (AT): 5′-CCGGATCAACTTCGACAAATACC-3′, (ST): 5′-CAGCCCCAGTCTTGTTTTTAGC-3′, S100A6 (NM_014624) (AT): 5′-GACTGCGACATAGCCCATCC-3′, (ST): 5′-TGACATACTCCTGGAAGTTCACC-3′, NDRG1 (NM_006096) (AT): 5′-GGCAAGAGAGGCTGAGTACG-3′, (ST): 5′-TTTCCGCTGCAAAGTTACAA-3′, HBB (NM_000518) (AT): 5′-GATGCTCAAGGCCTTTCATA-3′, (ST): 5′-GCAACCTCAAACAGACACCA-3′. Control experiments without reverse transcription were performed to detect DNA contamination. PCR was performed using the same primers for sense or antisense cDNAs and amplicons were analyzed by electrophoresis on 1.5% agarose gels. The SAGE libraries used to illustrate Figures 1 and 3 are referenced in Supplementary Table 1 online.
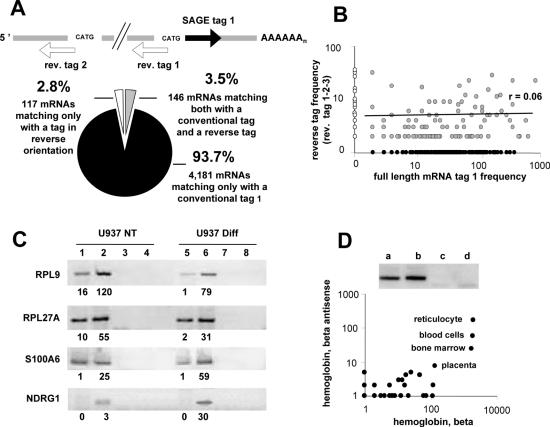
Figure 1.
Analysis of SAGE tags mapping to the reverse complement of known transcripts. (A) Conceptual scheme and tag distribution in a U937-cell library (15). (B) Scatter plot showing sense versus antisense frequencies; the regression coefficient was calculated for 146 duplexes (gray dots). (C) Strand-specific RT–PCR of selected transcripts using RNAs from untreated (NT) and differentiated (Diff) U937 cells prepared as for SAGE libraries (15). Tag frequencies are indicated below. First-strand cDNA synthesis initiated with antisense (lanes 1 and 5) and sense-specific primers (lanes 2 and 6). Controls without reverse transcriptase for DNA contamination (lanes 3, 4, 7 and 8). (D) Strand-specific RT–PCR of beta-globin (HBB) mRNA antisense (lane a), sense (lane b) and controls without reverse transcriptase (lanes c and d) using reticulocytes RNA. Scatter plot of globin tag levels in the reticulocyte library, compared to levels in 260 SAGE libraries.
Figure 3.
Comparative expression levels of SAGE tags mapping on to transcripts sharing complementary J-tags. Each dot corresponds to one SAGE library. (A) Expression levels of selected pairs in 260 SAGE libraries for cis-encoded, widely expressed genes. (B) Tissue-specific cis-encoded genes. (C) Tissue-specific trans-encoded genes including pseudogenes expressed in inverted orientation. (D) Co-expression of sense and antisense TNNI3 transcripts. (E) Alu elements inserted in the 3′UTR of multiple mRNAs.
Tag-to-gene mapping
Identification of regular SAGE tags was performed as previously described (15). The original Preditag® software (Skuld-Tech, Montpellier, France) used to predict the conventional SAGE tags was modified to register virtual tags matching with the reverse complement of the sequences.
Collection of J-tag sequences
UniGene unique human sequences (Hs.seq.uniq.Z, release # 162) were retrieved from NCBI (ftp://ftp.ncbi.nih.gov/repository/UniGene/). Information concerning UniGene clusters (Hs.seq.uniq and Hs.data file) were parsed and registered in a relational database collecting tag sequences and related data. Parsers were written in C on a Unix operated Workstation. Tables of virtual tags were managed using Microsoft Access functions. J-tag sequences resulting from the flow chart in Figure 2 are available as Supplementary Material online. Assignment of transcripts to chromosomal loci was performed using Map Viewer (http://www.ncbi.nih.gov/mapview/) and repeated sequences were retrieved from Repbase (http://www.girinst.org/).
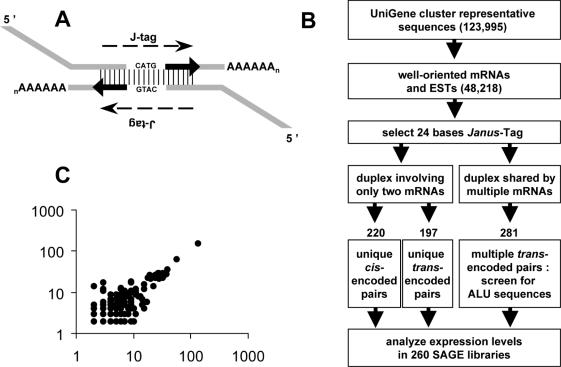
Figure 2.
Search for complementary mRNAs pairs in the UniGene collection of cluster-representative sequences. (A) Illustration of the ‘Janus’ tags concept. (B) Flow chart and results of the selection. (C) Numbers of well-oriented transcripts (from UniGene) sharing an Alu-matching J-tag in sense (x-axis) and antisense (y-axis) orientation.
SAGE library collections
We collected more than 13 million experimental tags from 260 publicly available human SAGE libraries retrieved from http://www.ncbi.nlm.nih.gov/SAGE/, ftp://ftp.ncbi.nih.gov/pub/sage/seq/ and http://www.prevent.m.u-tokyo.ac.jp/SAGE.html. These data were assembled to build a matrix giving the expression levels of 618 750 unique tags.
RESULTS
Analysis of SAGE tags mapping on to the reverse complement of known transcripts
When investigating unmatched tags revealed by SAGE in human U937 cells (15), we found tags matching with the reverse complement of mRNAs, either on the conventional 3′ site or on more internal sites (Figure 1A). Regular and reverse tags were simultaneously observed for 146 mRNAs. Such data might suggest a technical artifact: following digestion of cDNAs by the endonuclease creating SAGE anchoring sites, tags are extracted from the most 3′ end fragment, but, if incompletely removed, the other fragments could generate oppositely orientated tags. A statistical correlation would then be observed between regular and reverse tag frequencies. We did not find this correlation (Figure 1B), while orientation-specific RT-PCR revealed natural antisense transcripts (Figure 1C). We found also beta-globin (HBB) reverse tags in a SAGE study of reticulocytes and here again RT–PCR revealed antisense transcripts (Figure 1D). Screening other libraries showed globin mRNA reverse tags essentially in samples which might contain reticulocytes (unfractionated leucocytes, bone marrow, placenta). These data were corroborated by the description of HBB antisense RNA in mouse erythroid tissues (19) and by the recent study of a new form of thalassemia involving the antisense transcription of the alpha-globin (HBA) locus (20). Taken together, these data favored a natural origin for the reverse tags.
Collection of virtual SAGE tags capable of mapping on to complementary mRNA pairs
To test this hypothesis on a large scale, we needed to raise a collection of reliable pairs of complementary sequences, together with their virtual SAGE tags. Our strategy was to extract 24-nt sequences, including 10 nt on each side of potential anchoring sites (Figure 2A) and to search for complementary pairs within this collection of virtual ‘Janus’ tags (J-tags). Human transcript sequences registered in databanks range from well-annotated mRNAs to poorly characterized ESTs. For this study, we selected only sequences with at least 8A at their 3′ end and/or a polyadenylation signal in the last 300 nt and, because the anchoring sites of overlapping transcripts can be distant, we scanned six consecutive sites starting from the 3′ ends. We obtained 698 pairs of complementary J-tags by applying this strategy to 48 218 well-oriented sequences representative of UniGene clusters retrieved from NCBI (Figure 2B). Of 281 pairs displaying multiple matches (see Supplementary Table 2 online), several were discarded as being remnants of cloning vectors, and 73% were identified as Alu sequence fragments by using RepBase Update (21) (http://www.girinst.org/). It is noteworthy that all Alu-matching J-tags were found in both orientations in UniGene transcripts (Figure 2C).
For 417 pairs, each member referred to a unique UniGene cluster. Analysing their chromosomal location showed 197 pairs mapping in trans on remote loci and 220 pairs in cis on contiguous, oppositely oriented genes (see Supplementary Tables 3 and 4). A similar search of J-tags in previous collections of cis-oriented genes (3,4), yielding 232 pairs, showed that 198 pairs retrieved from UniGene were new. Merging these lists thus provided 430 pairs of cis-oriented genes (see Supplementary Table 5).
Mining SAGE datasets for expression of antisense transcripts
Having raised these lists of putative pairs of tags, we drew scatter plots to visualize their co-expression in a collection of 260 published SAGE libraries. Selected cases are illustrated in Figure 3 and more examples are provided in the Supplementary Table 6 online. This analysis revealed sense–antisense pairs proceeding either from cis-oriented genes or remote loci. The first row (Figure 3A) shows pairs of tags matching with cis-encoded, widely expressed genes, which may be involved in cell growth control, with both partners co-expressed in most libraries. Other tags from cis-encoded genes were more tissue-specific, with co-expression restricted to a small subset of libraries (Figure 3B). The third row shows examples of trans-encoded genes (Figure 3C). This category includes retrotransposed elements such as pseudogenes expressed in inverted orientation (22). As a whole, these data show the efficiency of computational searches in SAGE datasets for assessing the expression of new sense–antisense candidates. In addition to this strictly in silico approach, mining SAGE data was also efficient for studying the expression of antisense transcripts previously detected by experiment. The case of cardiac troponin I (TNNI3) illustrates a situation in which the existence of an antisense transcript was initially detected by RT–PCR and northern blotting (23), while the lack of information in databanks precluded its detection by computational search. Having derived the putative antisense SAGE tag from the normal TNNI3 sequence, we successfully detected it in only one SAGE library prepared from heart tissue (Figure 3D). This result, corroborating the initial report of its presence into the myocardium, showed that the expression of the sense–antisense duplex was actually restricted to this tissue. The sensitive detection of sense–antisense pairs requires the registration in separate files of the subset of tags matching with multiple UniGene clusters. We found that 88% of tags shared by 10 or more clusters mapped on to Alu elements. Depending on the subtype of Alu sequence, they exhibited either similar frequencies in both orientations or a predominant expression in one sense, but numerous duplexes involving Alu repeats were present in all libraries (Figure 3E).
DISCUSSION
In the present work, we used two computational strategies for investigating the expression of antisense transcripts through analysis of large SAGE datasets. The first approach consisted in mapping unknown SAGE tags on to the reverse complement of well-annotated transcripts. Its main interest is to detect antisense transcripts de novo even when complementary sequences are absent in expression databanks but experimental confirmation is also required. The advantage of the second approach, using 24-nt J-tags to assemble pairs of conventional tags, is to reduce the statistical risks of dealing with fortuitous, biologically irrelevant duplexes, as previously calculated for LongSAGE 21-nt tags (24). Its limitation is that it allows to screen only transcripts for which sequence data are presently available. The two strategies will converge only when the genome is fully annotated and most of the transcripts identified, which is not the case at present: when selecting virtual tags of cis-oriented genes, we found different pairs of tags in three collections of expressed sequences and only a small number common to all of them. This small overlap illustrates the difficulty of extracting representative expressed sequences from >4 million ESTs of unequal quality (2): in some instances, RNAs and ESTs actually mapping on to cis-oriented genes were inadequately registered in a unique UniGene cluster; conversely, as polyadenylation on different sites generates transcripts of different lengths, the resulting ESTs, which are not easily recognized as belonging to the same transcriptional unit, were used to create distinct clusters (25,26). In spite of these problems, we selected a large number of candidates for mining SAGE data and we observed their co-expression in a diversity of cells and tissues. Understanding the biological implication of these novel findings will require experimental investigation in each particular case.
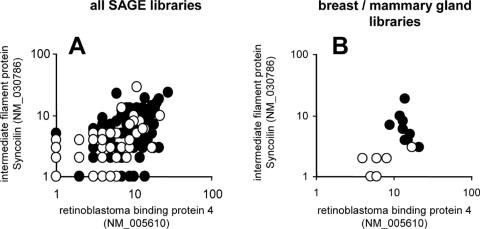
A major difficulty lies in the existence on interspersed repetitive elements, mostly represented in humans by Alu sequences, present in the 3′UTR of multiple mRNAs (27). Since they are inserted in both orientations, they can be associated in a large number of duplexes. In the present work, we calculated that Alu-matching J-tags could potentially form 27 215 combinations, contrasting with the 417 pairs of unique transcripts in the same UniGene built. Apart from the biological question of their existence and fate in vivo, abundant Alu duplexes may reduce the rate of identification of unique duplexes of similar size and stability when using experimental hybridization methods to capture dsRNAs in vitro (10). When mining SAGE data, this problem is solved in silico by sorting the tags mapping on repetitive elements and registering them in separate files. Computational searches can then be focused on unique sense–antisense pairs with high sensitivity, allowing the detection of a single tag in a unique library. Mining SAGE data then allows the analysis of co-expression of sense–antisense pairs of transcripts identified in individual case studies, with the unique opportunity to perform retrospective investigation of the whole collection of publicly available SAGE libraries. For example, the present work revealed that oppositely oriented SAGE tags mapping respectively on syncoilin (NM_030786) and retinoblastoma binding protein 4 (RBBP4, NM_005610) transcripts are co-expressed in numerous SAGE libraries (Figure 4A). Now, isolating the subset of libraries built from breast tissues reveals that breast tumors do not display the same ratio of sense–antisense transcripts as the corresponding normal cells (Figure 4B). It then becomes possible to investigate in more detail the value of this observation in terms of diagnosis or to analyze the underlying molecular mechanisms and the role of RBBP4 in the progression of the tumor. This kind of application will benefit from the increased availability of SAGE libraries built in parallel with normal tissue samples and their pathological counterparts.
Figure 4.
Comparative expression levels of SAGE tags mapping on to syncoilin and RBBP4 cis-encoded, oppositely oriented transcripts. White dots, normal tissue samples; black dots, cancer tissue samples. Comparative expression levels in 260 SAGE libraries (A) as in Figure 3, and (B) in libraries prepared from breast normal and cancer samples.
Up to now, the strategy described in the present work was limited by the unequal quality of EST collections. However, progress in genome annotation is providing an increasing number of candidates. A recent analysis of orientation-reliable transcripts shows that >20% of human transcripts have the potential to form sense–antisense pairs, now amenable to SAGE analysis of their expression (28).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Aurélie Baudet, Catherine Bisbal and Florence Ottones for their comments and insight concerning this manuscript. This work was supported by grants from the Centre National pour la Recherche Scientifique, Programme Bioinfo Inter-EPST and Ligue Nationale contre le Cancer.
REFERENCES
- 1.Vanhee-Brossollet C. and Vaquero,C. (1998) Do natural antisense transcripts make sense in eukaryotes? Gene, 211, 1–9. [DOI] [PubMed] [Google Scholar]
- 2.Shendure J. and Church,G.M. (2002) Computational discovery of sense-antisense transcription in the human and mouse genomes. Genome Biol., 3, RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehner B., Williams,G., Campbell,R.D. and Sanderson,C.M. (2002) Antisense transcripts in the human genome. Trends Genet., 18, 63–65. [DOI] [PubMed] [Google Scholar]
- 4.Yelin R., Dahary,D., Sorek,R., Levanon,E.Y., Goldstein,O., Shoshan,A., Diber,A., Biton,S., Tamir,Y., Khosravi,R. et al. (2003) Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol., 21, 379–386. [DOI] [PubMed] [Google Scholar]
- 5.Kampa D., Cheng,J., Kapranov,P., Yamanaka,M., Brubaker,S., Cawley,S., Drenkow,J., Piccolboni,A., Bekiranov,S., Helt,G. et al. (2004) Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res., 14, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiyosawa H., Yamanaka,I., Osato,N., Kondo,S. and Hayashizaki,Y. (2003) Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res., 13, 1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavorgna G., Dahary,D., Lehner,B., Sorek,R., Sanderson,C.M. and Casari,G. (2004) In search of antisense. Trends Biochem. Sci., 29, 88–94. [DOI] [PubMed] [Google Scholar]
- 8.Fahey M.E., Moore,T.F. and Higgins,D.G. (2002) Overlapping antisense transcription in the human genome. Comp. Functional Genomics, 3, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikaido I., Saito,C., Mizuno,Y., Meguro,M., Bono,H., Kadomura,M., Kono,T., Morris,G.A., Lyons,P.A., Oshimura,M. et al. (2003) Discovery of imprinted transcripts in the mouse transcriptome using large-scale expression profiling. Genome Res., 13, 1402–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosok O. and Sioud,M. (2004) Systematic identification of sense–antisense transcripts in mammalian cells. Nat. Biotechnol., 22, 104–108. [DOI] [PubMed] [Google Scholar]
- 11.Velculescu V.E., Zhang,L., Vogelstein,B. and Kinzler,K.W. (1995) Serial analysis of gene expression. Science, 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 12.Gunasekera A.M., Patankar,S., Schug,J., Eisen,G., Kissinger,J., Roos,D. and Wirth,D.F. (2004) Widespread distribution of antisense transcripts in the Plasmodium falciparum genome. Mol. Biochem. Parasitol., 136, 35–42. [DOI] [PubMed] [Google Scholar]
- 13.Patankar S., Munasinghe,A., Shoaibi,A., Cummings,L.M. and Wirth,D.F. (2001) Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of anti-sense transcripts in the malarial parasite. Mol. Biol. Cell, 12, 3114–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones S.J., Riddle,D.L., Pouzyrev,A.T., Velculescu,V.E., Hillier,L., Eddy,S.R., Stricklin,S.L., Baillie,D.L., Waterston,R. and Marra,M.A. (2001) Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res., 11, 1346–1352. [DOI] [PubMed] [Google Scholar]
- 15.Piquemal D., Commes,T., Manchon,L., Lejeune,M., Ferraz,C., Pugnere,D., Demaille,J., Elalouf,J.M. and Marti,J. (2002) Transcriptome analysis of monocytic leukemia cell differentiation. Genomics, 80, 361–371. [DOI] [PubMed] [Google Scholar]
- 16.Boon K., Osorio,E.C., Greenhut,S.F., Schaefer,C.F., Shoemaker,J., Polyak,K., Morin,P.J., Buetow,K.H., Strausberg,R.L., De Souza,S.J. et al. (2002) An anatomy of normal and malignant gene expression. Proc. Natl Acad. Sci. USA, 99, 11287–11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beutler E. and Gelbart,T. (1986) The mechanism of removal of leukocytes by cellulose columns. Blood Cells, 12, 57–64. [PubMed] [Google Scholar]
- 18.Brun A., Skadberg,O., Hervig,T.A. and Sandberg,S. (1994) Phenotyping autologous red cells within 1 day after allogeneic blood transfusion by using immunomagnetic isolation of reticulocytes. Transfusion, 34, 162–166. [DOI] [PubMed] [Google Scholar]
- 19.Volloch V., Schweitzer,B. and Rits,S. (1996) Antisense globin RNA in mouse erythroid tissues: structure, origin, and possible function. Proc. Natl Acad. Sci. USA, 93, 2476–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tufarelli C., Stanley,J.A., Garrick,D., Sharpe,J.A., Ayyub,H., Wood,W.G. and Higgs,D.R. (2003) Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet., 34, 157–165. [DOI] [PubMed] [Google Scholar]
- 21.Jurka J. (2000) Repbase update: a database and an electronic journal of repetitive elements. Trends Genet., 16, 418–420. [DOI] [PubMed] [Google Scholar]
- 22.Goncalves I., Duret,L. and Mouchiroud,D. (2000) Nature and structure of human genes that generate retropseudogenes. Genome Res., 10, 672–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podlowski S., Bramlage,P., Baumann,G., Morano,I. and Luther,H.P. (2002) Cardiac troponin I sense–antisense RNA duplexes in the myocardium. J. Cell. Biochem., 85, 198–207. [PubMed] [Google Scholar]
- 24.Saha S., Sparks,A.B., Rago,C., Akmaev,V., Wang,C.J., Vogelstein,B., Kinzler,K.W. and Velculescu,V.E. (2002) Using the transcriptome to annotate the genome. Nat. Biotechnol., 20, 508–512. [DOI] [PubMed] [Google Scholar]
- 25.Pauws E., van Kampen,A.H., van de Graaf,S.A., de Vijlder,J.J. and Ris-Stalpers,C. (2001) Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res., 29, 1690–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iseli C., Stevenson,B.J., de Souza,S.J., Samaia,H.B., Camargo,A.A., Buetow,K.H., Strausberg,R.L., Simpson,A.J., Bucher,P. and Jongeneel,C.V. (2002) Long-range heterogeneity at the 3′ ends of human mRNAs. Genome Res., 12, 1068–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deininger P.L. and Batzer,M.A. (2002) Mammalian retroelements. Genome Res., 12, 1455–1465. [DOI] [PubMed] [Google Scholar]
- 28.Chen J., Sun,M., Kent,W.J., Huang,X., Xie,H., Wang,W., Zhou,G., Shi,R.Z. and Rowley,J.D. (2004) Over 20% of human transcripts might form sense–antisense pairs. Nucleic Acids Res., 32, 4812–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.