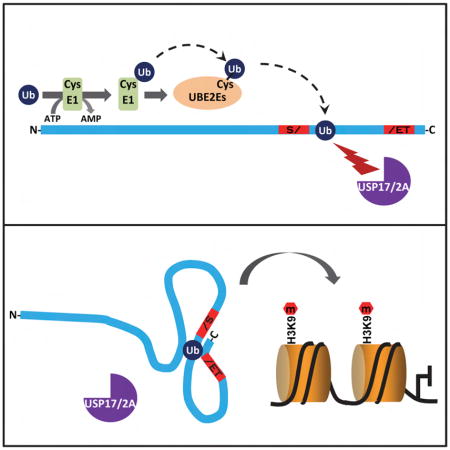
SUMMARY
Ubiquitination typically occurs through the sequential action of three enzymes catalyzing ubiquitin activation (E1), conjugation (E2) and ligation (E3) and regulates diverse eukaryotic cellular processes. Although monoubiquitination commonly confers nondegradative activities, mechanisms underlying its temporal and spatial regulation and functional plasticity still remain largely unknown. Here we demonstrate that SETDB1, a major histone H3K9 methyltransferase is monoubiquitinated at the evolutionarily conserved lysine-867 in its SET-Insertion domain. This ubiquitination is directly catalyzed by UBE2E family of E2 enzymes in an E3-independent manner while the conjugated-ubiquitin (Ub) is protected from active deubiquitination. The resulting constitutive lysine-867 monoubiquitination is essential for SETDB1’s enzymatic activity and endogenous retrovirus silencing in murine embryonic stem cells. Furthermore, the canonical hydrophobic patch on the conjugated-Ub is critical for Ub protection and function. Together, our findings highlight an E3-independent mechanism for monoubiquitination and reveal mechanistic details of SETDB1’s enzymatic activity and the functional significance of its SET-Insertion.
Graphical Abstract
INTRODUCTION
Ubiquitin (Ub) is a 76-amino acid small protein modifier which can be conjugated to substrates through a cascade of three classes of enzymes (Komander and Rape, 2012). In this process, C-terminal glycine of Ub is first covalently linked to the catalytic cysteine of an E1 enzyme for activation. Ub is next passed to an E2 conjugating enzyme to form E2~Ub thioester which further cooperates with an E3 ligase to transfer Ub to substrate. Ub can be conjugated to substrates as mono- or poly-ubiquitination through different mechanisms and the diversity of ubiquitination topologies is specified by different combinations of E2 and E3 enzymes (Wenzel et al., 2011; Ye and Rape, 2009). While Ub lysine48-linked chains normally target substrates for proteasomal degradation, monoubiquitination often confers a variety of nondegradative functions (Chen and Sun, 2009; Jackson and Durocher, 2013; Komander and Rape, 2012). Various Ub binding domain (UBD)-containing proteins can recognize distinct Ub chains and translate Ub signals to downstream effects (Dikic et al., 2009; Husnjak and Dikic, 2012). Many monoubiquitinated endocytic regulators also contain a functional UBD which couples substrate recruitment for their self-ubiquitination. This process is defined as coupled monoubiquitination (Hoeller et al., 2007; Polo et al., 2002; Ramanathan and Ye, 2012; Sigismund et al., 2004). Furthermore, different Ub conjugations are counteracted by deubiquitinases (DUBs) in cells (Komander et al., 2009; Reyes-Turcu et al., 2009). Therefore, monoubiquitination is precisely regulated at multiple molecular layers. However, mechanisms underlying these regulations still remain elusive.
As a fundamental player in chromatin structure and function, histone lysine methylation is catalyzed by SET domain-containing histone methyltransferases (HMTases) with an exception of DOT1L (Martin and Zhang, 2005). Among them, SETDB1 is a principal H3K9 HMTase that contributes to euchromatic gene silencing (Dodge et al., 2004; Schultz et al., 2002). SETDB1 has been implicated in various biological processes including development (Bilodeau et al., 2009; Yuan et al., 2009), endogenous retroviruses (ERV) silencing (Liu et al., 2014; Matsui et al., 2010), neural disease (Ryu et al., 2006) and cancer (Ceol et al., 2011; Fei et al., 2015). The intrinsic histone methylation activity of SETDB1 requires its catalytic SET domain and two adjacent Cys-rich pre-SET and post-SET domains (Schultz et al., 2002). The SET domain normally comprises two conserved portions that are separated by an insertion (SET-Insertion) with variable length contributing to substrate recognition (Xiao et al., 2003). However, the SET domain of SETDB1 is interrupted by a large 347aa insertion conserved across different organisms (Schultz et al., 2002). The function of this interposed sequence is completely unknown.
Here, we demonstrate that the SET-Insertion of SETDB1 is monoubiquitinated at lysine-867 by UBE2E family of E2 enzymes independent of E3 ligases. The conjugated-Ub is protected from deubiquitination through the hydrophobic patch on Ub and SETDB1 motifs. Importantly, K867 monoubiquitination serves as an integral part of SETDB1’s enzymatic activity and is crucial for mediated H3K9 methylation and endogenous retrovirus silencing in murine embryonic stem cells.
RESULTS
SETDB1 is Monoubiquitinated at Lysine 867
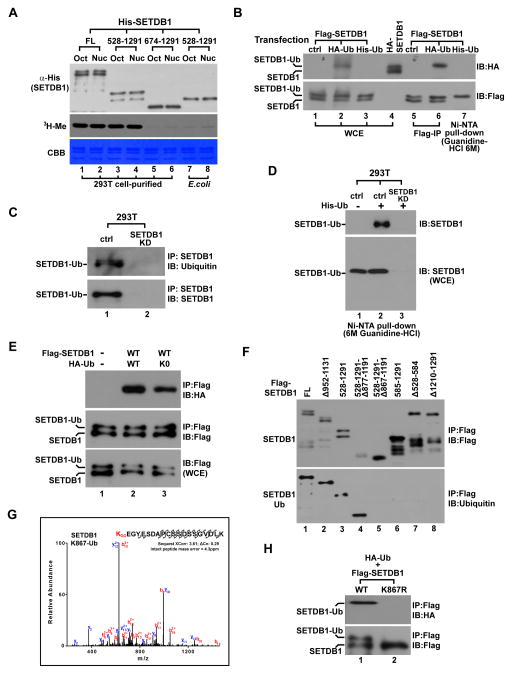
Since the importance of SETDB1’s domain structure is not fully understood, we first examined in vitro catalytic activity of SETDB1. Immunoprecipitation (IP)-purified SETDB1 displayed robust histone methylation activity comparable to G9A and GLP (Figure S1A). This activity requires the putative methyl-CpG-binding domain (MBD) but is not affected by deleting N-terminal 527aa. However, the same deletion purified from bacteria failed to methylate histones in parallel (Figure 1A). Given that 293T cell-purified SETDB1 showed a unique doublet on western blot whereas bacteria-purified protein lacked the higher molecular weight band, we wondered whether SETDB1 is subjected to ubiquitination. After co-expressing Flag-SETDB1 with HA-Ub in 293T cells, we detected robust ubiquitination at the same molecular weight as the top band of HA-SETDB1. We also co-expressed Flag-SETDB1 with His-Ub and performed a Ni-NTA pull-down assay under the denaturing condition. Western blot specifically detected Flag-SETDB1 (Figure 1B), confirming His-Ub is covalently conjugated onto SETDB1.
Figure 1. SETDB1 is monoubiquitinated at K867.
(A) Western blot (top), autoradiogram (middle) and gel image (bottom) of in vitro histone methylation (HMT) assay. His-tagged SETDB1-FL or different indicated deletions were purified from transfected 293T cells or bacteria using Ni-NTA agarose and was incubated with histone octamer (Oct) or mononucleosome (Nuc) for reaction.
(B) Immunoblot (IB) of Flag- or HA-tagged SETDB1 after it was co-expressed with HA- or His-tagged Ub in 293T cells. Flag-SETDB1 was IPed using Flag antibody while His-SETDB1 was purified by Ni-NTA pull-down under the denaturing condition. WCE represents whole cell extract.
(C) Endogenous SETDB1 was IP-purified from control and SETDB1 stable knockdown (SETDB1-KD) 293T cells followed by immunoblot analysis using indicated antibodies.
(D) His-Ub was expressed in control and SETDB1-KD 293T cells followed by Ni-NTA pull-down under the denaturing condition. WCE and purified proteins were next analyzed by immunoblot (IB) using SETDB1 antibody.
(E) Flag-SETDB1 was co-expressed with HA- Ub-wt or Ub-K0 in 293T cells followed by IP-western analysis using indicated antibodies.
(F) Flag-tagged SETDB1- FL or different deletions were IPed from transfected cells and analyzed by western blot using Flag or Ub antibody.
(G) LC-Mass Spectrometry analysis of SETDB1 ubiquitination. After Flag-SETDB1-FL was expressed in 293T cells, IPed and resolved by SDS-PAGE, the top ubiquitinated protein band was excised for LC-Mass. Arrows are the fragment ions that confirm K867 as the ubiquitination site.
(H) Flag-SETDB1-wt or K867R was co-expressed with HA-Ub in 293T cells followed by IP-western analysis.
See also Figure S1.
On western blot, endogenous SETDB1 predominantly showed the same molecular weight as ubiquitinated Flag-SETDB1 and was recognized by Ub antibody (Figures 1C and S1B). To confirm it is also ubiquitinated, we expressed His-Ub in control or SETDB1 stable knockdown (SETDB1-KD) 293T cells (Figure S1C) for similar assay. Ni-NTA agarose specifically captured endogenous SETDB1 under the denaturing condition (Figure 1D), suggesting that endogenous SETDB1 is predominantly ubiquitinated. As replacing Ub-wt with Ub-K0 mutant (all lysines to arginines) did not affect SETDB1 ubiquitination (Figure 1E), we reasoned that no poly-Ub chain was formed. SETDB1 ubiquitination is impaired when we deleted 528–584aa, 867–877aa or 1210–1291aa (Figure 1F). However, LC-mass spectrometry analysis of IP-purified Flag-SETDB1 (Figure S1D) identified the ubiquitination at lysine-867 (K867) (Figure 1G) while mutagenesis study further confirmed K867 is the solo ubiquitination site (Figure 1H). Together, our findings suggest that endogenous SETDB1 is constitutively monoubiquitinated at K867 (K867Ub1).
UBE2E Family of E2 Enzymes Catalyze SETDB1 Ubiquitination Independent of E3s
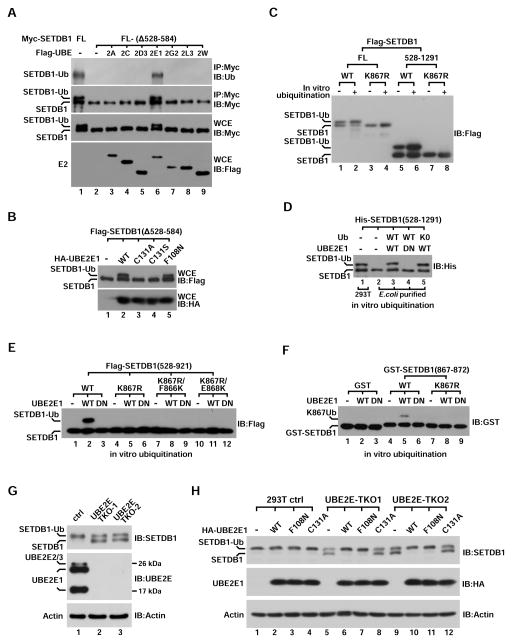
Given that Flag-SETDB1 was ubiquitinated in an Ub dosage dependent manner (Figure S2A), we reasoned that overexpression of cognate E2 could promote SETDB1-K867Ub1 by draining more Ub to SETDB1. Thus, we co-expressed 34 different E2s with SETDB1(Δ528–584), a non-ubiquitinated deletion (Figure 1F) in 293T cells to screen for such enzymes. Among them, three highly homologous UBE2E family members UBE-2E1, 2E2 and 2E3 rendered robust ubiquitination signal (Figures 2A, S2B and S2C), which was abolished when we mutated the catalytic cysteine of UBE2E1 (C131A, C131S) (Figure 2B). It has been recently proposed that UBE2Es function as Ub conjugating enzymes for TDP43 ubiquitination via an E3-dependent mechanism (Hans et al., 2014). We next generated a F108N mutation on UBE2E1 which is supposed to disable the canonical E2–E3 interaction (Hoeller et al., 2007). Similar to C131A mutation, F108N mutation significantly impeded UBE2E1-mediated TDP43 ubiquitination when they were co-expressed in 293T cells (Figure S2D). However, it displayed no obvious effect on SETDB1 ubiquitination (Figure 2B), suggesting that SETDB1-K867Ub1 requires enzymatic activity of UBE2Es but could occur independent of E3s. To test this possibility, we incubated recombinant UBE2E1 with IP-purified Flag-SETDB1- FL or (528–1291) deletion for an in vitro ubiquitination assay. Without any E3, UBE2E1 significantly increased ubiquitination on both proteins but not on their K867R mutants (Figure 2C). We also purified recombinant SETDB1(528–1291) from bacteria for the same in vitro assay while western blot and LC-Mass analyses confirmed that UBE2E1 can directly catalyze SETDB1-K867Ub1 (Figures 2D and S2E–S2F).
Figure 2. UBE2E enzymes catalyze SETDB1-K867Ub1 independent of E3.
(A) Myc-tagged SETDB1- FL or (Δ528–584) deletion was co-expressed with different Flag-tagged E2s in 293T cells followed by IP-western analysis using indicated antibodies. Protein expression was indicated by western blot of whole cell extract (WCE).
(B) Western blot analysis of WCE derived from 293T cells co-expressing Flag-SETDB1(Δ528–584) and UBE2E1-wt or its different mutants.
(C) Flag-tagged SETDB1- FL or (528–1291) (wt or K867R) was IP-purified from transfected cells and then incubated with recombinant UBE2E1 for in vitro ubiquitination. Products were analyzed by western blot using Flag antibody.
(D) Immunoblot of bacteria-purified His-SETDB1(528–1291) after it was in vitro ubiquitinated with Ub-wt or Ub-K0 by recombinant UBE2E1-wt or C131A (DN).
(E) IP-purified Flag-SETDB1(528–921)-wt or three indicated mutants were ubiquitinated in vitro by recombinant UBE2E1-wt or C131A (DN) and analyzed by anti-Flag western blot.
(F) Anti-GST western blot analysis of purified GST-SETDB1(867–872) fusion protein (wt or K867R) after it was in vitro ubiquitinated by UBE2E1-wt or C131A (DN).
(G) WCEs derived from control and two UBE2E triple-knockout (TKO) 293T cells were analyzed by immunoblot with indicated antibodies. TKO1 and TKO2 cells were derived from different UBE2E2/3 sgRNAs.
(H) Control and two UBE2E-TKO 293T cells were rescued by transient expression of HA-UBE2E1- wt or different mutant. WCEs were then analyzed by immunoblots using indicated antibodies.
See also Figure S2.
In a similar in vitro ubiquitination assay, replacing the adjacent F866 or E868 with lysine did not salvage ubiquitination on SETDB1-K867R mutant (Figure 2E), suggesting the high selectivity of K867 by UBE2E1 is unlikely due to inaccessibility of other lysines. We also purified several SETDB1 deletions for in vitro ubiquitination to determine whether any internal SETDB1 motif possesses E3-like function. However, only a short conserved stretch of 6-aa (867–872aa) is absolutely necessary for UBE2E1-catalyzed ubiquitination (Figures S2G and S2H). We next fused this 6-aa with GST while the resultant fusion protein was also ubiquitinated at K867 by UBE2E1 in vitro (Figure 2F). Given that such a small fragment unlikely forms a functional E3-ligase or UBD, our results demonstrate that UBE2Es directly conjugate Ub to SETDB1-K867. This E3-independent mechanism is likely different from reported coupled monoubiquitination which requires an functional UBD on substrates (Hoeller et al., 2007; Polo et al., 2002; Ramanathan and Ye, 2012; Sigismund et al., 2004).
To confirm that UBE2E enzymes catalyze SETDB1-K867Ub1 in vivo, we employed the CRISPR/CAS9 technique and knocked-out three UBE2E enzymes sequentially in 293T cells (Figures S2I and S2J). A significant reduction (>50%) of SETDB1-K867Ub1 was readily detected from two UBE2E triple-knockout (TKO) clones (Figure 2G), demonstrating that UBE2E enzymes are also responsible for SETDB1-K867Ub1 in vivo. To rule out possible off-targeting effects, we further rescued UBE2E-TKO 293T cells with UBE2E1- wt or its mutants. While expression of UBE2E1-wt or E2–E3 interaction deficient F108N mutant completely restored endogenous SETDB1-K867Ub1, expression of catalytic inactive C131A mutant displayed no effect (Figure 2H). Therefore, we conclude that UBE2E family of E2 enzymes directly catalyze SETDB1-K867Ub1 in vitro and in vivo through an E3 independent mechanism.
Conjugated-Ub on SETDB1-K867 Is Protected from Active Deubiquitination
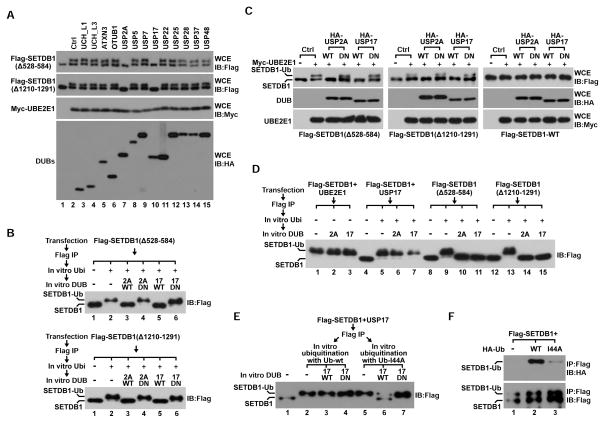
While protein ubiquitination is dynamically balanced by DUBs in cells (Reyes-Turcu et al., 2009), endogenous SETDB1 predominantly exists as a ubiquitinated form in seven cell lines derived from different tissues (Figure S3A). However, ubiquitination was not detected on SETDB1(Δ528–584) when it was rescue-expressed in SETDB1-KD cells. As the same protein was robustly ubiquitinated by over-expressed UBE2E1-wt (Figure S3B), we asked whether such a deletion is more sensitive to deubiquitination. In favor of this possibility, we detected distinctly higher level of SETDB1-K867Ub1 when cells co-expressing UBE2E1 and SETDB1(Δ528–584) were lysed with SDS-containing buffer or IPH buffer containing DUB inhibitor N-Ethylmaleimide (NEM). We had similar observation on SETDB1(Δ1210–1291) and control histone H2A but not on SETDB1-FL (Figure S3C). We next co-expressed above two SETDB1 deletions with UBE2E1 and 13 different DUBs in 293T cells while western blot analysis reveals that UBE2E1-catalyzed SETDB1-K867Ub1 was abolished by USP2A or USP17 (Figure 3A). To confirm it is due to active deubiquitination, we ubiquitinated two SETDB1 deletions in vitro and incubated them with recombinant USP2A or USP17 for an in vitro deubiquitination assay. While wild-type USP2A or USP17 readily eliminated ubiquitination on both SETDB1 deletions, their catalytic inactive mutants failed to do so (Figure 3B). Together, our results demonstrate that USP2A and USP17 actively counteract UBE2E1-catalyzed SETDB1-K867Ub1.
Figure 3. SETDB1-K867Ub1 is protected from deubiquitination.
(A) Flag-SETDB1-(Δ528–584) or (Δ1210–1291) was co-expressed together with UBE2E1 and 13 DUBs in 293T cells while WCE derived from transfected cells was analyzed by western blot using indicated antibodies.
(B) IP-purified Flag-SETDB1-(Δ528–584) and (Δ1210–1291) deletions were ubiquitinated in vitro by UBE2E1 and then incubated with recombinant USP2A [2A-wt or C276A (DN)] or USP17 [17-wt or C89S (DN)] for in vitro deubiquitination. Products were analyzed by western blot using Flag antibody.
(C) Flag-SETDB1-wt, (Δ528–584) or (Δ1210–1291) was expressed at endogenous level in SETDB1-KD 293T cells together with UBE2E1 and USP2A or USP17 (wt or DN). WCE derived from different transfected cells was analyzed by western blot using indicated antibodies.
(D) IP-purified Flag-SETDB1- wt, (Δ528–584) or (Δ1210–1291) was in vitro ubiquitinated by UBE2E1 and then incubated with recombinant USP2A or USP17 for in vitro deubiquitination. Products were analyzed by anti-Flag western blot.
(E) Flag-SETDB1 was co-expressed with USP17 in 293T cells while minimally ubiquitinated Flag-SETDB1 was IP-purified and further ubiquitinated in vitro with Ub-wt or Ub-I44A by UBE2E1. Products were then purified and incubated with recombinant USP17 (wt or DN) for in vitro deubiquitination followed by western blot analysis.
(F) Flag-SETDB1 was co-expressed with HA-Ub-wt or I44A in 293T cells and analyzed by IP-western using indicated antibodies.
See also Figure S3.
Furthermore, we co-expressed UBE2E1 with USP2A or USP17 in SETDB1-KD 293T cells, in which SETDB1- wt, (Δ528–584) or (Δ1210–1291) was respectively rescue-expressed at endogenous level. Although UBE2E1 ubiquitinated both SETDB1 deletions, USP17 or USP2A readily eliminated K867Ub1 and this deubiquitination requires their enzymatic activity. In contrast, USP2A or USP17 did not affect K867Ub1 on SETDB1-wt regardless of UBE2E1 (Figure 3C), indicating that two deleted SETDB1 motifs are involved in protection of K867Ub1 from active deubiquitination. After over-expressing Flag-SETDB1 and USP17 under optimized conditions, we generated and IP-purified Flag-SETDB1 with minimal K867Ub1 (Figure S3D). This protein and above two SETDB1 deletions were next ubiquitinated in vitro by UBE2E1 and then applied to in vitro deubiquitination. Similarly, USP2A and USP17 eliminated ubiquitination on SETDB1 deletions but not on wild-type protein (Figure 3D), demonstrating that K867Ub1 is protected by multiple SETDB1 motifs including 528–584aa and 1210–1291aa. Although Ub-wt and Ub-I44A was conjugated onto SETDB1 similarly in vitro, K867Ub1-I44A was more sensitive to USP17-catalyzed deubiquitination (Figure 3E). Furthermore, Flag-SETDB1 was ubiquitinated similarly with Ub-wt or Ub-I44A when they were co-expressed in cells. However, conjugated Ub-I44A was significantly less than Ub-wt, suggesting it was rapidly deubiquitinated and replaced by endogenous Ub (Figure 3F). Given that the I44-centered hydrophobic patch is involved in most Ub interactions (Husnjak and Dikic, 2012), we reasoned that K867Ub1 protection could be achieved by multiple Ub-SETDB1 interactions mediated by this hydrophobic patch.
K867Ub1 Is Essential for SETDB1’s Catalytic Activity and Function
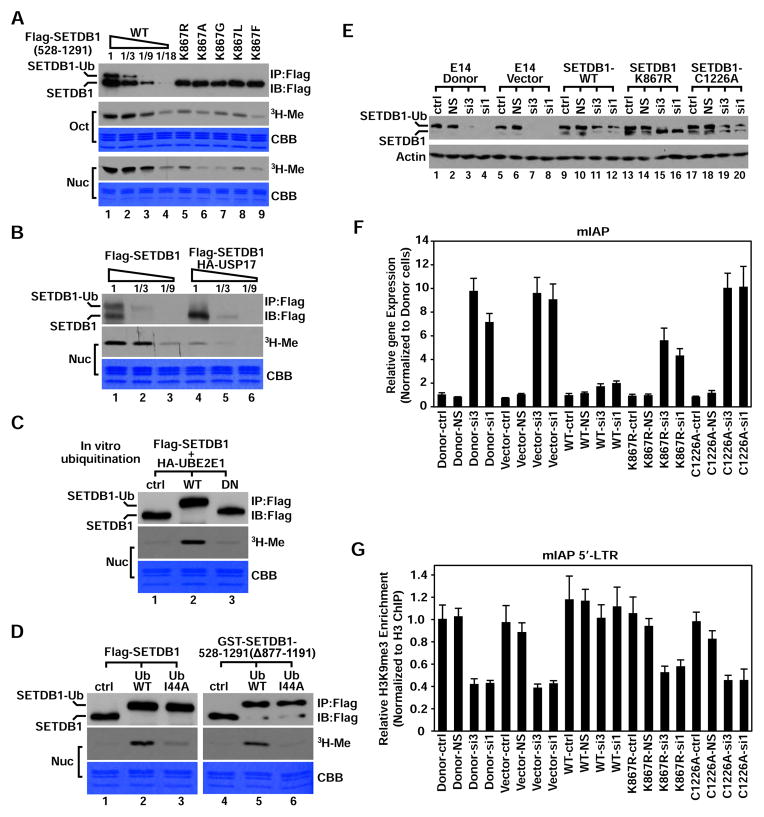
The observation that MG132 treatment has no effect on SETDB1 protein level suggests K867Ub1 did not lead to protein degradation (Figures S3E and S3F). Intriguingly, mutation of K867 severely reduced SETDB1-catalyzed histone methylation without affecting substrate specificity, methylation site or states (Figures 4A and S4A–S4C). We thus tested whether K867Ub1 promotes SETDB1’s enzymatic activity. Comparing with ubiquitinated protein, USP17-deubiquitinated SETDB1 showed significantly reduced histone methylation activity (Figure 4B), which was greatly restored by UBE2E1-mediated in vitro ubiquitination (Figure 4C), supporting that K867Ub1 is crucial for SETDB1’s enzymatic activity. Although the similar restoration of enzymatic activity was also observed on SETDB1(Δ528–584) after K867Ub1 was installed, UBE2E1-enforced K867Ub1 on SETDB1’s catalytic inactive mutant C1226A, Y1265F or Δ1275–1291 failed to do so (Figures S4E–S4G). Therefore, both K867Ub1 and the intact SET domain are necessary for SETDB1’s catalytic activity. Moreover, Ub-wt conjugated SETDB1-FL and a bacteria-derived SETDB1 deletion displayed significant higher enzymatic activity than Ub-I44A conjugated proteins (Figure 4D), suggesting that the canonical hydrophobic patch of K867Ub1 is also important for SETDB1’s catalytic activity.
Figure 4. K867Ub1 is fundamental to SETDB1’s enzymatic activity and ERV silencing.
(A) Immunoblot, gel image (CBB) and autoradiogram (3H-Me) of in vitro HMT assay. Flag-SETDB1(528–1291)-wt or different K867 mutants was IP-purified from transfected cells and incubated with histone octamer (Oct) or mononucleosome (Nuc) for reaction. Flag-SETDB1(528–1291)-wt was also titrated.
(B) Control or USP17-deubiquitinated Flag-SETDB1 was IP-purified, titrated and then incubated with mononucleosome (Nuc) for HMT assay.
(C) Minimally ubiquitinated Flag-SETDB1 was incubated with UBE2E1-wt or C131A (DN) for in vitro ubiquitination. Products were purified and incubated with mononucleosome for the same HMT assay.
(D) Purified Flag-SETDB1 (left) or GST-SETDB1-528–1291(Δ877–1191) (right) were in vitro conjugated with either Ub-wt or Ub-I44A by UBE2E1 and applied for the similar HMT assay with mononucleosome.
(E–F) Control E14 mES cells or cells stably expressing SETDB1-WT, -K867R or -C1226A were treated with Setdb1 siRNA (si3 and si1) or control siRNA (NS) for 96 hours. For different cells, WCE was analyzed with western blot (E). Expression of mIAP element was analyzed by real-time RT-PCR (normalized to GAPDH) (F) Relative enrichment of H3K9me3 on mIAP 5′-LTR was analyzed by chromatin-IP (ChIP) coupled real-time PCR (normalized to H3 enrichment) (G). In panel F and G, error bars represent Standard Deviation of technical triplicate.
See also Figure S4.
Given that Setdb1 and the catalyzed H3K9 trimethylation (H3K9me3) are required for ERV silencing during early embryogenesis (Matsui et al., 2010), we next assessed the importance of K867Ub1 in this process. Considering Setdb1 is critical for mouse embryonic stem (mES) cell pluripotency (Bilodeau et al., 2009; Yuan et al., 2009), we first stably expressed SETDB1-wt, K867R or C1226A in E14 mES cells and then knocked-down endogenous Setdb1 (Figure 4E). After siRNA treatment, ERV IAP and MusD elements were derepressed in control mES cells but remained silenced in SETDB1-wt expressing cells. The similar derepression was also observed in cells where endogenous Setdb1 was replaced by SETDB1-K867R or C1226A mutant (Figures 4F and S4H). Although SETDB1-wt and its mutants displayed similar localization in the 5’LTR region of both ERVs, H3K9me3 level reduced significantly in Setdb1 knockdown cells expressing either SETDB1-K867R or C1226A mutant as did in control cells (Figures 4G and S4I–S4K). Therefore, these findings demonstrate that disabling K867Ub1 did not affect SETDB1 targeting to ERV genes in mES cells but impaired the catalyzed H3K9me3 and thus abolished gene silencing.
DISCUSSION
Our report demonstrates that a principal H3K9 HMTase SETDB1 is monoubiquitinated at K867. Different from classic ubiquitination pathway, SETDB1-K867Ub1 is catalyzed by UBE2E E2 enzymes in an E3-independent manner in vitro and in vivo. Meanwhile, UBE2E1-conjugated SETDB1-K867Ub1 also exhibits several features different from previously reported coupled monoubiquitination (Hoeller et al., 2007). First, UBD binding disabled Ub-I44A is conjugated to SETDB1-K867 in vitro as efficiently as Ub-wt by UBE2E1. Second, UBE2E catalyzes SETDB1-K867Ub1 with high site specificity, which is different from promiscuous selectivity on substrate lysines in the coupled ubiquitination reaction. Third, evolutionarily conserved 6-aa in SETDB1 (867–872aa) including the ubiquitination site is necessary and sufficient for K867Ub1, which is unlikely sufficient for Ub binding. However, this 6-aa SETDB1 peptide is ubiquitinated less efficiently than longer proteins (Figure 2), suggesting that other domains are also required for the robust reaction. As interactions between ubiquitin moiety and SETDB1 are suggested by our results, the Ub binding by SETDB1 could be an additional contributing factor for the efficacy of SETDB1 ubiquitination, reminiscent of the coupled-monoubiquitination (Hoeller et al., 2007). Although the exact mechanism is under investigation, one possibility is that these 6-aa contribute to positioning the reactive E2~Ub thioester to the attacking K867 in the correct orientation and thus enables the direct Ub transfer. Alanine screening identified three residues around the ubiquitination site, including E864, Y870 and E871 might be involved in this process (Figure S4L).
Importantly, K867Ub1 plays a fundamental role in SETDB1’s enzymatic activity, likely through an in cis mechanism. As SETDB1’s SET domain harbors an unusually large insertion, it makes biological sense that the conjugated-K867Ub1 could contribute to the integrity of the functional SET domain to render robust catalytic activity. It has been shown that SET-Insertion varies in sequence across different HMTases but appears relatively conserved in the structure and acts as a substrate docking platform (Wu et al., 2010; Xiao et al., 2003). Therefore, K867Ub1 could also confer the correct conformation of SETDB1’s SET-Insertion to facilitate histone binding and in turn promote methylation. Consistent with its necessity in SETDB1’s enzymatic activity, the conjugated-Ub is protected from active deubiquitination and thus exists constitutively on endogenous SETDB1. We also demonstrate that the I44-centered canonical hydrophobic patch on the conjugated-Ub is essential for both Ub protection and function. Given that this hydrophobic patch is the “binding hot spot” on Ub moiety (Husnjak and Dikic, 2012), our findings suggest that it forms intra-molecular interactions with multiple regions of SETDB1 to enable Ub protection and augment SETDB1’s catalytic activity. While most post-translational modifications are dynamically regulated on the target proteins, our findings also highlight the constitutive role of K867Ub1, representing a paradigm shift of ubiquitination functions.
EXPERIMENTAL PROCEDURES
Cell culture, Transfection and Antibodies
E14 mES cells were cultured in gelatin-coated dish with DMEM with 15% FBS, 0.1mM NEAA, 1mM GlutaMAX (Gibco), 55μM β-Mercapoethanol (Sigma) and 1000 units/ml recombinant LIF (Millipore). Other cell lines were cultured as previously described (Kokura et al., 2010). Transient transfection was carried out using polyethylenimine (PEI) (Polysciences). Transfection of siRNA (Dharmacon) was performed using RNAi-Max (Invitrogen). Details and targeting sequences are described in Supplemental Data.
Constructs and Antibodies
Expression vectors for human UCH_L1, UCH_L3, ATXN3, OTUB1, USP5, USP17, USP22, USP25, USP37, USP48 (Sowa et al., 2009) and TDP43 (Yang et al., 2010) were obtained from Addgene. Other vectors were described in (Kokura et al., 2010). USP2A and USP7 cDNAs were from Jiandong Chen (Moffitt). cDNA of E2 enzymes were generated by PCR and subcloned into pCAG vector. Mutations were generated by overlapping PCR and verified by sequencing. The following antibodies were used: Ubiquitin (Enzo), β-actin (Sigma), histone H3 (Abcam), H3K9me3 (Abcam), Flag (Sigma), Myc (Santa Cruz), HA (Covance), GST (Covance), His (Covance), UBE2E (Boston Biochem). SETDB1 antibody was described in (Wang et al., 2003).
CRISPR/Cas9 Knockout
CRISPR/Cas9 vectors pX459 (Ran et al., 2013) and lentiCRISPR-v2-puro (Sanjana et al., 2014) were from addgene. The PAC gene in LentiCRISPR-V2-puro vector was replaced by BSR gene to generate Blasticidin resistant vector lentiCRISPR-v2-Bla. sgRNA targeting human UBE2E1 was cloned into pX459 while sgRNAs targeting human UBE2E2 and UBE2E3 were cloned into lentiCRISPR-v2-puro and lentiCRISPR-v2-Bla respectively. All sgRNAs target the core catalytic region of UBE2Es. For triple knockout, pX459-UBE2E1-sgRNA vector was transfected into 293T cell followed by serial dilution in 96-well plate to obtain single clones. Propagated colonies were screened using the surveyor assay (IDT) while PCR products from positive clones were cloned into T-vector (Promega) for sequencing. Ten transformants were sequenced for each clone while clones with frame-shift/premature stop codon on both alleles were subjected to western blot analysis. The validated UBE2E1 knockout 293T cell clone were next infected with Lenti-UBE2E2-sgRNA-puro and Lenti-UBE2E3-sgRNA-bla simultaneously followed by extensive selection with 2μg/ml puromycin and 20μg/ml blasticidin. Knockout efficiency was examined by western blot.
Ubiquitination Assay
The transfection-based assay was carried out as previously described (Stegmeier et al., 2007) with modifications. Briefly, transfected cells were lysed in IPH buffer (50mM Tris-Cl pH8.0, 0.5% NP-40) containing 150mM NaCl with sonication and Micrococcal Nuclease (Worthington) digestion. Flag- or Myc-tagged SETDB1 was IPed, intensively washed with IPH buffer containing 1000mM NaCl and resolved by SDS-PAGE followed by western blot analysis. Ubiquitination of endogenous SETDB1 was analyzed under the same condition after IP with anti-SETDB1 antibody.
For denaturing Ni-NTA pull-down assays, transfected cells were lysed with denaturing lysis buffer (0.1M Na2HPO4/NaH2PO4 pH8.0, 6M Guanidinium-HCl, 5mM Imidazole, 0.1% NP40) and incubated with Ni-NTA agarose (Qiagen). After intensive washing with denaturing wash buffer (25mM Tris-Cl, pH8.0, 8M Urea, 0.1% NP40), beads-bound proteins were resolved by SDS-PAGE followed by western blot analysis.
The in vitro assays were also carried out as previously described (Stegmeier et al., 2007) with modifications. IPed Flag-SETDB1 or purified recombinant GST- or His-SETDB1 was incubated with 100nM E1 (Enzo), 1 μM E2 (UBE2E1), 30μM ubiquitin (Enzo and Boston Biochem), 4mM ATP in the in vitro ubiquitination Buffer (25mM Tris-Cl pH7.6, 2.5mM MgCl2, 0.1mM DTT) at 30°C for 30min followed by western blot analysis. If the proteins will be applied to HMT assays, in vitro ubiquitination was carried out for 30 min at 22°C to preserve SETDB1’s enzymatic activity. After ubiquitination reaction, SETDB1-bound beads were separated from reactant and washed with BC1000 buffer (40mM HEPES-HCl pH7.9, 10% Glycerol, 1M NaCl) before being applied for in vitro HMT assay or in vitro deubiquitination assay.
In Vitro Deubiquitination Assay
We followed the previously reported protocol (Li et al., 2002; Stegmeier et al., 2007) with modifications. Briefly, Flag-SETDB1 ubiquitinated in vitro or in cells was purified by Flag-IP and then incubated with 1μg DUBs in 15μl BC50 buffer (40mM HEPES-HCl pH7.9, 50mM NaCl, 10% Glycerol) for reaction at 30°C for 30min with rotation.
Supplementary Material
Acknowledgments
This work was supported by NIH grant (R01CA172774) and Florida Department of Health grant (3KN02) to J.F. We thank Victoria Izumi and Dr. John M Koomen at the Moffitt Cancer Center Proteomics Core for the LC Mass Spectrometry analyses. We are extremely grateful to Dr. Xiaodong Cheng at Emory University for his help on mass spectrometry and suggestions.
Footnotes
Supplementary data, other detailed experimental procedures, sequence information for PCR, Chromatin IP, knockdown and knockout assays are described in Supplementary Data.
Author Contributions: L.S. and J.F. conceived the project, designed the experiments, analyzed data and wrote the manuscript. L.S did the experiments.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes & development. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Molecular cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nature reviews. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Molecular and cellular biology. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Shang K, Zhang J, Chuai S, Kong D, Zhou T, Fu S, Liang Y, Li C, Chen Z, et al. Histone methyltransferase SETDB1 regulates liver cancer cell growth through methylation of p53. Nat Commun. 2015;6:8651. doi: 10.1038/ncomms9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans F, Fiesel FC, Strong JC, Jackel S, Rasse TM, Geisler S, Springer W, Schulz JB, Voigt A, Kahle PJ. UBE2E ubiquitin-conjugating enzymes and ubiquitin isopeptidase Y regulate TDP-43 protein ubiquitination. J Biol Chem. 2014;289:19164–19179. doi: 10.1074/jbc.M114.561704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I. E3-independent monoubiquitination of ubiquitin-binding proteins. Molecular cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annual review of biochemistry. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Molecular cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Kokura K, Sun L, Bedford MT, Fang J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. The EMBO journal. 2010;29:3673–3687. doi: 10.1038/emboj.2010.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nature reviews. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annual review of biochemistry. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- Liu S, Brind’Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y, Lorincz MC. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes & development. 2014;28:2041–2055. doi: 10.1101/gad.244848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nature reviews. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- Ramanathan HN, Ye Y. Cellular strategies for making monoubiquitin signals. Critical reviews in biochemistry and molecular biology. 2012;47:17–28. doi: 10.3109/10409238.2011.620943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annual review of biochemistry. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Lee J, Hagerty SW, Soh BY, McAlpin SE, Cormier KA, Smith KM, Ferrante RJ. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & development. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S, Polo S, Di Fiore PP. Signaling through monoubiquitination. Current topics in microbiology and immunology. 2004;286:149–185. doi: 10.1007/978-3-540-69494-6_6. [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER, 3rd, Li MZ, Hannon GJ, Sorger PK, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG, Zhang Y. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Molecular cell. 2003;12:475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. The Biochemical journal. 2011;433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, Allali-Hassani A, Campagna-Slater V, Vedadi M, Arrowsmith CH, et al. Structural biology of human H3K9 methyltransferases. PloS one. 2010;5:e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Wilson JR, Gamblin SJ. SET domains and histone methylation. Current opinion in structural biology. 2003;13:699–705. doi: 10.1016/j.sbi.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Yang C, Tan W, Whittle C, Qiu L, Cao L, Akbarian S, Xu Z. The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PloS one. 2010;5:e15878. doi: 10.1371/journal.pone.0015878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nature reviews. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B, Ng HH. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes & development. 2009;23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.